Abstract
We developed a two-layer, four-channel PCR-capillary electrophoresis microdevice that integrates nucleic acid amplification, sample cleanup and concentration, capillary electrophoretic separation and detection for multiplex analysis of four human respiratory viral pathogens influenza A, influenza B, coronavirus OC43, and human metapneumovirus. Biotinylated and fluorescently labeled double-stranded (ds) DNA amplification products are generated in a 100-nL PCR reactor incorporating an integrated heater and a temperature sensor. After amplification, the products are captured and concentrated in a crosslinked acrylamide gel capture matrix copolymerized with acrydite-functionalized streptavidin-capture agents. Thermal dehybridization releases the fluorescently labeled DNA strand for capillary electrophoresis injection, separation and detection. Using plasmid standards containing the viral genes of interest, each target can be detected starting from as few as 10 copies/reactor. By employing one-step reverse transcription PCR amplification, the device can detect RNA analogues of all four viral targets with detection limits in the range of 25–100 copies/reactor. The utility of the microdevice for analyzing samples from nasopharyngeal swabs is demonstrated. Combining size-based separation with four-color detection, this platform provides excellent product discrimination, making it readily extendable to higher-order multiplex assays. This portable microsystem is also suitable for performing automated assays in point-of-care diagnostic applications.
Keywords: microfabrication, microfluidics, infectious disease detection, point-of-care diagnostics, lab-on-a-chip, viral detection, acute respiratory disease
Introduction
Accurate and rapid detection of the pathogens responsible for infectious human respiratory disease is crucial for outbreak control, disease surveillance, and vaccine development, as well as for appropriate and efficient use of antibiotics and antiviral therapies [1–3]. Considering the wide range of existing and emerging microbial pathogens causing respiratory illnesses, many of which begin with similar influenza-like symptoms [4], multiplex diagnostic assays i.e., assays that simultaneously screen for many pathogens, are imperative for clinical medicine and public health [3, 5–7]. Multiplex polymerase chain reaction (PCR), in which distinct primer pairs are used to amplify more than one target sequence in a single reaction, is one of the most appealing methods in molecular diagnostics because of its high sensitivity, rapid turnaround time and high throughput [6–10]. Through careful primer design and the use of hot start polymerase enzymes, high specificity and sensitivity can be achieved [6, 11]. A high degree of multiplexing, ranging from 2- to 22-plex for detection of respiratory pathogens including bacteria and viruses, has been reported [1, 5, 7, 12–16].
Advancements in miniaturization and microfluidic integration technology provide a pathway for the widespread practice of multiplex PCR diagnostics in clinical settings. By integrating multiple processes onto a single microfabricated device, rapid, low-volume, automated and sensitive assays can be performed [17–22]. Because of the superior performance offered by integrated microdevices, a variety of pathogens including the respiratory pathogen Bordetella pertussis [23], SARS-coronavirus [24], dengue-2 virus [25], influenza A virus [26] , and BK virus [27] have already been successfully detected with on-chip approaches. Nevertheless, most of these assays have been limited to single- or duplex analysis [23–27] performed on preliminary PCR-capillary electrophoresis (CE) microdevices, which do not include sample treatment and processing [20, 23–27]. Recently, our group developed an integrated PCR-CE device with a pre-amplification immunomagnetic bead-based cell capture for highly specific and sensitive detection of E. coli [28]. In addition, we have reported a novel integrated PCR-CE device with gel-phase oligonucleotide affinity capture for post-amplification cleanup [29]. By copolymerizing oligonucleotide capture probes in a photodefinable cross-linked polyacrylamide matrix directly between the PCR reactor and the CE separation channel, PCR products complementary to those probes are specifically hybridized and preconcentrated. This integrated capture/injection technique exhibits a three- to five-fold increase in signal intensity and a six-fold increase in resolution compared to that using externally polymerized, non-crosslinked capture matrices [29]. The technique, however, is limited in its ability to scale up for multiplex analysis because of the complexity in designing unique and specific capture probes with similar thermodynamic properties for multiple targets.
To expand the multiplexing capability of our devices to the level needed for practical pathogen detection, we developed a universal gel capture purification method that relies on the biotinstreptavidin recognition [30]. PCR products, one strand biotinylated and the other fluorescently labeled, are captured in their native double-stranded (ds) state by a photopolymerized streptavidin capture gel, followed by electrokinetic washing and thermal melting of the bound DNA. The flourophore-labeled strands are released and injected for electrophoresis, leaving the biotinylated strands bound to the capture gel. This technique has previously been used to demonstrate on-chip sample cleanup for forensic short tandem repeat analysis [30, 31], but is applied here for the first time to multiplex pathogen detection in an integrated format.
We present a new two-layer integrated PCR-CE microdevice with streptavidin/biotin-mediated post-amplification sample processing technology for rapid and parallel detection of respiratory viruses responsible for acute respiratory diseases (ARD). The pathogens include influenza A virus (FLUAV), human metapneumovirus (hMPV), human coronavirus OC43 (hCoV-OC43), and influenza B virus (FLUBV). Coupled with the multi-channel capillary array electrophoresis portable scanner (McCAEPS) instrument previously developed in our group [32], the entire process of nucleic acid amplification, capture and CE analysis is streamlined and automated, thus minimizing contamination and sample loss between steps.
Experimental Section
Microdevice design and fabrication
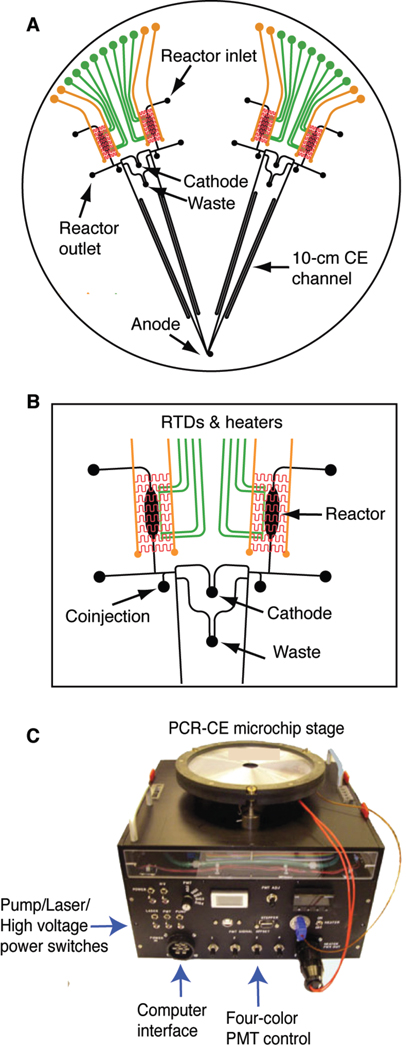
The integrated microdevice shown in Figure 1A contains four identical units forming two symmetrical doublet structures similar to the PCR-CE device previously developed in our group [33, 34]. In addition to PCR amplification and CE separation, the new microdevice is designed to accomplish post-amplification cleanup/concentration, as well as inline injection of amplified products. Each genetic analysis system consists of a 100-nL reaction chamber, an integrated heater and resistive temperature detector (RTD), a capture and injection region containing a 500-µm long double T-junction, and a 10-cm long CE separation channel. The expanded view of the analyzing units is shown in Figure 1B. To facilitate automated operation using the portable microcapillary array electrophoresis (µCAE) instrument (Figure 1C), the two doublets are designed to share common waste and cathode ports, while all four lanes share a common anode.
