Abstract
The tetraspanin CD63 resides in late endosomes, lysosomes, secretory vesicles, and at the plasma membrane, and it moves among these compartments. We find that CD63 is present also in tubulovesicular elements, the intracellular compartments that contain the H,K-ATPase in unstimulated gastric parietal cells. The H,K-ATPase β-subunit and CD63 colocalize in parietal cells and form a complex that can be coprecipitated. The β-subunit and CD63 also interact when they are coexpressed in COS-7 cells. Furthermore, expression with CD63 induces the redistribution of the β-subunit from the cell surface to CD63+ intracellular compartments. Immunofluorescence and biochemical experiments reveal that this redistribution occurs by enhanced endocytosis of H,K-ATPase β-subunit complexed with CD63. Coexpression of the β-subunit with mutant CD63 polypeptides demonstrates that the enhanced internalization of the β-subunit depends on the capacity of CD63 to interact with adaptor protein complexes 2 and 3. These data indicate that CD63 serves as an adaptor protein that links its interaction partners to the endocytic machinery of the cell and suggest a previously uncharacterized protein-trafficking role for the tetraspanins.
The tetraspanin family is composed of ≥28 membrane proteins that span the bilayer four times and share specific sequence homologies and structural features. Many of these proteins are expressed at high levels in membranes of a wide variety of cell types (1, 2). Despite the abundance and prevalence of the tetraspanins, their individual functions remain largely unknown. It has been proposed that tetraspanins may serve as molecular facilitators, collecting proteins together to improve the stability and activity of signaling complexes (3). The tetraspanins have been implicated also in membrane fusion, cell motility, and cell aggregation (2).
The tetraspanin CD63 is a highly glycosylated protein that is widely but not ubiquitously expressed (4). Although CD63 is typically regarded as a resident of late endosomes and lysosomes, it is present also in secretory vesicles as well as at the plasma membrane, and it cycles among these compartments (2, 5–7). This tetraspanin has numerous interaction partners, including other tetraspanins, such as CD82 (2); the MHC class II molecules HLA-DR, HLA-DM, and HLA–DO (8); several integrins (9); and phosphatidylinositol 4-kinase (10, 11). CD63 also contains a tyrosine-based motif on its extreme C terminus. Tyrosine-based motifs in the cytoplasmic domains of membrane proteins are recognized by clathrin adaptor complexes and play important roles in endocytosis, lysosomal targeting, and basolateral targeting (12). The tyrosine-based motif in CD63 mediates its interaction with the μ-subunits of adaptor protein complexes 2 and 3 (AP-2 and AP-3) (13). AP-2 and AP-3 participate in clathrin-mediated endocytosis and lysosomal targeting, respectively (12, 14). The subcellular localization of CD63 and its interaction with the adaptor complexes and phosphatidylinositol 4-kinase suggest that this tetraspanin may be involved in protein trafficking; however, there is little direct experimental support for this proposal (9).
The gastric H,K-ATPase is a heterodimeric ion pump that is present in parietal cells of the stomach and is responsible for the secretion of gastric acid (15, 16). In unstimulated parietal cells, H,K-ATPase resides in intracellular storage compartments called tubulovesicular elements (TVEs). On secretagogue stimulation, the TVEs fuse with the apical plasma membrane, delivering their stores of H,K-ATPase to the cell surface and thus initiating acid secretion into the lumen of the stomach (17). Previous studies have demonstrated that information present in the H,K-ATPase β-subunit (HKβ) is required for pump reinternalization (18).
We asked whether CD63 is associated with TVEs, which constitute unique regulated secretory and endocytic compartments. In this article we show that CD63 is resident in TVEs, and we find that the association between this tetraspanin and the HKβ promotes the internalization of the pump subunit. This work demonstrates a functional role for CD63 in membrane trafficking and suggests that this tetraspanin may play a role in the recycling of plasma membrane components to their appropriate intracellular compartments.
