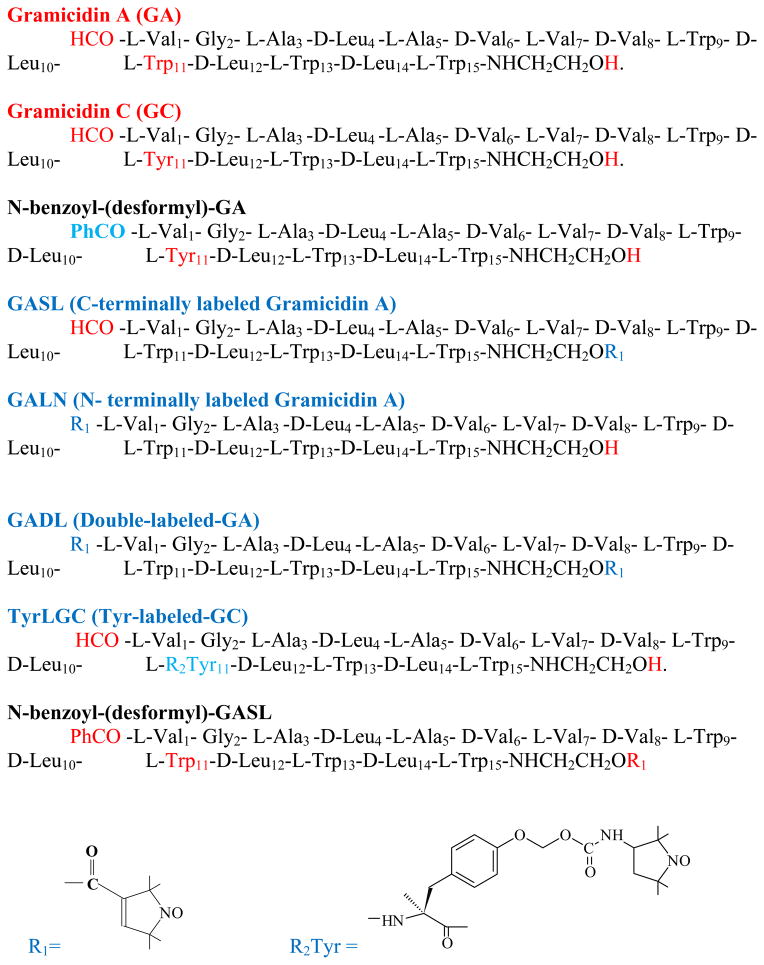
Abstract
Channel and non-channel forms of gramicidin A (GA) were studied by ESR in various lipid environments using new mono- and double- spin labeled compounds. For GA channels we demonstrate here how Pulse Dipolar ESR can be used to determine the orientation of the membrane-traversing molecule relative to the membrane normal and to study subtle effects of lipid environment on the interspin distance in the spin labeled gramicidin channel. To study non channel forms of gramicidin, Pulse Dipolar ESR was used first to determine interspin distances corresponding to monomers and double helical dimers of spin labeled GA molecules in the organic solvents trifluoroethanol and octanol. The same distances were then observed in membranes. Since detection of non-channel forms in the membrane is complicated by aggregation, we suppressed any dipolar spectra from intermolecular interspin distances arising from the aggregates by using double-labeled GA in a mixture with excess unlabeled GA. In hydrophobic mismatching lipids (Lβ phase of DPPC) gramicidin channels dissociate into free monomers. The backbone structure of the monomeric form is similar to a monomeric unit of the channel dimer. In addition to channels and monomers the double helical conformation of gramicidin is present in some membrane environments. In the gel phase of saturated phosphatidylcholines the fraction of double helices increases in the following order: DLPC<DMPC<DSPC<DPPC. The equilibrium DHD/monomer ratio in DPPC was determined. In membranes the double helical form is present only in aggregates. In addition, we studied the effect of N-terminal substitution in the GA molecule upon channel formation. This work demonstrates how Pulsed Dipolar ESR may be utilized to study complex equilibria of peptides in membranes.
Keywords: Spin labeling, Gramicidin A, Pulse Dipolar ESR, Hydrophobic Mismatch
INTRODUCTION
Membrane proteins make up approximately one-third of known proteins. In cells membrane proteins and peptides are responsible for a variety of important functions, such as transport of ions and molecules across the membrane, membrane fusion, signaling, antibiotic activity etc. Despite their abundance and importance the information on the structure and behavior of membrane proteins and peptides is limited compared to water soluble proteins. Since crystallization of membrane proteins or membrane protein complexes is still challenging, and resolving the structure even of a relatively small membrane protein by NMR is often complicated, alternative spectroscopic techniques, like fluorescence and ESR are often indispensible for their study.
Pulse Dipolar ESR spectroscopy 1 in combination with site-directed spin labeling (SDSL) 2 has emerged as a powerful method for studying the structure and conformational dynamics of proteins 3,4. The “triangulation” approach to protein mapping 1,4 is based on obtaining a network of distance constraints from a set of spin-labeled sites such that they uniquely define the coordinates of all (or most) of the sites. At the same time, complementary studies using CW EPR can provide information on molecular motion and conformational dynamics at conditions relevant to protein or peptide function 5–11. Gramicidin channels have served as prototypical channels in the development of many physical approaches towards understanding channel or membrane protein structure and function 12,13. While gramicidin is readily available and this relatively small molecule can be easily chemically modified, the gramicidin channels have the structural and functional features expected for more complex membrane proteins 13. This includes conformational polymorphism 14. In our present study, using gramicidin as an example of such a “mini protein”, we demonstrate the advantages of simultaneous application of several ESR techniques to assign the multiple conformational states.
Gramicidins are a family of linear hydrophobic pentadecapeptide antibiotics produced by the soil bacterium Bacillus brevis during sporulation 15. The primary interest in gramicidin lies in the ability of the peptide to form ion channels in lipid membranes in its head-to-head dimer (HHD) conformation 13. The gramicidin channel specifically conducts monovalent cations. The gramicidin sequence consists of alternating L- and D- amino acids 16 (fig. 1). This pattern allows gramicidin to adopt conformations which cannot exist in an all L-peptide. It folds as a β-helix with the side chains projecting from the exterior surface of a cylindrical tube formed by the peptide backbone 13. Since many such folding patterns are possible, this leads to its conformational polymorphism 14. Gramicidin can adopt a number of conformations depending on the environment. In organic solvents gramicidin can take at least seven double helical (DH) structures with different helical pitch, stagger and orientation 13,14,17. Some of these structures were resolved by X-ray diffraction or NMR: the ππ5.6 left-handed antiparallel DH structure crystallized from ethanol, PDB:1ALZ 18,19; the ππ7.2 right-handed antiparallel DH structure crystallized from CsCl/methanol or from acetic acid, PDB:1AV2 20; the ππ5.7 left-handed parallel DH structure crystallized from CaCl2/methanol, PDB: 1MIC 21; the ππ6.4 left-handed antiparallel DH structure from KSCN/methanol, PDB:1GMK 22. The spin-labeled versions of some of these structures appear in Fig. 3.
Fig. 1.
Sequences (from N- to C- terminus) of Gramicidin A, Gramicidin C and their chemical modifications used in this work..
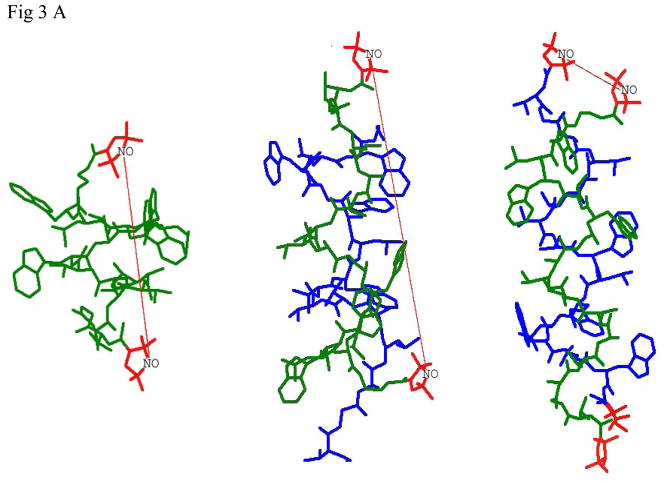
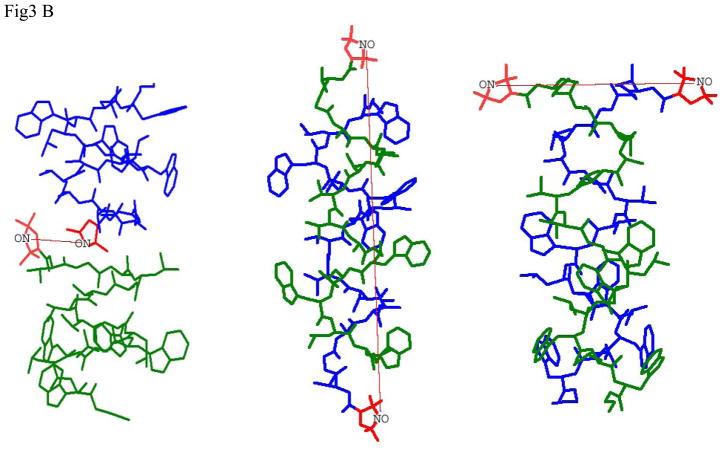
Fig 3.