Figure 1.
(A) Layout of the integrated PCR-CE microdevice. Each of the four analyzers is comprised of an RTD temperature sensor (green) and integrated heater (red and orange) for thermal cycling, a 100-nL PCR reaction chamber, etched structures for analyte capture and purification, and 10-cm long CE separation channels with common cathode and waste structures. (B) Magnified view of one reactor pair with temperature control elements and capture/purification structures. (C) The portable 12 ×12 ×10” multi-channel capillary array electrophoresis portable scanner (McCAEPS) instrument contains all necessary elements for microchip control and operation as well as fluorescence excitation and detection.
Microchannel features are patterned photolithographically on the surface of a four-inch glass wafer and sealed by thermally bonding a second wafer to form enclosed structures. The top wafer contains Ti/Pt heaters fabricated on its top-side and the etched fluidic structure containing reactors, capture regions, and separation channels on the bottom, while the bottom wafer contains microfabricated Ti/Pt RTDs on its top side. The microfabrication procedure of the microdevice follows the protocols developed previously [33–35]. Briefly, the fluidic layer is created from a 500-µm thick Borofloat glass wafer coated with a 2000-Å layer of amorphous silicon on one side and 200-Å Ti/ 2000-Å Pt on the other side. Fluidic structures are etched on the amorphous silicon side to a depth of 40 µm using 49% hydrofluoric (HF) acid. Heaters are fabricated on the Ti/Pt side of the same wafer by electroplating gold leads followed by ion beam etching of the Ti/Pt to form the serpentine heater elements. All access holes are then drilled using a CNC mill. The RTD layer is formed using a 762-µm Borofloat glass wafer coated with 200-Å Ti and 2000-Å Pt. Features are formed by etching the coated glass wafer in hot aqua regia (3:1 HCl: HNO3) for 5 min. The two wafers are then permanently bonded together in a vacuum furnace at 650 °C for 6 hr.
Sample preparation
The multiplex PCR panel was adapted from a 22-plex MassTag PCR assay [1]. Gene targets, primer sequences, and expected product fragment sizes are listed in Table 1. Primer sequences were designed based on conserved genomic regions so that each pathogen target could be robustly detected. To ensure stringent multiplex assays, the primers chosen had been verified to have similar melting temperatures with minimal cross-hybridization interactions to avoid primer-dimers. To enable streptavidin/biotin-mediated capture and cleanup, the forward primers were fluorescently labeled at the 5’ end with FAM, TAMRA, or JOE, while the 5’-end of the reverse primers were functionalized with biotin (Integrated DNA Technologies (IDT), Coralville, IA). Each PCR mix was prepared to a volume of 25 µL, containing up to 8 µL of up to four different types of plasmid standards, 0.2 U of HotStarTaq DNA polymerase, 0.5 mM MgCl2, 0.5 mM dNTPs, 2.5 µL of multiplex PCR buffer containing 6 mM MgCl2, pH 8.7 (Qiagen, Valencia, CA), 2.5 µL of Q-solution (Qiagen), and additional 1.25 µL of multiplex master mix containing HotStar Taq DNA polymerase, multiplex PCR buffer, and dNTP mix (Qiagen), and 0.2, 0.4, 0.8, and 0.8 µM (each) concentrations of the forward and reverse primers for FLUBV, hMPV, hCoV-OC43, and FLUAV, respectively. These primer ratios and concentrations were optimized to give similar product yield for all targets at the same starting concentrations. The same assay conditions were applied for standards and swab samples, allowing successful detection of the respective targets. Standard plasmids containing the gene target sequence were prepared from virus or assembled from synthetic polynucleotides. The plasmid standards were diluted in 2.5 ng/µL human placenta DNA to mimic random nucleic acid background present in clinical samples, and stored at −20 °C. cDNA standards generated from nasopharyngeal swabs (NPS) that contained the respective agents were used to assess performance of the final assay. Briefly, total nucleic acid from 250 µL NPS suspended in 1 ml standard viral transport media had been extracted using a NucliSENS easyMAG system (Biomerieux, Inc.). Reverse transcription was performed with 10 µL of total nucleic acid using Superscript II with hexamer priming according to the manufacturer’s protocol (Invitrogen, Carlsbad, CA). Three µL cDNA sample were added to the PCR cocktail to a final volume of 20 µL. Off-chip reactions were performed in parallel to the on-chip experiments using an Eppendorf Mastercycler thermocycler following the same thermocycling protocol. The amplified product was analyzed by conventional capillary electrophoresis using an Applied Biosystems 96 capillary 3730 xl DNA Analyzer. The final sample mixture was prepared using 9:1:1 ratio of PCR Hi-Di™ Formamide (Applied Biosystems): unpurified PCR sample: 500 GeneScan™ ROX size standard (Applied Biosystems).
Table 1.