Materials and Methods
Cloning. Rabbit HKβ (cDNA provided by G. Sachs, University of California, Los Angeles) was subcloned into the mammalian expression vector pcDNA3.1 (Invitrogen). CD63 was amplified by PCR from a human kidney library (Clontech) and inserted into pcDNA3.1. A FLAG tag was introduced on the N terminus of CD63, and the CD63-YEVI construct was generated by PCR. The CD63-AEVM construct was created by using the QuikChange kit (Stratagene) according to the manufacturer's instructions. Primer sequences are available on request. All PCR products were sequenced.
Immunohistochemistry. Sprague–Dawley rats were anesthetized, and the internal organs were fixed as described by Biemesderfer et al. (19). Stomachs were cut at 4 μm on a CM 3050S cryostat (Leica Microsystems, Bannockburn, IL). Tissue was incubated with the anti-H,K-ATPase α-subunit (HKα) polyclonal antibody (pAb) HK9 and the anti-CD63 mAb ME491 (BD Biosciences; 1:100), followed by Alexa Fluor 594- and 488-conjugated IgG (Molecular Probes; 1:200) (20). Samples were visualized with an LSM 410 laser scanning confocal microscope (Zeiss). Images are the product of 8-fold line averaging.
Cell Culture and Transfection. COS cells were grown in a 5% CO2/95% air humidified incubator at 37°C and in α-MEM (GIBCO/BRL) supplemented with 10% FBS, 2 mM l-glutamine, 50 units/ml penicillin, and 50 μg/ml streptomycin. cDNA transfection of COS cells was performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Assays were performed 48 h after transfection.
Immunofluorescence. Transfected COS cells were fixed in 3% paraformaldehyde, permeabilized in dilution buffer [PBS containing 10% (vol/vol) goat serum, 2% (wt/vol) saponin, and 10 mM glycine], and incubated for 1 h with anti-HKβ antibody (gift from J. Forte, University of California, Berkeley; 1:100), aquaporin-4 (AQP4) antibody (gift from P. Deen, University of Nijmegen, Nijmegen, The Netherlands; 1:200), or anti-FLAG antibody (Sigma; 1:200), followed by incubation with FITC- or rhodamine-conjugated secondary antibody (1:100) for 1 h. Cells were visualized by confocal microscopy using an extended depth-of-focus macro to combine images from planes of focus separated by 1 μm each. Images are the product of 4-fold line averaging.
For immunofluorescent internalization experiments, all manipulations were performed at 4°C unless otherwise noted. Transfected COS cells were incubated in anti-HKβ mAb (1:50) for 1 h. Cells were covered with 37°C medium and incubated at 37°C for 40 min. Cells were returned to 4°C, incubated for 45 min in Cy5-conjugated goat anti-mouse IgG (1:10), and blocked with unconjugated goat anti-mouse IgG (1:10) for 1 h. Cells were fixed and permeabilized as described above.
Biochemical Procedures. Samples were resolved with SDS/10% PAGE and transferred to nitrocellulose for incubation with anti-HKβ antibody (1:1,000), anti-AQP4 antibody (1:3,000), anti-FLAG mAb (1:2,000), and anti-CD63 antibody H5C6 (obtained from the National Institute of Child Health and Human Development-supported Developmental Studies Hybridoma Bank, University of Iowa, Iowa City; 1:500), anti-Lamp-1 antibody H4A3 (Research Diagnostics, Flanders, NJ; 1:1,000), or biotin-conjugated tomato lectin (Sigma; 0.2 mg/ml) by using standard protocols. Blots were then incubated with peroxidase-conjugated antibody or peroxidase-conjugated streptavidin and visualized with enhanced chemiluminescence (ECL; Amersham Biosciences).
For coimmunoprecipitations, cells were incubated in lysis buffer (5 mM MgCl2/150 mM NaCl/25 mM Hepes, pH 7.4) supplemented with 2% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) and 1% Brij 96 or 1% Triton X-100. The lysate was cleared by centrifugation (10,000 × g for 30 min at 4°C) and incubated with 2 μg of anti-FLAG pAb or mAb and immobilized protein A or G agarose beads (Pierce) overnight at 4°C. Beads were washed, incubated in SDS/PAGE sample buffer containing 100 mM DTT, and heated to 65°C for 10 min.