- Labels at different termini of double labeled gramicidin A (GADL):
- right-handed, β6.3 - helical channel structure PDB:1MAG with RSS=20.0Å (left), this distance is observed in lipid membranes and for TFE solution of GADL
- the ππ5.6 left-handed antiparallel DH structure, PDB:1ALZ. RSS=31.1Å (center), this distance can be observed in octanol. Interaction with spins on the second GA molecule or in the aggregate is suppressed by using mixtures of GADL with excess unlabeled GA A substantial fraction of this conformation can be detected in mismatching lipid environment.
- the same PDB:1ALZ structure for GADL without dilution with unlabeled GA. The short interspin distance manifests itself in broad features of CW spectra.
- Labels on N-termini of different GA molecules (GALN):
- right-handed, β6.3 - helical structure PDB:1MAG should show short well defined interspin distance of RSS=7.5Å (left), this short distance can be detected by CW EPR in DLPC
- the ππ5.6 left-handed antiparallel DH structure, PDB:1ALZ. RSS=40.6Å (center), a broad distribution of distances approximately around this number is observed in octanol.
- the ππ5.7 left-handed parallel DH structure, PDB: 1MIC. RSS=20.2Å (right). This distance has not been detected for GALN, as well as a distance of ~13.5Å corresponding to this conformation for GASL.
Despite this remarkable conformational polymorphism of DH structures in organic solvents, in matching lipid bilayer membranes or in bilayer-like environments the predominant conformation of the GA molecule is not double helical, but a head-to-head dimer (HHD). It is an antiparallel formyl-NH-terminal-to-formyl-NH-terminal dimer 23–25 formed by right-handed, β6.3 – helical subunits, which are held together by six intermolecular hydrogen bonds, PDB:1MAG. The exclusion of tryptophan residues from the lipid bilayer and their propensity to locate at the membrane interface is seen as an important factor stabilizing HHD compared to DH in the bilayer 13,26. It has been shown that replacement of tryptophan residues by phenylalanines or their chemical modification causes an increase in the fraction of non-channel forms in the membrane. Similar effects were observed in mismatching lipids, with the lipid bilayer thickness substantially larger than the HHD length, so that tryptophan residues on the opposite sides of the dimer cannot be easily located at the interfaces of each membrane leaflet 14,27,28.
In our earlier study 29 we used spin-labeling ESR to detect the formation and dissociation of the gramicidin channel in response to changes in the lipid environment, in particular to hydrophobic mismatch between the bilayer thickness and the length of the gramicidin channel. Variation of the bilayer thickness, as in a series of lipids or as a result of a phase transition in the same lipid, manifests itself in emergence/disappearance of distinct spin pairs with a well-resolved dipolar ESR signal as well as some effects in the CW ESR spectra indicative of channel formation/dissociation, see also 30. Pulse Dipolar ESR enables us to detect subtle effects of lipid environment on gramicidin channels, and to clearly distinguish the different GA species. In this work we continue our study of the gramicidin channel using spin labeled gramicidin derivatives. Our objective is to describe the physical chemistry associated with equilibria amongst the various GA species that exist within the membrane.
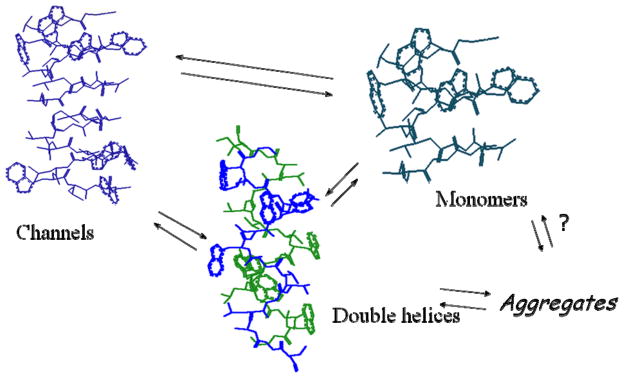
One of the important issues we address is the nature of non-channel forms of gramicidin, which are less well studied in the membrane environment than the channel form. Evidently, free GA monomers can exist in a membrane environment 31. Traditionally, the kinetics of channel formation studied by single-channel conductance is described in terms of a monomer/dimer equilibrium 32–34. On the other hand, most studies of the equilibrium of gramicidin conformers in the membrane upon changes in the membrane composition 35,36 do not discuss the possible presence of monomers. Though the fraction of free monomers is usually insignificant, as follows from the values of dimerization constants in most fluid phases 32,33,37, it could increase with the change in the membrane composition or in the gel phase. Moreover, there are indications that GA monomers participate in the interconversion of different dimer forms of GA 28. Possible conformation/aggregation states of GA in the membrane environment and the equilibria that could exist between them are shown in fig. 2.
Fig 2.
The equilibrium of possible conformation states of gramicidin in phospholipid membranes includes HHDs (channels), free monomers and a number of double helical conformations. These conformational states may exist either as free entities or aggregates.
In this study we demonstrate how ESR spectroscopy can simultaneously monitor various forms of spin-labeled gramicidin molecules, monomers and dimers, in the membrane environment. Pulse Dipolar ESR offers a unique opportunity to study the conformation of spin-labeled peptides and proteins 8 using distances between spin labels introduced at chosen positions. In our previous work 29 we detected the Pulsed Dipolar ESR spectrum for the channel form (HHD) and measured the corresponding interspin distance. At that time, we showed that, in the Lβ phase of DPPC, GASL is not grouped in isolated pairs with a well-defined interspin distance, but shows signs of aggregation instead. This evidence of aggregation is consistent with an AFM study on supported saturated phosphatidylcholines in the gel phase 38. This AFM study also detected aggregation and suggested a basic aggregation unit, most probably a hexamer of GA, which can be observed even at low GA concentrations. The assumption that HHD exist mainly as separate entities, whereas the non-channel forms of gramicidin aggregate, could resolve the controversy between the data of AFM microscopy in the gel phase and the data of other methods, obtained mainly for the fluid phase 39–41.
However, an exact assignment of the aggregating non-channel form of spin labeled GA at these conditions could present a challenge for Pulse Dipolar ESR. The dipolar spectrum under conditions of aggregation will be unresolved and weakened by multiple interactions between electron spins at various distances existing in the aggregate. In this case a different approach should be used. Instead of measuring the distance between spin labels attached at the same position on both GA molecules in the dimer, we spin labeled two different sites on the same gramicidin molecule. The distance between the two intra-molecular spin labels is used as a fingerprint of a particular conformer (Fig. 3). A change in the distance indicates a change in the conformational state of the gramicidin molecule itself in the membrane. To exclude interactions between spin labels on different monomers of the same dimer or different dimers (in the case of aggregation), the initial double spin-labeled gramicidin was mixed with excess unlabeled gramicidin.
In addition to double labeled GA, we synthesized and studied by CW and Pulse Dipolar ESR a number of mono-labeled gramicidin derivatives. They allowed us to first assign the gramicidin conformation in octanol, in order to better interpret the results obtained with double labeled GA in membranes, and to understand the effect of the N-terminal substitution in the GA molecule on channel formation.