Primer sequences for multiplex respiratory virus detection
| Pathogen/Gene target | Primer | Tm (°C) | Sequence | Size (bp) |
|---|---|---|---|---|
| FLUBV/Hemagglutinin protein (HA gene) | BHA-U188-1 | 52 | 5’-FAM-AGA CCA GAG GGA AAC TAT GCC-3’ [35] | 159 |
| BHA-L347-1 | 54 | 5’-Biotin-CTG TCG TGC ATT ATA GGA AAG CA-3’ [35] | ||
| hMPV/Nucleocapsid protein (N gene) | MPV01.2 | 52 | 5’-TAMRA-AAC CGT GTA CTA AGT GAT GCA CTC-3’ [36] | 212 |
| MPV02.2 | 60 | 5’-Biotin-CAT TGT TTG ACC GGC CCC ATA A-3’ [36] | ||
| hCoV-OC 43/Nucleocapsid protein (N gene) | Co43-270F | 55 | 5’-JOE-TGT GCC TAT TGC ACC AGG AGT-3’ [1] | 238 |
| Co43-508R | 54 | 5’-Biotin-CCC GAT CGA CAA TGT CAG C-3’ [1] | ||
| FLUAV/Matrix protein (M gene) | AM-U151 | 56 | 5’-FAM-CAT GGA ATG GCT AAA GAC AAG ACC-3’ [35] | 246 |
| AM-L397 | 55 | 5’-Biotin-AAG TGC ACC AGC AGA ATA ACT GAG-3’ [36] | ||
For each one-step RT-PCR reaction, a 20 µL RT-PCR master mix is composed of 0.5 µL of OneStep RT-PCR enzyme mix containing Ominiscript Reverse Transcriptase, Sensiscript Reverse Transcriptase, and HotStarTaq DNA Polymerase (Qiagen), 5 µL of OneStep RT-PCR buffer containing 12.5 mM MgCl2 (Qiagen), 0.2 U of HotStarTaq DNA polymerase, 0.5 mM MgCl2, 0.5 mM dNTPs, 2.5 µL of multiplex PCR buffer containing 6 mM MgCl2, pH 8.7 (Qiagen), 5 µL of Q-solution (Qiagen), and 10 U of RNase inhibitor (Promega, Madison, WI).
To prevent the formation of secondary structure during the reaction, the RNA template and primer mixture was prepared separately: the 5 µL primer-template mixture contains 2.5 µL of the RNA standards and 2.5 µL of 0.5, 2.0, 1.5, and 2.0 µM (each) forward and reverse primers for FLUBV, hMPV, hCoV-OC43, and FLUAV, respectively. The primer-template mixture was heat-denatured at 75 °C for 5 min then immediately chilled on ice to allow the primers to bind to the RNA templates. Finally, the RT-PCR master mix was added to the primer-annealed templates. In vitro transcribed RNA standards were diluted in 2.5 ng/µL yeast t-RNA (Sigma) to prevent RNA degradation due to frequent freeze-thaw cycles and stored aliquoted and stored aliquoted at −80 °C. Materials and methods employed for customized DNA ladder synthesis can be found in the supporting information (Section I).
Matrix synthesis
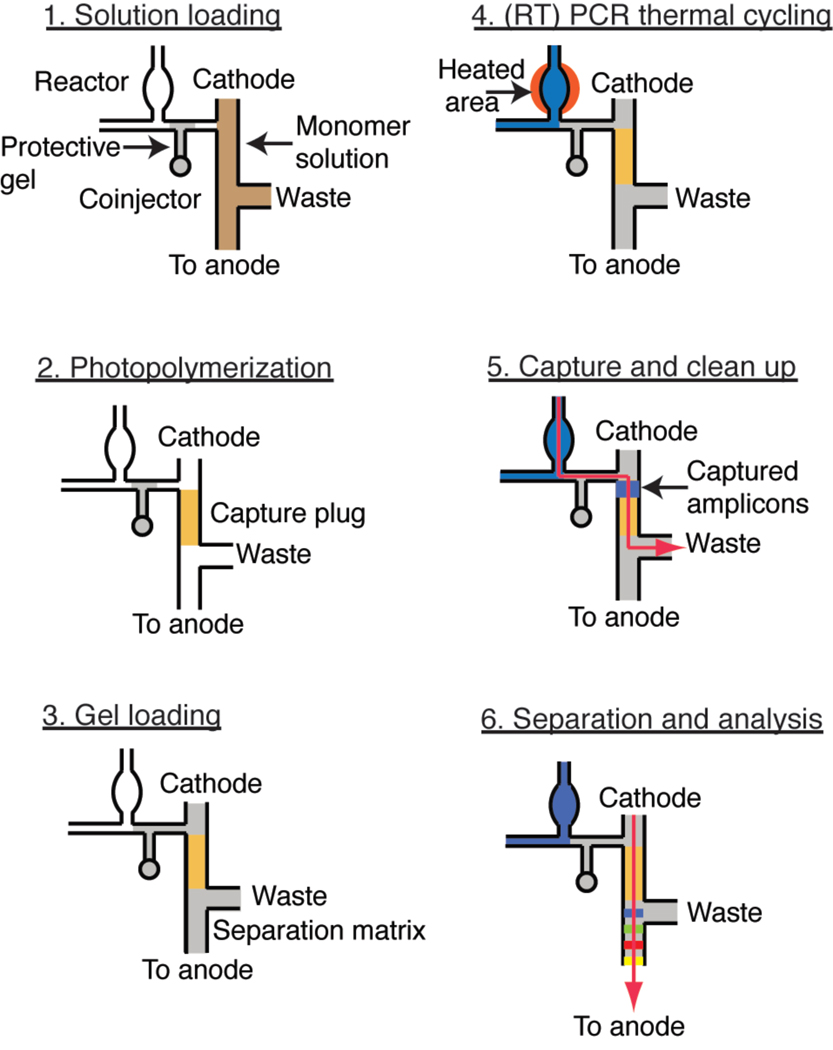
A 5% linear polyacrylamide (LPA) gel with 6.15 M urea in 1× TTE (50 mM Tris, 50 mM TAPS acid and 2 mM EDTA) was used as a sieving matrix for all separations. The synthesis protocol of the gel was reported previously [38]. The crosslinked polyacrylamide capture gel was prepared in situ by co-photopolymerizing a 19:1 acrylamide/bis monomer mix with a 5’ acrydite-modified streptavidin capture probe. Following the previously developed protocol [30], a 500 µL solution was prepared to 5 % (v/v) bis-acrylamide, 6.15 M urea, 1× TTE, 2 µg/µL streptavidin-acrylamide, 0.0006 % riboflavin (w/v) and 0.125 % TEMED (v/v) in an opaque 2-mL scintillation vial with Teflon closure (National Scientific, Rockwood, TN). This solution was used to form a 500-µm capture gel plug within the double-T junction in the dynamically coated microchannel as illustrated in Figure 2. To prevent the monomer solution from contaminating the reactor with PCR and RT-PCR inhibitors such as EDTA and acrylamide monomers, 8% LPA was first loaded from the coinjector into the channel separating the reactor from the capture and separation region (Figure 2, step 1). The monomer solution was then introduced into the microchannels. Using an inverted microscope equipped with a mercury arc lamp (Nikon) centered on 365 nm [29], the double-T junction was irradiated (10 mW/cm2) for 5 min, polymerizing the solution in this region. After photopolymerization, unreacted monomer solution was removed and the microchannels were rinsed with deionized water (Figure 2, step 2).