For tomato lectin pull-downs, 200-μg aliquots of hog TVEs (gift from C. T. Okamoto, University of Southern California, Los Angeles) were lysed in 1 ml of lysis buffer containing either 2% CHAPS or 1% Triton X-100. Samples were centrifuged at 10,000 × g for 30 min at 4°C. Streptavidin-conjugated agarose beads (Pierce) were charged with 0.33 mg of biotin-conjugated tomato lectin for 1 h at room temperature. TVE lysate was added to the washed beads, and the mixture was incubated overnight at 4°C. Beads were then washed, and precipitated proteins were eluted as described above.
Cell-Surface Biotinylation. Manipulations were performed at 4°C unless otherwise noted. COS cells cotransfected with the HKβ and CD63 were grown in 10-cm tissue culture-treated dishes. COS cells transfected with the HKβ alone were grown in six-well tissue culture-treated dishes. Cells were biotinylated as described (21). For internalization assays, cells were placed in medium heated to 37°C and allowed to incubate at 37°C. Cells were removed from the incubator at the appropriate time, washed with ice-cold PBS supplemented with 1 mm MgCl2 and 0.1 mm CaCl2, incubated three times for 20 min each at 4°C in freshly mixed 2-mercaptoethanesulfonic acid, sodium salt (MesNa), solution (100 mM MesNa/100 mM NaCl/1 mM EDTA/0.2% BSA/50 mM Tris, pH 8.6), and quenched with 120 mM iodoacetic acid. Cells that were not subjected to the MesNa strip were incubated only with iodoacetic acid.
Samples from cells cotransfected with both the HKβ and CD63 were immunoprecipitated with 5 μg of anti-FLAG pAb in 2% CHAPS lysis buffer. The beads were then agitated in 100 μl of elution buffer (300 mM Tris/3% SDS, pH 6.8) for 30 min at 25°C and for 10 min at 37°C. The supernatant was removed, and 10% (vol/vol) was reserved. Remaining supernatant was diluted 30-fold with Triton X-100 lysis buffer and incubated overnight at 4°C with streptavidin-conjugated agarose beads. Precipitated proteins were eluted from the beads with sample buffer as described above.
Cells transfected with the HKβ alone were incubated in 1% Triton X-100 lysis buffer for 30 min and rotated overnight at 4°C with streptavidin-conjugated agarose beads. Precipitated proteins were eluted as described above. The entire volume of each sample was loaded in one well of an SDS/polyacrylamide gel. The intensity of the HKβ signal was quantified with a GS-800 densitometer (Bio-Rad).
Results
We first assessed whether CD63 and the H,K-ATPase reside in the same subcellular compartment in gastric parietal cells. Previous work has shown that rat parietal cells express high levels of CD63 (22). Immunofluorescence microscopy on frozen sections of rat stomach indicate that CD63 is present in most cells in rat stomach, including parietal cells (Fig. 1A). CD63 partially colocalizes with the HKα in parietal cells. Additionally, we analyzed a highly purified TVE preparation from hog stomach (a kind gift from C. T. Okamoto). H,K-ATPase contributes ≈50% of the protein content in similar preparations (23). Western blot analysis indicates that CD63 is abundantly present in this purified preparation of TVEs (Fig. 1B).
Fig. 1.
CD63 and H,K-ATPase localization and association in parietal cells. (A) Rat stomach cryosections costained with anti-HKα pAb (green) and anti-CD63 mAb (red). (Scale bar is 50 μm.) (B) TVE samples from the 27% (6 μg) and 32% (14 μg) sucrose interfaces (kind gift from C. T. Okamoto) were immunoblotted with anti-HKβ mAb or anti-CD63 mAb (35). (C) Purified preparations of hog TVEs (13.3 μg) blotted with anti-HKβ mAb or biotinylated tomato lectin. (D) Purified TVEs incubated in 2% CHAPS (CH) or 1% Triton X-100 (Tr) lysis buffer. TVEs were incubated with streptavidin beads that were (+ lectin) or were not (–lectin) preconjugated with biotinylated tomato lectin. Precipitated material was immunoblotted with anti-CD63 mAb. The lane labeled TVE lysate contains 25 μg of lysate.