MATERIALS AND METHODS
Materials
All lipids were obtained from Avanti Polar Lipids, Inc. (Birmingham, AL), the spin labels 2,2,5,5 – tetramethyl – 3 – pyrrolin – 1 - oxyl – 3 – carboxylic acid and 3-(2-Iodoacetamido)-PROXYL, cesium carbonate and solvents were purchased from Sigma (St. Louis, MO); Gramicidin, DMAP (4-dimethylamino pyridine), DCC (N, N′ – Dicyclohexylcarbodiimide), were purchased from Fluka. Gramicidin C and gramicidin C free gramicidin A were obtained as described by 42. C-terminus spin labeled gramicidin A (GASL) was synthesized as in 29 and additionally purified by preparative TLC.
Synthesis of new spin-labeled gramicidin derivatives
All chemical modifications of gramicidins A or C used this work are shown in Fig. 1. The detailed procedures for their synthesis and are given in Supporting Information.
Sample preparation
Sample preparation for multilamellar vesicles and ISDU aligned membranes is described in 29
Preparation of aligned membranes by evaporation
10 μl of 100 mg/ml chloroform solution of lipid/GALN mixture (100:1 molar ratio) was uniformly spread over a 4×20 mm glass slide. The glass slide was dried in a horizontal position in a stream of dry nitrogen to obtain a layer of dry lipid uniformly distributed over the layer. The lipid mixture was then hydrated at 100% humidity and 45°C for an hour and put into hermetically sealed standard ESR tube with a piece of water-soaked tissue paper outside of the sample area.
CW ESR measurements
ESR spectra were recorded on a Bruker EMX spectrometer at a frequency of 9.55 GHz under standard conditions. The field sweeps were calibrated with a Bruker ER 035 gaussmeter. The microwave frequency was monitored with a frequency counter. Dry nitrogen gas flow under the control of a Varian temperature controller was used to stabilize the temperature.
Pulse Dipolar ESR
Four-pulse DEER experiments were conducted at 60 K as described in 43 using a home-built Ku-band (17.35 GHz) pulse ESR spectrometer set to operate in a dual-amplifier DEER configuration or a single-amplifier mode. The data analysis was based on the L-curve Tikhonov regularization method to reconstruct distances 44. In general, our results showed narrow distributions (except in organic solvents) enabling accurate assessments of most probable distances (i.e. the maximum of each distance distribution). Typical uncertainties in these distances are ±0.5Å.
Theoretical estimates of interspin distances for different conformations of spin-labeled gramicidin molecules were made by the HYPERCHEM 7.5 program based on the corresponding PDB structures. After the addition of the nitroxide fragment the geometry optimization was carried out using the Molecular Mechanics Method (MM+)
RESULTS AND DISCUSSION
Spin labeled derivatives of gramicidin in octanol and trifluoroethanol
We first obtained resolved Pulse Dipolar spectra from one of the DH structures in an organic glass. A variety of double helical (pore) structures are known for gramicidin in different organic solvents 14. Individual (and different) X-ray 18,21,22,45 and NMR 46–48 structures were obtained for GA from ethanol, methanol, propanol, acetic acid etc. In a wide range of organic solvents and concentrations, under ion-free conditions, four double helical conformations exist in equilibrium with each other as noted in Introduction. The transition between the conformations is relatively slow. In dioxane, for example, the four species can be separated chromatographically, but on standing will convert back to the equilibrium mixture 17. O’Boyle and Wallace 49 showed that in octanol gramicidin A exists predominantly in the form of the left-handed antiparallel double-helix (PDB:1ALZ; cf. Fig. 3).
Conveniently for low temperature ESR, octanol forms a good glass, and it has been traditionally used as a model for a membrane environment 50. Pulse Dipolar ESR shows, for GASL in octanol, a resolved dipolar spectrum with an interspin distance of ~ 30.4Å (Table 1). Molecular mechanics (MM) calculations based on the ππ5.6 left-handed antiparallel structure (18; also referred as 1ALZ) which is assumed predominant (~ 90%) in octanol 49, are in good accord with this distance. That is for the 1ALZ structure one predicts 31.2 Å between nitroxide nitrogens and 33.4 Å between corresponding oxygens (see Supporting Information, Supplement Fig. 1). Unfortunately this interspin distance is very close to the value obtained for the HHD channel form of GASL in DMPC 29. This similarity of the distances indicates that spin labeled gramicidin compounds other than GASL are necessary to distinguish the DH form from the HHD form based on their interspin distances.
Table 1.
Interspin distances measured by pulse ESR for various spin-labeled derivatives of gramicidin in octanol and TFE and their estimates based on RCSB structures
| Measured in octanol | Measured in TFE | 1ALZ estimate | 1BDW estimate | 1MIC estimate | HHD estimate | |
|---|---|---|---|---|---|---|
| GASL | 30.4 | -- | 31.2 | 30.3 | 13.5 | 30.9 |
| GADL | 31.6* | 20.0 | 32.0 | 31.2 | 33.8 | 20.0 |
| GALN | 36–42 | -- | 40.6 | 32.0 | 20.2 | 7.5 |
| TyrLGC | 22.0 | -- | 21.6 | ~10 | 29.6 | 36.8 |
Interspin distances measured in octanol for a number of gramicidin derivatives are shown in Table 1. Although for some spin labeled gramicidin derivatives the measured distance may match several possible conformers, only the ππ5.6 antiparallel structure (1ALZ) is generally consistent with all distances determined for all spin labeling positions. For double-labeled gramicidin the distance corresponds to two spin labels on the same gramicidin molecule and was measured for a 1:20 mixture of labeled compound and unlabeled gramicidin. For single labeled gramicidin derivatives the distance given in the Table was measured between spin labels on different gramicidin molecules in the dimeric structure. For double labeled GADL, Table 1 also shows the interspin distance in trifluoroethanol (TFE), a solvent in which gramicidin assumed to be exclusively in monomeric form 51–53.
GADL in TFE shows a single interspin distance of 20 Å. This distance is exactly the same as to estimates for a GADL in a HHD (Fig 3A), and to the predominant distance observed for GADL in a variety of lipid phases (see below). A single folded conformation, most likely β6.3 left-handed monomer, was previously suggested for GA in solutions of TFE and DMSO 53 or for the trace GA amounts present in water 54. (For DMSO, however, this coiled conformation is reported in rapid equilibrium with the disordered state having local random-coil character 55). Thus we conclude that the observed interspin distance of 20 Å in TFE is consistent with a single β6.3 left-handed monomeric backbone conformation similar to half of a HHD.
GASL in the channel form: orientation of the interspin vector relative to the membrane normal
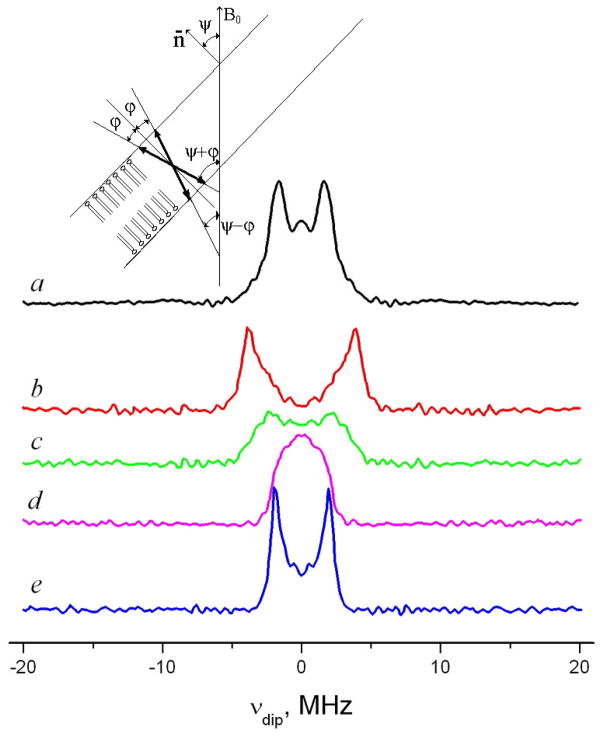
The channel form of gramicidin has been previously detected and studied by ESR in lipid model membranes using GASL 29. GASL in the gel phase of DMPC (but not DPPC) showed a well resolved dipolar spectrum from spin pairs. In vesicles it translates into a Pake doublet, which represents a sum of the dipolar splitting 2D = 2D0 (3cos2 θ − 1), where , over all orientations, θ of the interspin distance relative to B0 with appropriate weighting factors. In aligned membranes with the membrane normal parallel to B0 (corresponding to Ψ=0, cf. fig. 4) one expects a dipolar spectrum from a single orientation of the interspin vector relative to B0 with splitting D = D0(1−3cos2φ), where φ is the angle between the interspin vector and the membrane normal. For other orientations Ψ of the membrane normal relative to the magnetic field, there is partial averaging, which depends on φ. However, for φ=0, one will again observe just a single orientations which is θ=Ψ. In the case of Ψ=90° the dipolar splitting will be exactly half of the splitting in the Ψ=0° orientation. For the other values of φ the dipolar spectrum will be a superposition of orientations lying in the range between Ψ+ φ and Ψ−φ (fig. 4), so that the spectrum for orientations other than Ψ=0° will be broadened and its peak-to-peak width for Ψ= 0° will be less than the double width for Ψ=0° 56. Since for GASL in the aligned DMPC (fig. 4) membrane the Ψ=0° orientation shows a dipolar splitting which is exactly double of that observed for Ψ=90°, the direction of the interspin vector in this case should be very close to the membrane normal. We also point out the absence of noticeable line broadening for the Ψ=90° orientation compared to Ψ=0°, consistent with the conclusion that φ=0°.