Figure 2.
Schematic of microdevice preparation and operation. (1) To avoid contamination of the reactor, a 5% linear polyacrylamide (LPA) gel (gray) is loaded into the coinjector prior to introducing the bis/acrylamide monomer solution (brown). (2) A streptavidin-copolymerized 5% bis/acrylamide gel plug 40 µm deep × 120 µm wide × 500 µm long is formed within the microchannel through in situ photopolymerization (gold). (3) The plug is sandwiched by a separation matrix manually loaded from both anode and cathode. (4) The assay is started by filling the reactor with PCR (or RT-PCR) cocktail (blue), immediately followed by thermocycling. (5) After the amplification is completed, the sample is electrophoresed (red arrow) through the capture plug, where biotinylated PCR products are captured. (6) The captured dsDNA is dehybridized at 67 °C, releasing the fluorescently labeled ss-DNA for size-based separation at 300 V/cm.
Microdevice preparation
The Ti/Pt RTDs were calibrated following methods previously described [33]. To passivate the glass surface, all microchannels were first treated with 1M HCl for 15 min. The channels were then rinsed with deionized water prior to treatment with 0.25% w/v polyDuramide dynamic coating solution [39]. After an hour, the dynamic coating solution was evacuated and the channels were rinsed with water and dried by vacuum.
To prepare the microchip for a PCR-based assay, photopolymerized capture plugs were created following the protocol described above. A 0.5×2 cm piece of PDMS (0.25-inch thick, Bisco Silicones, Carol Stream, IL) with a 3 mm-diameter opening was first placed over each access hole of the microdevice to serve as a longer reservoir preventing solution evaporation during the run. The separation matrix was then loaded from the anode to waste and from the cathode to the coinjection reservoir, sandwiching the capture plug previously synthesized (Figure 2, step 3). The reactor wall was passivated by treatment with 1% w/v heat-denatured bovine serum albumin (BSA) at room temperature for 10 min to prevent enzyme adhesion during the reaction. The reactor was thoroughly rinsed with nuclease-free water before the introduction of PCR solution.
Microdevice operation
Each reactor was filled by placing PCR (or RT-PCR) cocktail at the reactor inlet and applying vacuum briefly at the reactor outlet (Figure 2, step 4). Immediately after loading, a drop of 5% LPA was added at both ports to completely seal the reactor. Then the microchip was transferred to a portable detection instrument containing all electrical and detection interfaces for subsequent analysis steps. Amplification was initiated with a 10-min polymerase activation at 95 °C, followed by 15 step-down cycles consisting of a 10-sec denaturation step at 95 °C, a 20-sec annealing step starting at 65 °C with a reduction of 1 °C per cycle, and a 30-sec extension at 72 °C. Following the step-down process, the final cycle (annealing at 50 °C) was repeated for 35 cycles followed by a final extension at 72 °C for 5 min. The thermocycling protocol for the one-step RT-PCR is similar, only with an additional step of cDNA synthesis at 42 °C for 20 min before the polymerase activation.
To perform post-amplification sample cleanup and concentration, the sample was electrophoresed from the reactor to the waste at room temperature under an electric field of 25 V/cm for 10 min (Figure 2, step 5). While biotinylated dsDNA products were retained by the streptavidin-copolymerized capture matrix, excess salts, dNTPs and unreacted forward primers were washed toward the waste. Following thermal melting of retained dsDNA products at 67 °C, the fluorescently labeled strands were directly injected into the separation channel at a separation field of 300 V/cm between the cathode and the anode (Figure 2, step 6). Sample detection was performed using laser-induced fluorescence 1 cm from the anode. After each run, the gels were pushed out with water. The microchannels and reactors were cleaned with piranha (3:1 H2SO4: H2O2) at 65 °C for 10 min, followed by a 5 min water rinse.
Instrumentation and data processing
The 12 × 12 × 10” McCAEPS instrument (Figure 1B) contains an optical system including a 488-nm diode laser (75 mW, Sapphire 488-75, Coherent, Santa Clara, CA), an objective and a four-color PMT (Hamamatsu H9797, Bridgewater, NJ) for fluorescence detection [35]. In addition, the device features pneumatic controls, electronics for temperature control, and four high voltage power supplies for CE. Thermal cycling is controlled through a LabVIEW program (National Instruments, Austin, TX) with a proportional/integral/differential (PID) module. More detail on the instrument can be found at http://www.cchem.berkeley.edu/ramgrp/scanner. Fluorescence crosstalk was corrected using BaseFinder v.6.1 (Giddings Lab, University of North Carolina), and processed traces were analyzed using PeakFit v4. (Systat Software, Inc., San Jose, CA). Fragment analysis data obtained from the conventional Applied Biosystems instrument was processed by GelQuest (SequentiX, Germany).
Results and discussion
Microdevice and assay design
The two-layer glass microdevice is designed for simple operation and automation. The doublet structure with common cathode, waste, and anode structures is employed to reduce the number of access holes, minimizing the number of electrical connections during electrophoresis and facilitating easier and faster gel loading. Unlike four-layer integrated PCR-based microdevices previously developed, this microdevice contains no removable PDMS-glass microvalves or pumps. The reactors are sealed by adding ~500 µL of a viscous 5% LPA gel over the reactor inlets and outlets. To prevent the reaction cocktail from leaking out of the reactor during thermocycling, excess (~500 µL) 5% linear polyacrylamide (LPA) gel is added to the coinjectors. The absence of PDMS microvalves vastly simplifies chip preparation and operation. By removing the need to apply an active pressure to seal the microvalves, we eliminate the potential for displacement of the PCR solution out of the reactor during thermocycling due to air leakage through the porous PDMS membrane.
Furthermore, the sequence-independent streptavidin/biotin-mediated capture process employed here provides a universal target capture technique applicable to a variety of multiplex assays. In contrast to the oligonucleotide-affinity capture format employed in previous work [29, 40], streptavidin/biotin-based capture eliminates complications in designing sequence-specific capture probes with similar thermodynamic properties, as well as the tedious process of optimizing capture conditions. Because of the denaturing condition introduced by 6.15 M urea in both capture and separation matrices, full-strand melting and release can be performed at a moderate temperature of 67 °C. As a result, the potential for failed electrophoresis due to acrylamide gel breakdown is avoided [41].