To investigate a possible in situ interaction between CD63 and the H,K-ATPase, we performed pull-down experiments using biotinylated tomato lectin. Previous studies have shown that this lectin binds specifically and uniquely to the HKβ in gastric preparations (24–27). We blotted a TVE preparation with anti-HKβ mAb and biotinylated tomato lectin to confirm that the lectin associates specifically with the glycosylated HKβ (Fig. 1C). We then used biotinylated tomato lectin to isolate proteins that interact with the HKβ in the TVE preparation. Western blot analysis indicates that CD63 is precipitated by tomato lectin (Fig. 1D), demonstrating that CD63 and the H,K-ATPase associate in situ and suggesting that this association may be physiologically relevant. Pretreatment of the TVE preparation with 4% SDS, followed by a 20-fold dilution in lysis buffer, significantly reduced the amount of CD63 that was precipitated. The amount of the HKβ that was precipitated was not altered by this treatment (data not shown). These data suggest that the presence of CD63 in the pull-down is attributable to its association with the HKβ and not to a direct association between CD63 and tomato lectin.
To investigate the functional significance of the interaction between CD63 and the HKβ further, we transfected COS-7 cells with a cDNA encoding rabbit HKβ either singly or together with a cDNA encoding a FLAG-tagged CD63 construct (CD63-FL). We have demonstrated that the HKβ does not need to assemble with the HKα to reach the cell surface in transfected COS cells (20). We used a CD63 construct that carries a FLAG epitope tag on its N terminus because the C terminus of CD63 contains a tyrosine-based motif that is required for the intracellular trafficking of this tetraspanin (13).
COS cells cotransfected with the HKβ and CD63-FL were subjected to immunoprecipitation using an anti-FLAG pAb. Western blot analysis of the immunoprecipitated material indicates that the HKβ coprecipitates with CD63 (Fig. 2, HKβ + CD63-FL). The CD63-FL and HKβ proteins coimmunoprecipitate in a variety of detergents, including 2% CHAPS, 1% Brij 96, and 1% Triton X-100. The association between CD63 and the HKβ constitutes one of the few tetraspanin complexes described that survives solubilization in Triton X-100 and is thus likely to represent a direct interaction (28). The β-subunit has two forms: βm (mature), which is modified with complex carbohydrates, and βc (core), which is modified with high-mannose carbohydrates (29). Both forms coprecipitate with CD63, although the ratio of βc to βm in the precipitate varies as a function of the solubilization conditions.
Fig. 2.
The HKβ coprecipitates with CD63. COS cells were transfected with the HKβ alone, CD63-FL alone, or the HKβ and CD63-FL (HKβ + CD63-FL). Cells were lysed in 2% CHAPS, 1% Brij 96 (Br), or 1% Triton X-100 (Tr) and immunoprecipitated with anti-FLAG pAb. The second lane was immunoprecipitated from a mixture of cell lysates from cells separately transfected with CD63-FL and the HKβ. Precipitated proteins were blotted with anti-HKβ mAb or anti-Lamp-1 mAb.
The HKβ was not detected in the FLAG immunoprecipitations performed on cells transfected with the HKβ alone, demonstrating that the FLAG pAb does not interact with the β-subunit (Fig. 2, HKβ). The HKβ does not coimmunoprecipitate with FLAG pAb from a mixture (50:50, vol/vol) of lysates prepared from cells transfected singly with CD63-FL and the β-subunit, confirming that the interaction occurs in vivo (Fig. 2, second lane). Samples incubated with FLAG pAb do not precipitate the lysosomal protein Lamp-1, indicating that the interaction between CD63-FL and the HKβ is specific and not an artifact of incomplete solubilization of the CD63 compartment (Fig. 2).
We then examined whether the interaction between the HKβ and CD63 affects the subcellular localization of the β-subunit. The HKβ is present at the cell surface in transiently transfected COS cells (Fig. 3 A–C). There is a pronounced redistribution of the β-subunit to intracellular CD63-containing vesicles when COS cells are cotransfected with both the HKβ and CD63-FL (Fig. 3 D–F). To rule out the possibility that this altered localization could be a nonspecific effect of CD63 overexpression, we investigated the effect of CD63 expression on the distribution of AQP4. AQP4 is a water channel that is present in the basolateral plasma membranes of parietal cells. It does not undergo regulated endocytosis and exocytosis with the H,K-ATPase (30, 31). This channel does not coimmunoprecipitate with CD63-FL in a Triton X-100 lysate of cotransfected COS cells (Fig. 4A). When cotransfected cells are visualized by immunofluorescence confocal microscopy, AQP4 is not present in the CD63-containing compartment (Fig. 4B), indicating that CD63-induced redistribution to intracellular vesicles is specific for proteins that interact with this tetraspanin.