Fig. 4.
Pulse Dipolar Spectra of GASL in DMPC vesicles (a) and aligned DMPC membrane at different orientations of the magnetic field B0 relative to the membrane normal n̄ (b–e): b) Ψ=0°; c) Ψ=30°; d) Ψ=60°; e) Ψ=90°
Effect of lipid environment on the interspin distance in the GASL channel
Previously we found some variation in interspin distance observed for the channel form of GASL depending on the lipid phase 29. In the present work we carried out a more systematic study of this matter. Table 2 shows the interspin distances for several hydrophobic matching and mismatching lipids (i.e. length or “thickness” relative to GA channel). One can see that this distance varies by nearly 3Å with longer distances observed in thicker bilayers. Given the high accuracy of Pulse Dipolar ESR in these cases (i.e. ±0.5Å uncertainty cf. Methods Section, 29 compare for example with 57) this difference is significant. It is very likely that this variation reflects the response of the channel itself to the stress imposed by the rigid environment in the gel phase of mismatching lipids, but we cannot completely rule out that it is due to a different tilt of the nitroxide tethers towards the membrane surface, GASL in DMPC, DLPC and di-decanoyl-PC show within measurement error the same local polarity (2Azz at 77K is 67.6, 67.4 and 67.6G) which corresponds to a non-polar environment. If nitroxides of the spin labeled channel form had a tendency to locate at the membrane interface, they would report higher polarity in the bilayer of short-chain di-decanoyl-PC. Moreover, the same value of the local polarity for these three lipids points at the same position of the nitroxide relative to the neighboring lipid molecules. It is known that anchoring tryptophans which have a specific affinity for a well-defined site near the lipid carbonyl region 13,26,58 is an important factor for gramicidin channel stability. While channels can adjust to positive or negative hydrophobic mismatch, they cause thinning or thickening of the bilayer in their vicinity 59,60 by applying pressure on the lipid bilayer through these anchoring points. The spin label of GASL is attached above the pressure point. It should have the same depth position between tryptophan-anchoring carbonyls and phosphate groups in the boundary lipid and should directly report, via the interspin distance, the distance between anchoring tryptophans on the different ends of HHD.
Table 2.
Interspin distances measured for the GASL channel determined by pulse ESR in various lipids
| Lipid environment | Interspin distance, Å |
|---|---|
| GASL in DMPC membrane | 30.9 |
| GASL in DLPC membrane | 28.8 |
| GASL in di-decanoyl PC membrane | 28.6 |
| GASL in DPPC membrane quenched after an exposure at 40°C | 31.4 |
| GASL in DPPC membrane | No pairs detected |
| GASL in DSPC membrane | No pairs detected |
| GASL in DOPC membrane | 28.7 |
| GASL in POPC membrane | 31.0 |
| GASL in egg yolk lecithin | 30.2 |
Although the helical pitch of the channel form of GA has been shown to be the same in different lipids61 (4.7±0.2 Å/turn), our precision in distance measurements may allow us to observe subtle effects of lipid-induced deformation. Intuitively it seems logical that the gramicidin-induced force, which causes deformation of the lipid bilayer, implies lipid - peptide counterforce which induces some deformation of the channel. It has been widely accepted that transmembrane peptides are rigid and lipids are more flexible so that effect of this counterforce is negligible 62. However, recent 2H-NMR results revisit this topic. Long WALP and KALP peptides are shown to adapt themselves to mismatching thin lipid bilayers by introducing a kink into the membrane-spanning helix63. Whereas the protein backbone appears to be insensitive to the lipid bilayer in the fluid phase, which is 102–103 times softer than the embedded proteins 64, in the gel phase the situation can be different. In our case GASL responds to the deformation caused by hydrophobic mismatch in the gel phase of DPPC by gradual dissociation of existing channels 30. Also, compared to many other spectroscopic studies of lipid/gramicidin interactions 36,59,61, our Pulse Dipolar ESR experiments correspond to very high ratios of lipid/peptide, so channels exist as distinct entities surrounded by lipid bilayer at its usual thickness. Therefore, the counterforce imposed by the lipids on the peptide is not mitigated by effects of the network of channels, which tend to cause thinning of the bilayer.
Detection of channel and non-channel forms of gramicidin in membranes for other spin labeled gramicidin derivatives
The new gramicidin derivatives (i.e. those except for GASL) are expected to vary in their channel-forming potency (see below), but as shown below, are still able to form HHD at some conditions in matching membrane environments. In general, C-terminal spin-labeled derivatives of gramicidin (GASL and N-benzoyl GASL) in matching lipids give well-resolved dipolar spectra and show a well-resolved Pake doublet pattern in the frequency domain. The interspin distance determined from the splitting of this pattern varies in different lipid phases within an interval of 29–31Å.
As in GASL, the spin-label of TyrLGC does not directly affect the N-terminus of the gramicidin molecule and allows HHD formation. Furthermore, the spin-labeling position should leave the whole channel function intact, as for dansyl-gramicidin C, which was labeled with a fluorescent label at the same position and used for a simultaneous conductivity and fluorescence study 40. The behavior of TyrLGC in membranes is very similar to GASL 29, although, due to the longer nitroxide tethers the distance distribution is broader. Similar to GASL, for matching lipids (DLPC, DMPC) the reporter nitroxide group for the channel form shows lower polarity of the local environment, with higher polarity for mismatching DPPC and DSPC (see Supporting Information, Supplement Fig. 2). Also, in DPPC, with TyrLGC in the non-channel form, the Z axis of the magnetic tensors preferentially aligns parallel to the membrane normal, whereas for DMPC this Z-ordering is somehow distorted (Supporting Information, Supplement Fig. 3, compare to 29,30).
As an important advantage of TyrLGC compared to GASL is that the expected interspin distance in the DH conformation is substantially different from that of the HHD. As seen from Table 1, there is almost no difference between the interspin distances in the two conformations for GASL. Hence, one cannot completely rule out a possibility that the dipolar signal of GASL HHDs contains a fraction from double helices, which complicates the exact determination of the interspin distance for the HHD form. The dipolar spectrum from TyrLGC, however, also shows only one detectable signal in channel-forming lipids and no distinct spin pairs in DPPC and DSPC. The interspin distance for the signal is 35Å in DMPC and 36.5Å in DPPC quenched after a 10 min exposure at 40°C, consistent with the channel form (see Tab. 1). Pulse ESR detects no interspin distance in the range expected for DHD.