Multiplex amplification
Multiplex amplification of gene targets from FLUAV, FLUBV hMPV, and hCoV-OC43 was carried out on the integrated microdevice using plasmid standards, each containing the cloned respective viral target sequence. In the multiplex PCR experiments, each target was amplified from 100 copies of plasmid standard. The higher annealing temperature during the initial step-down process promotes assay specificity by ensuring that the primers only hybridize to their complementary target in these critical early cycles [42]. A high level of PCR discrimination is crucial in pathogen detection assays because the pathogen target templates are commonly present in much lower abundance than the ubiquitous host genomic and mRNA backgrounds. For this reason, a step-down PCR scheme was employed. Because a total of 50 cycles were performed, additional amounts of Taq DNA polymerase, enzyme-stabilizing MgCl2, and dNTPs were added to ensure that the reaction is not reagent-limited.
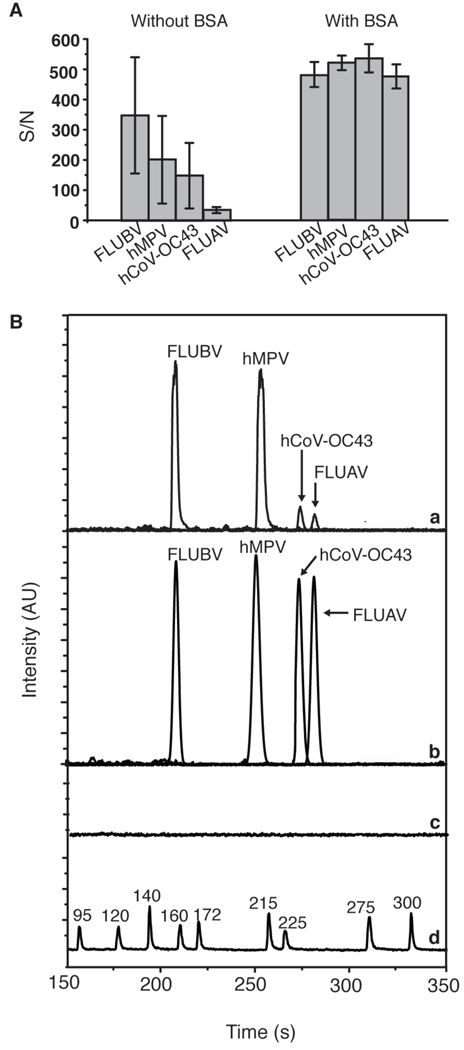
The effects of BSA passivation on PCR efficiency and robustness were examined by comparing amplifications in non-passivated and BSA-passivated microreactors. As indicated by the S/N ratios in Figure 3A, varying PCR yields with declining yields as a function of amplicon size were obtained when reactions were performed in a bare, untreated reactor (n = 3) using 100 copies of each target. In contrast, all four products were generated in comparable or greater amounts (n = 3) when reactions were performed in BSA-coated reactors. These results confirm that BSA passivation helps increase reaction yield by likely occupying sites on the glass reactor wall and preventing adherence of the polymerase [43]. This effect is also seen in the individual electropherograms of representative 4-plex PCR amplification in passivated and non-passivated reactors shown in Figure 3B for the 159-bp FAM-labeled FLUBV, 212-bp TAMRA-labeled hMPV, 238-bp JOE-labeled hCoV-OC43, and 246-bp FAM labeled FLUAV dsDNA amplification products. In a non-passivated reactor, the products are biased towards smaller fragments whereas all products were generated in approximately equal yield when the reactor was coated with BSA. A negative control verifies the absence of carry-over from run to run.
Figure 3.
On-chip multiplex PCR analysis of FLUBV, hMPV, hCoV-OC43, and FLUAV. (A) Bar graph shows the effects of BSA passivation on reaction yield obtained from multiplex on-chip PCR amplification (n=3). (B) Representative electropherograms of multiplex PCR products obtained from an uncoated reactor (a) and BSA-coated reactor (b). (c) Negative control containing no plasmid template. (d) Custom-made ROX-labeled sizing reference.
A ROX-labeled DNA ladder serving as a sizing reference was captured and injected in a similar fashion. The average resolution/base for the ladder between 95–300 nucleotides (nt) is 0.26 ± 0.08. The separation performance of our system characterized by peak capacity [44], defined as the maximum number of analytes detected by the system, can be calculated based on the separation of the DNA ladder. With the average number of theoretical plates of 2 × 104 plates/cm, the peak capacity in the 90 s- 300 s time interval was determined to be 126. However, since the peak width of the PCR products is significantly larger than that of the reference ladder peaks, the actual peak capacity when analyzing real samples will be lower. Nevertheless, this theoretical calculation suggests that our microsystem is highly suitable for multiplex analysis assays. Successful amplification and detection of the four viral targets using both size-based separation and four-color spectral discrimination establish the feasibility of the device and system for multiplexed viral detection.