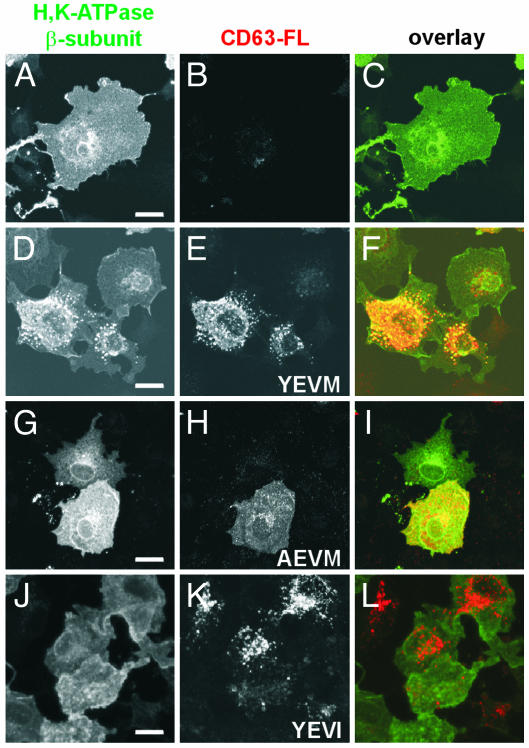
Fig. 3.
The HKβ is redistributed to CD63-positive intracellular vesicles. COS cells were transfected with the HKβ alone (A–C), the HKβ and CD63-FL including the wild-type tyrosine motif YEVM (D–F), the HKβ and CD63-AEVM-FLAG (G–I), or the HKβ and CD63-YEVI-FLAG (J–L). The β-subunit was detected with anti-HKβ mAb (green), and CD63-FL constructs were detected with an anti-FLAG pAb (red). (Scale bar is 10 μm.)
Fig. 4.
Colocalization to intracellular vesicles is specific for proteins that interact with CD63. (A) COS cells were transfected with AQP4 alone or were cotransfected with AQP4 and CD63-FL (AQP4 + CD63-FL). The cells were lysed in 1% Triton X-100 and immunoprecipitated with anti-FLAG mAb. The immunoprecipitated material and the cell lysates were probed with anti-AQP4 pAb. (B) COS cells were transfected with either AQP4 (green) or AQP4 and CD63-FL (red). AQP4 was detected with anti-AQP4 pAb, and CD63 was detected with anti-FLAG mAb. (Scale bar is 10 μm.)
To investigate the mechanisms of the CD63-mediated redistribution of the HKβ, we took advantage of recent work that has characterized the intracellular trafficking of CD63. Rous et al. (13) demonstrated that the tyrosine-based motif present at the extreme C terminus of CD63 interacts with the adaptor subunits μ2 and μ3. The μ2 protein is a member of the heterotetrameric AP-2 complex. This complex is present at the plasma membrane and links cargo proteins with the clathrin coat, mediating their endocytosis (14). The μ3 protein is a member of the heterotetrameric AP-3 complex. This complex participates in lysosomal targeting and is found both in the trans-Golgi network and in a vesicular compartment that partially colocalizes with endosomes (12). When the YEVM tyrosine-based motif of CD63 is mutated to AEVM, CD63 is unable to interact with either μ2 or μ3 and is primarily expressed on the cell surface (13). When the HKβ is coexpressed with the FLAG-tagged CD63-AEVM construct, both CD63-AEVM and the β-subunit are distributed on the cell surface (Fig. 3 G–I). Thus, the intracellular distribution of the HKβ is affected by trafficking signals embedded within the CD63 cytoplasmic tail.