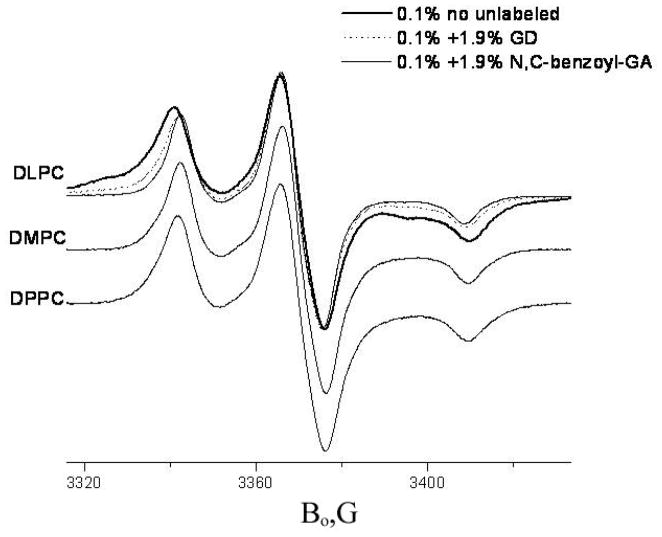
For two other studied spin labeled derivatives of gramicidin, GALN and GADL, the spin label is attached to the N-terminal end of the peptide molecule. The N-terminus of the GA molecule is directly involved in HHD formation (cf. Figs 1 and 2). Replacing the N-terminal formyl group of GA by a BOC group (di-tert-butyl dicarbonate) has been shown 31 to destabilize the dimeric channel compared with GA by 5-orders-of-magnitude. Even a simple formyl/acetyl substitution at the N-terminal end of the gramicidin molecule decreases the mean channel lifetime by a factor of 56 65. Double-labeled GADL and mono-labeled GALN have a bulky spin label at the N-terminal end. One can expect (and, indeed, observes) a substantial reduction in KD for GADL and GALN with a shift of the equilibrium to favor monomers. For a detailed discussion on the effects of replacing the N-terminal formyl in GA by a nitroxide (GALN) or a benzoyl group (N-benzoyl-(desformyl)-gramicidin and N-benzoyl-(desformyl)-GASL) see Supporting Information. Nevertheless, if the channel formation occurs, it should bring the N-terminal spin labels close together (see GALN in Tab. 1), and this could be easily detected by dipolar broadening in the CW ESR spectrum. Fig. 5 shows CW spectra of GALN in DLPC, DMPC and DPPC at 77K. Indeed, in DLPC, GALN shows a broad feature which can be assigned to the HHD form. This means that blocking the N-terminus with the spin label does not completely impair HHD formation.
Fig. 5.
CW spectra of GALN in different lipids, 77K. For DLPC the thick line shows 0.1% mol. GALN, dots 0.1% mol. GALN+1.9% mol. GD, thin solid line 0.1% mol. GALN + 1.9% mol. N, C-dibenzoyl GA.
Rigid biradicals in disordered systems yield a broad ESR signal, which is a superposition of signals from all the orientations of the interspin vector relative to the external magnetic field B0. However aligned samples afford greater resolution. Fig. 6 shows angular-dependent CW spectra of GALN in DLPC membrane aligned by the evaporation method at room temperature. Although an exact assignment of spectral lines is complicated, a simple inspection of the spectrum indicates a splitting picture typical for an ordered rigid biradical; (compare for example with the TEMPAD biradical 66 which has an interspin distance of ~ 8.5 Å). If B0 is directed parallel to the aligned membrane normal, one sees a single orientation of the interspin vector relative to B0 and therefore a well-resolved spectrum. If the interspin vector is not perpendicular to the membrane, other orientations of the membrane relative to B0 would show less resolved spectra, (cf. fig. 6). They will be a superposition of different dipolar orientations vs. B0, since gramicidin molecules can freely rotate in the membrane plane. The number of orientations participating in the superposition and their weight factors will depend on the angular orientation of the membrane relative to B0 and the angle between the membrane normal and the interspin vector (cf. fig 4).
Fig. 6.
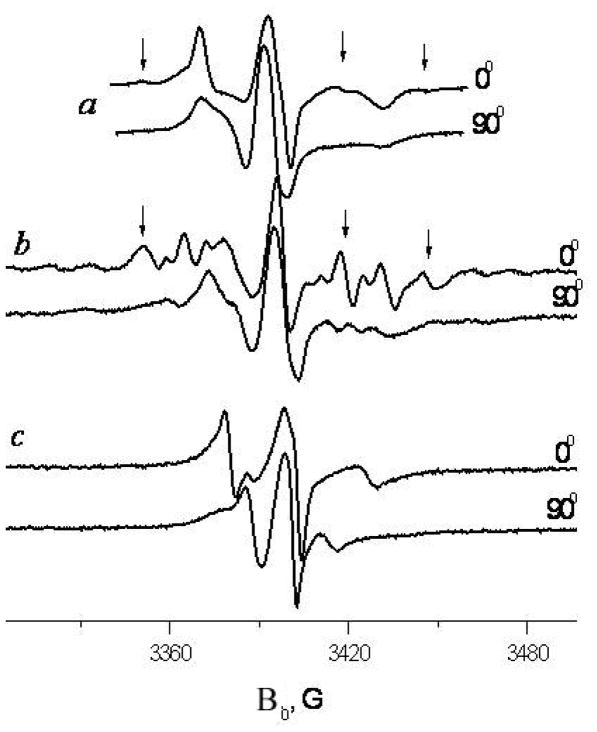
Spectra of GALN in two orientations in aligned membranes of a) DMPC (ISDU aligned, 4°C), b) DLPC and c) EYL (aligned by evaporation, 14°C). Corresponding spectral features are marked with arrows.
GADL has two spin labels in the molecule at both the N- and C- termini (cf. Fig. 1). Fig. 7 shows spectra of GASL, GALN and GADL in the aligned DPPC membrane. Spin labels at the C-terminus of GASL and N-terminus of GALN both show good Z-ordering. One can also see that the spectrum of GADL can, in general, be approximately described as a superposition of spectra for GALN and GASL.
Fig. 7.
Spectra of GASL, GALN and GADL in the ISDU aligned DPPC, 20°C, 0° orientation. Doted curve shows a 1:1 superposition of GASL and GALN spectra.
In DLPC, GADL forms HHDs similar to GALN; their formation also manifests itself in vesicles at 77K in a broad signal (not shown). As expected, since spin labels at the C-termini do not form close couples and should contribute a resolved rigid limit component to the spectrum, the fraction of the broad signal for GADL is approximately half the fraction for GALN. In contrast to DLPC, in DMPC vesicles GALN and GADL show insignificant, if any fraction of a broad signal as discussed above. This difference can be explained by hydrophobic mismatch between the DMPC bilayer and the channel length, which is sufficient to prevent formation of sterically-hindered GALN channels.
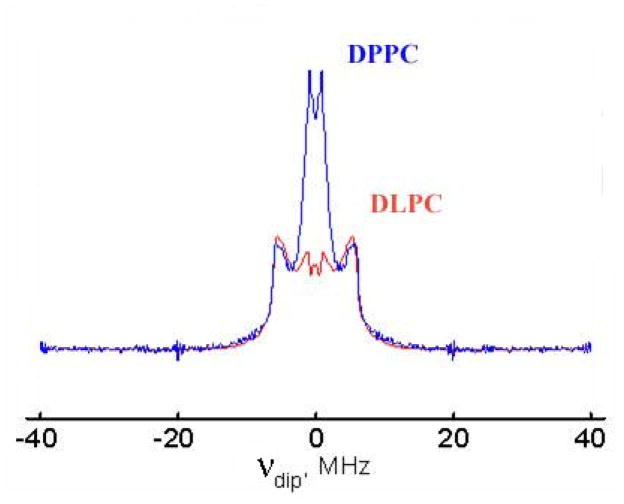
However, with further increase of the bilayer thickness, in DPPC and DSPC, GADL, but not GALN shows again substantial fraction of a broad singlet-like signal. This broad signal also disappears upon dilution with unlabeled GD. Since the signal does not appear for GALN, it cannot be explained by the proximity of N-terminal spin labels of different monomers in the channel form. We attribute this signal to a fraction of double helices. Fig. 8 shows Pulse Dipolar spectra of 0.1% GADL + 1.9% unlabeled GD in DPPC and DLPC. One can clearly see two components, which correspond to two different conformations of GADL. The interspin distance for the main fraction (20.0Å) is the same in both lipid phases. It also exactly coincides with the distance observed for the monomeric state in TFE. Yet, as discussed above, GADL in DLPC readily forms channel dimers, while the hydrophobic mismatch in DPPC destabilizes channels not only for sterically hindered GADL, but also for GASL having unblocked N-termini 29,30. The same interspin distance for these different lipid phases likely indicates similarity in the backbone structure of halves of HHD (DLPC) and free monomers (DPPC). This similarity between the monomeric closed state of GA and a half of HHD was previously suggested from circular dichroism and X-ray in-plane scattering studies for Boc-GA (tert-butoxycarbonyl-GA) 31.
Fig. 8.
Dipolar DEER spectrum of 0.1% GADL + 1.9% unlabeled GD in DLPC and DPPC. The interspin distance can be estimated from the Pake doublet splitting as 70.