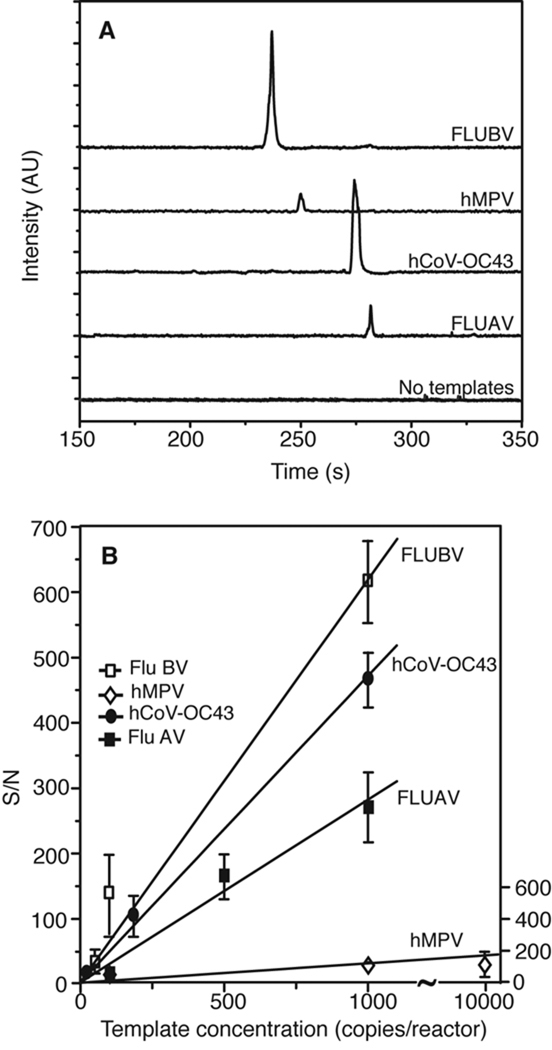
Assay sensitivity
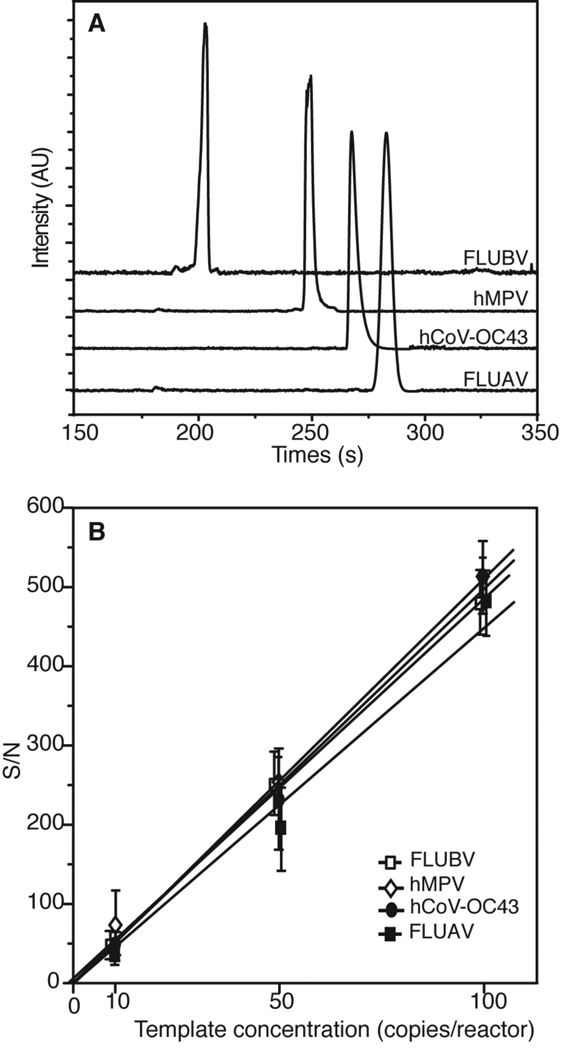
To determine the sensitivity of the microdevice for each virus in the multiplex panel, each individual plasmid was amplified separately in the presence of all primer pairs included in the panel. Representative electropherograms in Figure 4A show successful on-chip amplification from 100 copies/reactor. The detection limit of the system was determined in a similar fashion by amplifying each of the four templates following serial dilution to 100, 50, and 10 copies. The average S/N ratios are plotted Figure 4B. A linear increase of S/N for all targets indicates that the amplifications are performed in the exponential phase [45, 46]. The system demonstrates high sensitivity, with S/N > 10 for 10 copies (n = 3) of all four standard plasmids. The calculated detection limits of FLUBV, hMPV, hCoV-OC43, and FLUAV obtained by extrapolation of the linear regression for S/N = 3 are 0.65, 0.63, 0.60, and 0.66, respectively, indicating the feasibility of single-copy viral detection. In addition, the four templates exhibit comparable S/N for each starting concentration with a standard deviation of 26% for 100 copies, 13% for 50 copies, and 6% for 10 copies. This observation confirms that similar amplification efficiency was achieved for all targets at the optimized primer concentration ratios.
Figure 4.
Assay specificity and sensitivity determination. (A) Representative electropherograms show that each plasmid standard (100 copies each) exclusively generates the correct product in the presence of all four primer pairs. (B) Sensitivity determination: standard plasmid for FLUBV, hMPV, hCoV-OC43 and FLUAV are amplified at 100, 50, 10, and 0 copies/reactor (n=3 for each data point). The S/N ratios for each of the four products at the three starting template concentrations are shown.
Analysis of RNA samples
Because many viruses have RNA genomes, the capability of the microdevice to analyze RNA samples is crucial. Therefore, RT-PCR amplification using in vitro transcribed RNA standards was investigated, and the sensitivity of the assay in detecting each RNA target was determined. In the presence of all four primer pairs, each RNA standard was reverse-transcribed on-chip at 42 °C for 20 min and amplified in the same reactor using the previously described step-down PCR conditions. Representative electropherograms in Figure 5A show detection and identification of all transcripts at 100 copies/reactor. A negative RT-PCR control containing no templates verifies the absence of nonspecific priming. The sensitivity of the device in detecting each viral RNA transcript was determined by performing RT-PCR on serially diluted RNA standards in the range of 25–10,000 copies/reactor. Each standard was reverse-transcribed and amplified separately in the presence of all four primer pairs as described previously. Figure 5B shows a comparison of S/N for the amplified products over the range of starting template concentrations (n =3). While FLUBV and hCoV-OC43 can be detected down to 50 and 25 copies respectively, FLUAV and hMPV can be detected at 100 copies (with S/N > 5). As determined by extrapolation to S/N = 3, the calculated limits of detection for FLUBV, hCoV-OC43, FLUAV and hMPV are 4.8, 6.3, 10 and 167 copies, respectively. A lower sensitivity obtained from the RNA samples compared to their DNA counterparts resulting from the known limited efficiency of reverse transcription has been previously observed [1]. In particular, for a multiplex one-step RT-PCR where more than one virus-specific RT primer is present, the optimal choice of RT conditions can become more difficult than in hexamer primed reactions [6]. Nevertheless, the results obtained here demonstrate the ability of the microdevice to process RNA samples, which opens the door to other RNA-based applications such as detection and genotyping of other viruses, HIV viral load testing, and gene expression studies.
Figure 5.
Analysis of in vitro transcribed RNA standards processed on-chip using one-step RT-PCR. (A) Representative electropherograms shows that each RNA standard (100 copies each) can be accurately identified in the presence of all four primer pairs using the microdevice. A negative control containing no template verifies the absence of carryover between analyses. (B) Sensitivity analysis, as a function of initial RNA template concentrations, shows that the microdevice can detect hCoV-OC43, FLUBV, FLUAV, and hMPV RNA at 25, 50, 100, and 100 copies respectively with S/N > 5 (n=3).