When the YEVM sequence in the tail of CD63 is mutated to YEVI, the modified motif continues to interact with μ3 but no longer associates with μ2 (13). CD63-YEVI is localized primarily to the late endosomal–lysosomal compartment, most likely because the altered tetraspanin protein moves directly to this compartment after its initial biosynthetic passage through the Golgi (13). The small population of CD63-YEVI that reaches the plasma membrane exhibits impaired internalization because it can no longer interact with μ2 (13). When the HKβ and a FLAG-tagged CD63-YEVI construct are coexpressed in COS cells, much of the CD63-YEVI is localized to the late endosomal–lysosomal compartment; however, the β-subunit remains on the cell surface and is not redistributed to an intracellular compartment (Fig. 3 J–L). Coimmunoprecipitation analysis confirms that the HKβ can associate with both CD63-YEVI and CD63-AEVM (data not shown). These data suggest that for the HKβ to be distributed to a CD63-positive intracellular compartment, these proteins must interact at the cell surface and be endocytosed as a complex.
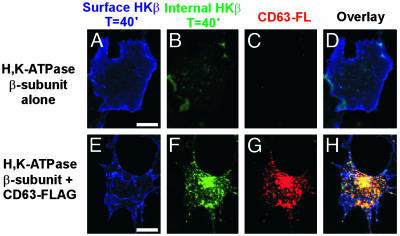
To determine whether the HKβ detected in intracellular vesicles corresponds to protein that was endocytosed from the surface of the cell, we observed the internalization of the β-subunit in the presence and absence of exogenous CD63 expression. COS cells were cotransfected with the HKβ and CD63-FL. The cells were then chilled to 4°C to stop endocytosis and surface labeled with the HKβ mAb. The labeled cells were incubated at 37°C for 40 min to allow endocytosis to resume. After the incubation, the cells were cooled to 4°C and surface labeled with Cy5-conjugated goat anti-mouse IgG. Cells were fixed and incubated with anti-FLAG pAb and were then labeled with both fluorescein isothiocyanate-conjugated goat anti-mouse IgG and rhodamine-conjugated goat anti-rabbit IgG. Thus, the HKβ that remained on the surface was visualized on the Cy5 channel, internalized HKβ was visualized on the fluorescein isothiocyanate channel, and CD63-FL was visualized on the rhodamine channel. This analysis demonstrated that internalized β-subunit colocalizes with CD63, suggesting that the pool of the HKβ that is colocalized with CD63 is derived through endocytosis from the cell membrane (Fig. 5 E–H). Cells that do not overexpress CD63 exhibit much less internalized β-subunit after a 40-min incubation (Fig. 5 A–D).
Fig. 5.
The HKβ is endocytosed from the cell surface into CD63-positive vesicles. COS cells were transfected with the HKβ (A–D) or with β-subunit and CD63-FL (E–H). Surface β-subunit is labeled with Cy5 (blue; A and E), internalized β-subunit is labeled with fluorescein isothiocyanate (green; B and F), and CD63-FL is labeled with rhodamine (red; C and G). (Scale bar is 10 μm.)
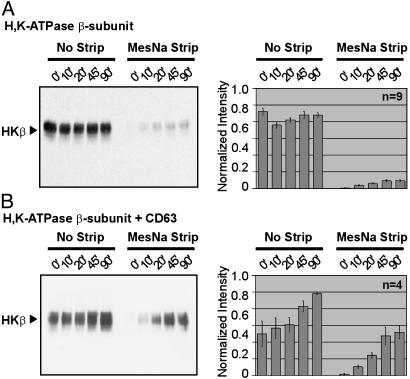
To quantitate the internalization of the HKβ and verify further that the HKβ and CD63 associate at the surface of the cell, we performed cell-surface biotinylation experiments. In the first experiment, COS cells were transfected with the β-subunit alone and biotinylated with biotin-SS-N-hydroxysuccinimide ester at 4°C. Duplicate samples were then incubated at 37°C for various lengths of time to allow endocytosis to resume. Cells were returned to 4°C, and one plate from each time point was stripped of the remaining cell-surface biotin by exposure to the membrane-impermeant reducing agent MesNa. The cells were lysed and incubated with streptavidin beads. Proteins that associated with the streptavidin beads were separated by SDS/PAGE, and the membranes were probed with the HKβ mAb (Fig. 6A). The fraction of β-subunit on the cell surface does not change appreciably as the 37°C internalization proceeds. After the first 10 min, a small fraction of the surface-labeled HKβ is sequestered inside the cell, and the ratio of surface to internalized HKβ remains high. These data suggest that the β-subunit not associated with CD63 either internalizes very slowly or is internalized but recycled quickly back to the surface, much like the transferrin receptor.