Double helices (31.6Å, as observed in octanol) are a minor component with the fraction changing in the saturated lipid series in the following order:
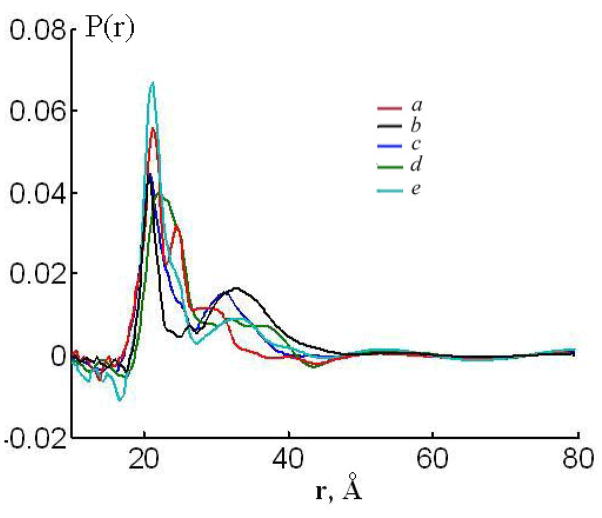
DLPC<DMPC<DSPC<DPPC. The distance distributions P(r) for GADL obtained for different lipid environments, saturated and unsaturated, are shown in Fig. 9. Consistent with previous observations 35, a measurable fraction of double helices is detectable by Pulse Dipolar ESR in unsaturated lipids phases, such as DOPC and egg yolk lecithin.
Fig. 9.
Distance distribution P(r) for 0.1% GADL + 1.9% GD obtained by the Tikhonov regularization with the regularization parameter determined by the L-curve criterion 44 in different lipid environment. a) DLPC, b) DPPC, c) DMPC, d) DOPC, e) EYLlecithin.
HHD/DH/monomers equilibrium
Now that we have shown how Pulse Dipolar ESR in combination with the several spin-labeled gramicidin derivatives enables us to distinguish the different conformers, we address the issue of equilibria amongst them. It has been shown in the literature 51,67,68, etc. that a substantial fraction of DH’s in some lipid environments (e.g. DMPC) example) may not be in equilibrium due to slow conversion of DH’s introduced from some organic solvents into HHD’s. It also has been shown that using trifluoroethanol as a cosolubilization solvent yields the β6.3-helical conformation (HHD or monomers), while incorporation from CHCl3/methanol mixtures gives a non-equilibrium mixture of HHD and DH conformations. Upon long incubation at 60° or sonication the system reaches equilibrium, which favors HHD’s 67,68. On the other hand, in some lipid environment a fraction of DH’s is assumed to be in equilibrium with channels 35,36.
To assign the fraction of double helices observed by pulse ESR to either conformers in equilibrium in the membrane or to a metastable state we incubated for 2 hrs at ~ 70°C and also compared samples prepared from TFE and CHCl3/Methanol solutions. Since we found that the ratio of spectral components corresponding monomers and DH dimers did not depend on the method of sample preparation, we infer that we obtained DH’s in equilibrium with monomers
This conclusion is in good accord with our CW data. Fig. 10 shows the dependence of the CW spectrum of GADL in DPPC on the GADL/DPPC ratio. One can see a substantial increase in the broad signal intensity. The fraction of the signal is substantially increasing (from <10% to ~ 66%) with an increase in the GADL/DPPC ratio from 0.1 to 2%. To determine its fraction the broad signal was approximated with a Gaussian singlet. As seen in fig. 10c, subtracting the broad signal from the initial GADL/DPPC signal gives reasonably good approximation for the signal for the 1:19 GADL/GD mixture in DPPC and a flat baseline. The broad signal fraction F was estimated from the double integrals for the broad Gaussian and the initial spectrum. Note that the fraction of the broad signal does not change upon sonication, which is a proven approach to obtain GA/membrane samples in thermodynamically stable form 67.
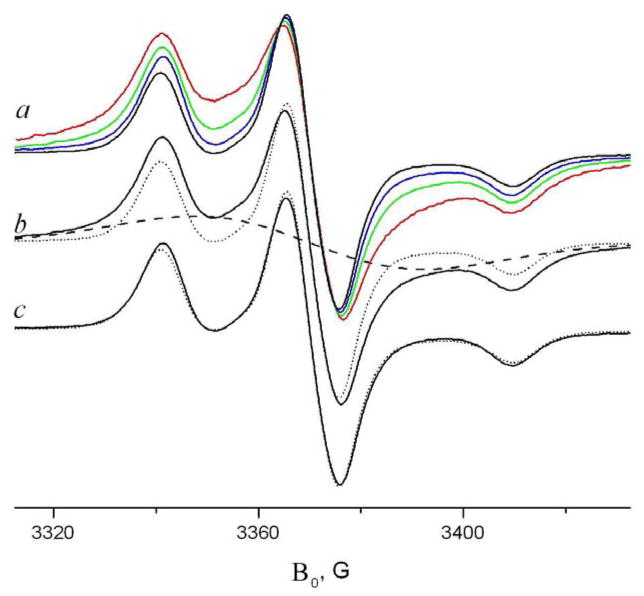
Fig. 10.
a) Concentration dependence of GADL spectrum in DPPC at 77K, % mol: 2% (red), 0.5% (green) 0.1% (blue) 0.1% GADL +1.9% GD (black) b) analysis of the spectrum with 0.5% GADL by separating the broad signal. The solid line is 0.5% GADL in DPPC, dashed line is a Gaussian approximation for the broad signal. c) solid line is the subtraction of the Gaussian approximation from the experimental spectrum. Both dotted lines (b, c) are 0.1% GADL + 1.9% unlabeled GD in DPPC
Since there are no HHDs in DPPC, (they cannot be detected for GASL – so GADL with impaired channel forming capacity certainly does not form them) the signal can be attributed either to aggregation of monomers or to double helices. However, both single labeled analogs of GADL, GASL and GALN, do not show any conspicuous broadening effects under the same conditions. This indicates distances between N-termini – N-termini or C-termini – C-termini of at least 20Å and therefore the broad signal cannot be explained by parallel DHDs. To assign the broad signal to aggregation one might suggest some kind of head-to-tail pattern in the aggregates of GADL monomers with a close N-terminus – C-terminus proximity. However, this possibility can also be ruled out. Our polarity data indicate that the N-terminal spin label (GALN) is located in the hydrophobic part of the membrane, whereas the C-terminus (GASL) is more exposed to the water phase (the corresponding 2Azz values at 77K are 66.9 and 70.6G for the 0.1% peptide/lipid ratio). Besides, gramicidin in DPPC at low concentrations (<0.1% GA/DPPC ratio) already exists in aggregates 29,38 and any further increase in concentration should not substantially affect the spectra. On the other hand, for DHDs (PDB:1ALZ) MMD simulations show close proximity (~ 9Å) between the N- and C-terminal spin labels of different GADL monomers (fig 3A). Based on this evidence we assign the broad signal to the same antiparallel double helical structures that were detected by Pulse Dipolar ESR for GADL/unlabeled GA mixtures. When double helices exist in equilibrium with monomers DH ↔ 2M, then [DH]=K [M]2, where K is the corresponding equilibrium constant. If [G] is the total concentration of GADL, then [G]=2[DH]+[M], and 2K[M]2+[M]−[G]=0
Which gives , thus
At 0.1% GADL in the membrane of the broad signal is 19% of the total spectrum which gives and 2[G]K=0.28. For a surface area per molecule of 0.59 nm2 in fluid DPPC 69 it gives K=5×1011 cm2/mol (compare to the values of dimerization constants for HHD formation, see Supporting Information) For 0.5% GADL (2[G]K=1.42) in equilibrium we can expect 44% of the spins to be contributing to the broad signal and ~ 66% for 2% molar ratio GADL/DPPC, in good agreement with our experimental estimates (45 and 68%).
Comparing fractions of double helical dimers for 1.9% GD+0.1%GADL determined by pulse ESR and for 2% GADL in DPPC by CW ESR one finds that K is about twice as great for the latter. The discrepancy might be due to effects of the labels at the N-terminus which in the monomeric conformation could perturb the bilayer (compare to the effect of tryptophan residues discussed below) and thus cause some energy cost shifting the equilibrium towards DHs.
A fraction of double helical dimers also well qualitatively explains the change in CW spectra for the system GALN/GD/DPPC or GASL/GD/DPPC with an increase in total gramicidin concentration. (see Supporting Information).