Demonstration of microdevice detection with nasopharyngeal swabs (NPS)
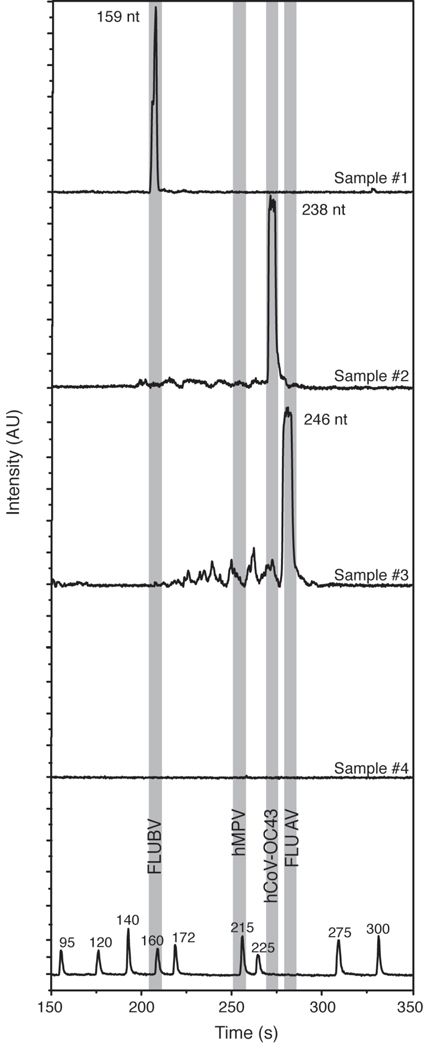
To evaluate the capability of the microsystem to analyze real diagnostic samples, four cDNA samples derived from NPS were PCR amplified and analyzed on-chip. Total nucleic acid extracts from NPS reverse-transcribed using random hexamers were used for multiplex amplification without further purification or concentration. Samples were aliquoted into two groups: the first group was processed on-chip in the same manner as the standard plasmids, while the second group was amplified and analyzed off-chip as a control.
The electropherograms in Figure 6 indicate successful detection of FLUBV, hCoV-OC43, and FLUAV samples, while one sample was negative for all four viruses tested. The identity of the pathogens was determined by matching the migration time of the product peaks with those obtained from the standards of known identity. Whereas the performance for FLUAV, FLUBV, hCoV-OC43 was excellent and the negative control yielded no amplification signal, detection of hMPV was not achieved with NPS samples (data not shown). The hMPV primer pair, as already indicated by results with in vitro transcribed RNA, appears to perform less efficiently than the other three primer pairs in this platform, and would need to be specifically redesigned for the microsystem application. Although nonspecific PCR noise in amplification #2 was minimal, the more pronounced background seen in amplification #3 may lead to false-positive or false-negative calls, especially when more primer pairs are introduced in the assay. For an expanded panel, the cut-off levels and the detection limit will need to be empirically redetermined and optimized.
Figure 6.
PCR-based analysis NPS derived cDNA with the PCR-CE microdevice. Total nucleic acids derived from nasopharyngeal swabs (NPS) and reverse transcribed into cDNA were amplified and analyzed on-chip. Electropherograms reveal that sample #1 is positive for FLUBV, #2 for hCoV-OC43, and #3 for FLUAV, while sample #4 is negative for all the four viruses tested. Shaded bands show the expected locations of FLUBV (159 nt), hMPV (212 nt), hCoV-OC43 (238 nt) and FLUAV (246 nt). The accuracy of the results was verified by running parallel PCR reactions off-chip.
Conclusions
We have developed a new user-friendly two-layer integrated PCR-CE microdevice for rapid multiplex detection of FLUBV, hMPV, hCoV-OC43, and FLUAV. The post-amplification purification process employs streptavidin/biotin-mediated recognition for simultaneous capture of multiple targets. The PCR amplification, sequence-independent product capture and concentration, and CE analysis on the automated portable genetic analysis instrument were streamlined to within two hours. The microdevice is capable of processing both DNA and RNA samples with high specificity and sensitivity in clinical samples. This novel integrated microdevice has potential as a platform for robust, automated, high-throughput and sensitive point-of-care diagnostics.
Supplementary Material
Acknowledgements
We thank Vishal Kapoor and Aaloki Shah for excellent technical assistance, as well as helpful suggestions and valuable discussions. We also acknowledge Dr. James Scherer for the design and fabrication of the portable genetic analyzer. Microfabrication was performed at the University of California Berkeley Microfabrication Laboratory. This work was supported by Samsung Electronics Co. Ltd., Award 20082064 and National Institutes of Health (NIH) Grants #CA77664 and #AI57158 (Lipkin, Northeast Biodefense Center).
References
- 1.Briese T, Palacios G, Kokoris M, Jabado O, Liu Z, Renwick N, Kapoor V, Casas I, Pozo F, Limberger R, Perezbrena P, Ju J, Lipkin WI. Emerg. Infect. Dis. 2005;11:310–313. doi: 10.3201/eid1102.040492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin B, Blaney KM, Malanoski AP, Ligler AG, Schnur JM, Metzgar D, Russell KL, Stenger DA. J. Clin. Microbiol. 2007;45:443–452. doi: 10.1128/JCM.01870-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woo PC, Chiu SS, Seto WH, Peiris M. J. Clin. Microbiol. 1997;35:1579–1581. doi: 10.1128/jcm.35.6.1579-1581.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelly H, Birch C. Aust Fam Physician. 2004;33:305–309. [PubMed] [Google Scholar]
- 5.Gröndahl B, Puppe W, Hoppe A, Kühne I, Weigl JA, Schmitt HJ. J. Clin. Microbiol. 1999;37:1–7. doi: 10.1128/jcm.37.1.1-7.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elnifro EM, Ashshi AM, Cooper RJ, Klapper PE. Clin. Microbiol. Rev. 2000;13:559–570. doi: 10.1128/cmr.13.4.559-570.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marshall DJ, Reisdorf E, Harms G, Beaty E, Mose MJ, Lee W, Gern JE, Nolte FS, Shult P, Prudent JR. J. Clin. Microbiol. 2007;45:3875–3882. doi: 10.1128/JCM.00838-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henrickson KJ. Pediatr. Infec. Dis J. 2004;23:S6–S10. doi: 10.1097/01.inf.0000108187.63151.ea. [DOI] [PubMed] [Google Scholar]
- 9.Whelen AC. Annu. Rev. Microbiol. 1996;50:349–373. doi: 10.1146/annurev.micro.50.1.349. [DOI] [PubMed] [Google Scholar]
- 10.Kehl SC, Kumar S. Clin. Lab. Med. 2009;29:661–671. doi: 10.1016/j.cll.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chou Q, Russel M, Birch DE, Raymond J, Bloch W. Nucleic. Acids. Res. 1992;11:1717–1723. doi: 10.1093/nar/20.7.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boivin G, Côté S, Déry P, Serres GD, Bergeron MG. J. Clin. Microbiol. 2004;42:45–51. doi: 10.1128/JCM.42.1.45-51.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leung TF, To MY, Yeung ACM, Wong YS, Wong GWK, Chan PKS. CHEST. 2010;137:348–354. doi: 10.1378/chest.09-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.the CAP study group. Miyashita N, Saito A, Kohno S, Yamaguchi K, Watanabe A, Oda H, Kazuyamag Y, Matsushima T. Respir. Med. 2004;6:542–550. doi: 10.1016/j.rmed.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 15.Stockton J, Ellis JS, Saville M, Clewley JP, Zambon MC. J. Clin. Microbiol. 1998;36:2990–2995. doi: 10.1128/jcm.36.10.2990-2995.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Elden LJR, Nijhuis M, Schipper P, Schuurman R, van Look AM. J. Clin. Microbiol. 2001;39:196–200. doi: 10.1128/JCM.39.1.196-200.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manz A, Graber N, Widmer HM. Sens. Actuators, B. 1990;1:244–248. [Google Scholar]
- 18.Dittrich PS, Tachikawa K, Manz K. Anal. Chem. 2006;78:3887–3907. doi: 10.1021/ac0605602. [DOI] [PubMed] [Google Scholar]
- 19.Liu P, Mathies A. Trends Biotechnol. 2009;27:572–581. doi: 10.1016/j.tibtech.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 20.Chen L, Manz A, Day PJR. Lab Chip. 2007;7:1413–1423. doi: 10.1039/b708362a. [DOI] [PubMed] [Google Scholar]
- 21.Bienvenue JM, Legendre LA, Ferrance JP, Landers JP. Forensic. Sci. Int. Genet. 2010;4:178–186. doi: 10.1016/j.fsigen.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 22.Ligler FS. Anal. Chem. 2009;81:519–526. doi: 10.1021/ac8016289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prakash AR, De la Rosa C, Fox JD, Kaler KVIS. Microfluid. Nanofluid. 2007;4:451–456. [Google Scholar]
- 24.Zhou ZM, Liu DY, Zhong RT, Dai ZP, Wu DP, Wang H, Du YG, Xia ZN, Zhang LP, Mei XD, Lin BC. Electrophoresis. 2004;25:3032–3039. doi: 10.1002/elps.200305966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang FC, Liao CS, Lee GB. Electroporesis. 2006;27:3297–3305. doi: 10.1002/elps.200600458. [DOI] [PubMed] [Google Scholar]
- 26.Pal R, Yang M, Lin R, Johnson BN, Srivastava N, Razzacki SZ, Chomistek KJ, Heldsinger DC, Haque RM, Ugaz VM, Thwar PK, Chen Z, Alfano K, Yim MB, Krishnan M, Fuller AO, Larson RG, Burke DT, Burns MA. Lab Chip. 2005;5:1024–1032. doi: 10.1039/b505994a. [DOI] [PubMed] [Google Scholar]
- 27.Kaigala GV, Huskins RJ, Preiksaitis J, Pang XL, Pilarski LM, Backhouse CJ. Electrophoresis. 2006;27:3753–3763. doi: 10.1002/elps.200600061. [DOI] [PubMed] [Google Scholar]
- 28.Beyor N, Yi L, Seo TS, Mathies RA. Anal. Chem. 2009;81:3523–3528. doi: 10.1021/ac900060r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thaitrong N, Toriello NM, Del Bueno N, Mathies RA. Anal. Chem. 2009;8:1371–1377. doi: 10.1021/ac802057f. [DOI] [PubMed] [Google Scholar]
- 30.Yeung SHI, Liu P, Del Bueno N, Greenspoon SA, Mathies RA. Anal. Chem. 2009;81:1371–1377. [Google Scholar]
- 31.Liu P, Scherer JR, Greenspoon SA, Chiesl TN, Mathies RA. Forensic. Sci. Int. Genet. doi: 10.1016/j.fsigen.2010.10.009. Submitted. [DOI] [PubMed] [Google Scholar]
- 32.Scherer JR, Liu P, Mathies RA. Rev. Sci. Instr. doi: 10.1063/1.3502457. Accepted. [DOI] [PubMed] [Google Scholar]
- 33.Liu CN, Toriello N, Mathies RA. Anal. Chem. 2006;78:5474–5479. doi: 10.1021/ac060335k. [DOI] [PubMed] [Google Scholar]
- 34.Toriello NM, Liu CN, Mathies RA. Anal. Chem. 2006;78:7997–8003. doi: 10.1021/ac061058k. [DOI] [PubMed] [Google Scholar]
- 35.Liu P, Seo TS, Beyor N, Shin KJ, Scherer JR, Mathies RA. Anal. Chem. 2007;79:1881–1889. doi: 10.1021/ac061961k. [DOI] [PubMed] [Google Scholar]
- 36.Schweiger B, Zadow I, Heckler R, Timm H, Pauli G. J. Clin. Microbiol. 2000;38:1552–1558. doi: 10.1128/jcm.38.4.1552-1558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mackay IM, Jacob KC, Woolhouse D, Waller K, Syrmis MW, Whiley DM, Siebert DJ, Nissen M, Sloots TP. J. Clin. Microbiol. 2003;41:100–105. doi: 10.1128/JCM.41.1.100-105.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruiz-Martinez MC, Berka J, Belenkii A, Foret F, Miller AW, Karger BL. Anal. Chem. 1993;65:2851–2858. doi: 10.1021/ac00068a023. [DOI] [PubMed] [Google Scholar]
- 39.Albarghouthi MN, Buchholz BA, Huiberts PJ, Stein TM, Barron AE. Electrophoresis. 2002;23:1429–1440. doi: 10.1002/1522-2683(200205)23:10<1429::AID-ELPS1429>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 40.Toriello NM, Liu CN, Blazej R, Thaitrong N, Mathies RA. Anal. Chem. 2007;79:8549–8556. doi: 10.1021/ac0712547. [DOI] [PubMed] [Google Scholar]
- 41.Muller G. Polymer Bulletin. 1981;5:31–37. [Google Scholar]
- 42.Don RH, Cox PT, Wainwright BJ, Baker K, Mattick JS. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang C, Xing D. Nucleic Acids Res. 2007;34:4223–4237. doi: 10.1093/nar/gkm389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grushka E. Anal. Chem. 1970;42:1142–1147. [Google Scholar]
- 45.Sachse K. In: Methods in molecular Biology: PCR Detection of Microbial Pathogens. J. Sachse K, Frey J, editors. NJ: Humana Press; 2003. pp. 10–12. [PubMed] [Google Scholar]
- 46.Lantz O, Bonney E, Griscelli F, Taoufik Y. In: Methods in Molecular Medicine: Quantitative PCR Protocol. Kochanowski B, Reischl U, editors. NJ: Humana Press; 1999. pp. 91–94. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.