Tryptophans and aggregation of DHDs
As follows from our results, the double helical form is always present in a mismatching lipid environment. Its presence can be directly detected for GADL by Pulse Dipolar ESR through the interspin distance fingerprint or inferred from CW spectra in the case of GASL or GALN. However, unlike HHD, single labeled gramicidin derivatives (GASL, GALN, TyrLGC) in the lipid membrane do not show distinct spin pairs with the interspin distance expected for the corresponding DH structure. It indicates a propensity of DHDs to aggregate. This aggregation could be caused by the tendency of tryptophan residues to get excluded from the hydrophobic part of the lipid bilayer and their specific affinity for a well defined site near the lipid carbonyl region at the membrane interface 58. While for HHD all tryptophans can be located at the membrane surface, in DH conformations of GA they are distributed more evenly along the length of the dimer with Trp9 and Trp11 near the membrane center 26. The energy cost of submerging the tryptophans in the middle of the membrane could be mitigated by aggregation, with stacking the tryptophan residues or locating them at the disordered vicinity of another GA molecule. The aggregation behavior of free GA monomers may be different in different lipid environment. However, for GALN in DMPC similar to described in Appendix of 29 experiments showed little signs of aggregation. This data implies that at least in some lipid environments monomers can exist as separate entities with a uniform distribution in the membrane plane.
CONCLUSIONS
Pulse Dipolar ESR spectroscopy and CW ESR were used to study the equilibrium of gramicidin conformations.
Several new spin-labeled derivatives of gramicidin were synthesized.
The interspin distances of their monomeric and double helical (PDB:1ALZ) forms were initially determined in octanol and TFE in order to recognize the forms in membranes.
All spin-labeled variants of gramicidin studied are able to form HHD in matching membrane environments. However, for N-terminal substituted derivatives the propensity to form HHD’s is substantially impaired.
The interspin vector between two spin labels attached at different C-termini of the gramicidin channel is perpendicular to the membrane normal. The corresponding interspin distance is lipid dependent. It varies by ~ 3Å and shows systematic increase with an increase in hydrophobic mismatch.
Non-channel forms, which include monomers and double helices, can be detected by ESR in mismatching lipid environment. There are indications that the monomers, which were for the first time reliably observed spectroscopically in a membrane environment, are similar to halves of the HHD.
Double helices (DH) appear from sample preparations in equilibrium with monomers and HHD. The DH form of studied spin labeled gramicidin derivatives detected by ESR in the membrane environment is an anti-parallel, most likely the ππ5.6 left-handed dimer. The fraction of double helices changes in saturated lipids as DLPC≤DMPC≪DSPC<DPPC. While HHDs and free monomers, at least in some lipid environments, exist as separate entities, double helices tend to aggregate. For channel-forming lipids, such as DLPC and DMPC, the DH fraction is insignificant.
In addition, we studied the effect of N-terminal substitution in the GA molecule upon channel formation.
Supplementary Material
Acknowledgments
We thank Prof. T. Begley for fruitful discussions on organic synthesis, Prof. O. Andersen for a critical reading of the manuscript. This work was supported by grants NIH/NIBIB R010EB003150 and NIH/NCRR P41RR 016292.
List of Abbreviations
- GA
Gramicidin A
- GASL
gramicidin A spin labeled at the C-terminus
- GADL
gramicidin A double spin labeled at both C- and N- termini
- GALN
gramicidin A spin labeled at the N-terminus
- TyrLGC
gramicidin C labeled at 11Tyr
- HHD
Head-to- Head dimer
- DH, DHD
double helices, double helical dimers
- DLPC
1,2-lauroyl-sn-glycero-3-phosphocholine
- DMPC
1,2-dimyristoyl-sn-glycero-3-phosphocholine
- DPPC
1,2-dipalmitoyl-sn-glycero-3-phosphocholine
- DSPC
1,2-stearoyl-sn-glycero-3-phosphocholine
- DOPC
1,2-oleoyl-sn-glycero-3-phosphocholine
- EYL
Egg Yolk Lecithin
- TFE
2,2,2 – trifluoroethanol
- ISDU
Isopotential Spin-Dry Centrifugation
Footnotes
Supporting Information available
They include detailed protocols for synthesis and purification of new gramicidin derivatives; a figure showing HHD and ππ5.6 left-handed antiparallel DH structures for GASL in comparison; ESR spectra of TyrLGC in multilamellar vesicles and aligned membranes; discussion on effects of N-terminal substitution in the GA molecule (formyl to the spin label or a benzoyl group) on the HHD dimerization constant and equilibrium of corresponding homodimers and heterodimers with unsubstituted GA; further discussion on detection of DHD by CW ESR. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Borbat PP, Mchaourab HS, Freed JH. JACS. 2002;124:5304. doi: 10.1021/ja020040y. [DOI] [PubMed] [Google Scholar]
- 2.Hubbell W, Altenbach C. Site-directed spin labeling of membrane proteins. In: White SH, editor. Membrane Protein Structure:Experimental Approaches. Oxford University Press; London: 1994. p. 224. [Google Scholar]
- 3.Steinhoff HJ. Biol Chem. 2007;385:913. doi: 10.1515/BC.2004.119. [DOI] [PubMed] [Google Scholar]
- 4.Borbat PP, Freed JH. Methods in Enzymology. 2007;423:52. doi: 10.1016/S0076-6879(07)23003-4. [DOI] [PubMed] [Google Scholar]
- 5.Freed JH. ESR and Molecular Dynamics, Ch.9. In: Eaton S, Berliner GELJ, editors. Biological Magnetic Resonance, v.24. Biomedical EPR, Part B: Methodology, Instrumentation and Dynamics. Vol. 24. 2005. p. 239. [Google Scholar]
- 6.Hubbell W, Altenbach C. Curr Opin Struct Biol. 1994;4:566. doi: 10.1016/j.sbi.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang Z, Freed JH. J Phys Chem B. 1999;103:6384. [Google Scholar]
- 8.Borbat PP, Costa-Filho AJ, Earle KA, Moscicki JK, Freed JH. Science. 2001;291:266. doi: 10.1126/science.291.5502.266. [DOI] [PubMed] [Google Scholar]
- 9.Lou Y, Ge M, Freed JH. J Phys Chem B. 2001;105:11053. [Google Scholar]
- 10.Sezer D, Freed JH, Roux B. JACS. 2009;131:2597. doi: 10.1021/ja8073819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Z, Fleissner M, Tipikin D, Liang Z, Moscicki JK, Earle KA, Hubbell W, Freed JH. J Phys Chem B. 2010;114:5503. doi: 10.1021/jp910606h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Busath DD. Annu Rev Physiol. 1993;55:473. doi: 10.1146/annurev.ph.55.030193.002353. [DOI] [PubMed] [Google Scholar]
- 13.Andersen OS, Koeppe REI, Roux B. Biological Membranes Ion Channels. Dinamics, Structure and Applications. Springer; New York: 2007. Gramicidin Channels: Versatile Tools; p. 33. [Google Scholar]
- 14.Wallace BA. J Struct Biol. 1998;121:123. doi: 10.1006/jsbi.1997.3948. [DOI] [PubMed] [Google Scholar]
- 15.Hotchkiss RD, Dubois RJ. J Biol Chem. 1940;132:791. [Google Scholar]
- 16.Sarges R, Witkop B. JACS. 1965;87 doi: 10.1021/ja01087a028. [DOI] [PubMed] [Google Scholar]
- 17.Veatch WR, Blout ER. Biochemistry. 1974;13:5257. doi: 10.1021/bi00723a002. [DOI] [PubMed] [Google Scholar]
- 18.Langs DA. Science. 1988;241:188. doi: 10.1126/science.2455345. [DOI] [PubMed] [Google Scholar]
- 19.Burkhart BM, Gassman RM, Langs DA, Pangborn WA, Duax WL. Biophys J. 1998;75:2135. doi: 10.1016/S0006-3495(98)77656-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burkhart BM, Li N, Langs DA, Pangborn WA, Duax WL. Proc Natl Acad Sci USA. 1998;95:12950. doi: 10.1073/pnas.95.22.12950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y, Tucker A, Wallace BA. J Mol Biol. 1996;264:757. doi: 10.1006/jmbi.1996.0675. [DOI] [PubMed] [Google Scholar]
- 22.Doyle DA, Wallace BA. J Mol Biol. 1997;266:963. doi: 10.1006/jmbi.1996.0837. [DOI] [PubMed] [Google Scholar]
- 23.Ketchem RR, Roux B, Cross TA. Structure (London) 1997;5:1655. doi: 10.1016/s0969-2126(97)00312-2. [DOI] [PubMed] [Google Scholar]
- 24.Arseniev AS, Lomize AL, Barsukov IL, Bystrov VF. Biol Membr. 1986;3:1077. [Google Scholar]
- 25.Townsley LE, Tucker WA, Sham S, Hinton JF. Biochemistry. 2001;40:11676. doi: 10.1021/bi010942w. [DOI] [PubMed] [Google Scholar]
- 26.Sun H, Greathouse DV, Andersen OS, Koeppe REI. J Biol Chem. 2008;283:22233. doi: 10.1074/jbc.M802074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Killian JA. BBA. 1998;1376:401. doi: 10.1016/s0304-4157(98)00017-3. [DOI] [PubMed] [Google Scholar]
- 28.Mobashery N, Nielsen CM, Andersen OS. FEBS letters. 1997;412:15. doi: 10.1016/s0014-5793(97)00709-6. [DOI] [PubMed] [Google Scholar]
- 29.Dzikovski BG, Borbat PP, Freed JH. Biophys J. 2004;87:3504. doi: 10.1529/biophysj.104.044305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dzikovski BG, Earle KA, Pachtchenko SV, Freed JH. J Magn Reson. 2006;179:273. doi: 10.1016/j.jmr.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 31.He K, Ludtke SJ, Wu Y, Huang HW, Andersen OS, Greathouse D, Koeppe RE., II Biophys Chem. 1994;49:83. doi: 10.1016/0301-4622(93)e0085-j. [DOI] [PubMed] [Google Scholar]
- 32.Bamberg E, Läuger P. J Membr Biol. 1973;11:177. doi: 10.1007/BF01869820. [DOI] [PubMed] [Google Scholar]
- 33.Rokitskaya TI, Antonenko YN, Kotova EA. BBA. 1996;1275:221. doi: 10.1016/0005-2728(96)00025-4. [DOI] [PubMed] [Google Scholar]
- 34.Lundbaek JA, Andersen OS. Biophys J. 1999;76:889. doi: 10.1016/S0006-3495(99)77252-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sychev SV, Barsukov LI, Ivanov VT. Eur Biophys J. 1993;22:279. doi: 10.1007/BF00180262. [DOI] [PubMed] [Google Scholar]
- 36.Zein M, Winter R. Phys Chem Chem Phys. 2000;2:4545. [Google Scholar]
- 37.Schönknecht G, Althoff G, Junge W. J Membr Biol. 1992;126:265. doi: 10.1007/BF00232323. [DOI] [PubMed] [Google Scholar]
- 38.Mou J, Czajkowsky DM, Shao Z. Biochemistry. 1996;35:3222. doi: 10.1021/bi9520242. [DOI] [PubMed] [Google Scholar]
- 39.Tank DW, Wu ES, Meers PR, Webb WW. Biophys J. 1982;40:129. doi: 10.1016/S0006-3495(82)84467-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Veatch WR, Mathies R, Eisenberg M, Stryer L. J Mol Biol. 1975;99:75. doi: 10.1016/s0022-2836(75)80160-4. [DOI] [PubMed] [Google Scholar]
- 41.Macdonald PM, Seelig J. Biochemistry. 1988;27:2357. doi: 10.1021/bi00407a017. [DOI] [PubMed] [Google Scholar]
- 42.Pepinsky RB, Feigenson GW. Anal Biochem. 1978;86:512. doi: 10.1016/0003-2697(78)90776-5. [DOI] [PubMed] [Google Scholar]
- 43.Upadhyay AK, Borbat PP, Wang J, Freed JH, Edmondson DE. Biochemistry. 2008;47:1554. doi: 10.1021/bi7021377. [DOI] [PubMed] [Google Scholar]
- 44.Chiang YW, Borbat PP, Freed JH. J Magn Reson. 2005;172:279. doi: 10.1016/j.jmr.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 45.Wallace BA, Ravikumar K. Science. 1988;241:182–187. doi: 10.1126/science.2455344. [DOI] [PubMed] [Google Scholar]
- 46.Arseniev AS, Barsukov LI, Bystrov VF. FEBS letters. 1985;180:33. doi: 10.1016/0014-5793(85)80702-x. [DOI] [PubMed] [Google Scholar]
- 47.Pascal SM, Cross TA. J Mol Biol. 1992;226:1101. doi: 10.1016/0022-2836(92)91055-t. [DOI] [PubMed] [Google Scholar]
- 48.Pascal SM, Cross TA. J Biomol NMR. 1993;3:495. doi: 10.1007/BF00174606. [DOI] [PubMed] [Google Scholar]
- 49.O’Boyle F, Wallace BA. Protein & Peptide Lett. 2003;10:9. doi: 10.2174/0929866033408246. [DOI] [PubMed] [Google Scholar]
- 50.Hansch C. Acc Chem Res. 1969;2:232. [Google Scholar]
- 51.Bouchard M, Auger M. Biophys J. 1993;65:2484. doi: 10.1016/S0006-3495(93)81300-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Killian JA. BBA. 1992;1113:391. doi: 10.1016/0304-4157(92)90008-x. [DOI] [PubMed] [Google Scholar]
- 53.Urry DW, Mayers DF, Haider J. Biochemistry. 1972;11:487. doi: 10.1021/bi00754a002. [DOI] [PubMed] [Google Scholar]
- 54.Jagannadham MV, Nagaraj R. Biopolimers. 2005;80:708. doi: 10.1002/bip.20293. [DOI] [PubMed] [Google Scholar]
- 55.Hawkes GE, Lian LY, Randall EW, Sales KD, Curzon EH. Eur J Biochemistry. 1987;166:437. doi: 10.1111/j.1432-1033.1987.tb13535.x. [DOI] [PubMed] [Google Scholar]
- 56.Barnes JP, Freed JH. Biophys J. 1988;75:2532. doi: 10.1016/S0006-3495(98)77698-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Larsen RG, Singel DJ. J Chem Phys. 1993;98:5134. [Google Scholar]
- 58.de Planque MRR, Kruijtzer JAW, Liskamp RMJ, Marsh D, Greathouse DV, Koeppe REI, de Kruijff B, Killian AJ. J Biol Chem. 1999;274:20839–20846. doi: 10.1074/jbc.274.30.20839. [DOI] [PubMed] [Google Scholar]
- 59.Harroun TA, Heller WT, Weiss TM, Yang L, Huang HW. Biophys J. 1999;76:937. doi: 10.1016/S0006-3495(99)77257-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nielsen CM, Goulian M, Andersen OS. Biophys J. 1998;74:1966. doi: 10.1016/S0006-3495(98)77904-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Katsaras J, Prosser RS, Stinson RH, Davis JH. Biophys J. 1992;61:827. doi: 10.1016/S0006-3495(92)81888-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Killian JA, Nyholm TKM. Curr Opin Struct Biol. 2006;16:473. doi: 10.1016/j.sbi.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 63.Daily AE, Greathouse DV, van der Wel PCA, Koeppe REI. Biophys J. 2008;94:480. doi: 10.1529/biophysj.106.097543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Andersen OS, Koeppe REI. Ann Rev Biophys Biomol Struct. 2007;36:107. doi: 10.1146/annurev.biophys.36.040306.132643. [DOI] [PubMed] [Google Scholar]
- 65.Szabo G, Urry DW. Science. 1979;203:55. doi: 10.1126/science.83000. [DOI] [PubMed] [Google Scholar]
- 66.Nakajima A. Bull Chem Soc Japan. 1973;46:1129. [Google Scholar]
- 67.LoGrasso PV, Moll F, III, Cross TA. Biophys J. 1988;54:259. doi: 10.1016/S0006-3495(88)82955-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Killian JA, Nicholson LK, Cross TA. BBA. 1988;943:535. doi: 10.1016/0005-2736(88)90387-2. [DOI] [PubMed] [Google Scholar]
- 69.Marsh D. Handbook of Lipid Bilayers. CRC Press; Boca Raton, FL: 1990. [Google Scholar]
- 70.Borbat PP, Freed JH. Double-Quantum ESR and Distance Measurements. In: Berliner LJ, Eaton GR, Eaton SS, editors. Biological Magnetic Resonance. Vol. 19. Academic/Plenum Publishers; New York: 2000. p. 383. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.