Fig. 6.
Internalization of the HKβ is enhanced by its association with CD63. (A) COS cells transfected with the HKβ alone were biotinylated with biotin-N-hydroxysuccinimide ester and incubated at 37°C to allow internalization. Plates were then cooled to 4°C, and one dish from each time point (in min) was subjected to a MesNa strip. Biotinylated material was collected with streptavidin-conjugated agarose beads. Three independent experiments were performed in triplicate. (B) COS cells cotransfected with the HKβ and CD63-FL were biotinylated with or without the MesNa strip as described above. All cells were lysed in 2% CHAPS and immunoprecipitated with anti-FLAG pAb. The immunoprecipitate was eluted and incubated with streptavidin-conjugated agarose beads. Four independent experiments were performed. The sample from each time point was analyzed by SDS/PAGE and immunoblotting with anti-HKβ mAb. (Right) Signal intensity from each band was quantified by densitometry.
We next sought to characterize the behavior of the HKβ that associates with CD63. COS cells were cotransfected with the HKβ and CD63-FL. The cells were biotinylated and stripped as described above. To isolate the population of the HKβ that is associated with CD63, the cell lysate was incubated with FLAG pAb and protein A-conjugated agarose beads. The immunoprecipitated material was eluted with a small amount of buffer containing 3% SDS, diluted 30-fold with 1% Triton X-100, and incubated with streptavidin beads. Proteins that associated with the streptavidin beads were separated by SDS/PAGE and probed with the HKβ mAb (Fig. 6B). For cells that were not subjected to a stripping protocol, the signal detected in this Western blot represents the total pool of surface-labeled β-subunit associated with CD63, including complexes that were still at the plasma membrane and complexes that had been internalized into vesicles. For cells that were subjected to a stripping protocol, the signal represents the population of β-subunit associated with CD63 that was present at the cell surface during biotinylation and was subsequently internalized and protected from the MesNa strip.
At the 0-min time point, biotinylated HKβ is readily detected in anti-FLAG immunoprecipitates from unstripped cells, demonstrating that the HKβ and CD63 are capable of association at the cell surface. All of the biotinylated HKβ recovered at the 0-min time point is sensitive to the MesNa strip. The quantity of CD63-associated, biotinylated β-subunit protected from the MesNa reagent increases as the cells are allowed to incubate at 37°C. In fact, the amount of the HKβ that is present on the surface and associated with CD63 at 0 min is approximately the same as the amount of HKβ that is associated with CD63 and protected from stripping at 45 min. These findings suggest that β-subunit associated with CD63 is internalized and does not return to the cell surface.
Discussion
In the present study we demonstrate a previously uncharacterized interaction between an ion pump subunit protein and the tetraspanin CD63. Our data suggest that the enhanced endocytosis of CD63-associated HKβ requires an interaction between these two proteins at the cell surface, initiating a process that may be mediated by sequential associations with AP-2 and AP-3. A diagram that illustrates this model is presented in Fig. 7. These data support a functional role for tetraspanin proteins as facilitators of intracellular trafficking.
Fig. 7.
Model for internalization of the HKβ by association with the tetraspanin CD63. CD63 may be an adaptor protein that facilitates the extended trafficking of associated proteins from the cell surface all of the way through to the late endosomal–lysosomal compartment by sequential interactions with AP-2 and AP-3. It is possible also that association with CD63 prevents the rapid recycling of associated proteins from internal compartments to the cell surface.
Our data show that CD63 may participate in protein trafficking by inducing functional associations between the HKβ and clathrin adaptor complexes. CD63 might serve to link clathrin adaptors and the β-subunit, or it might act to favor the formation or stabilization of direct HKβ–clathrin adaptor interactions. The β-subunit accumulates at the cell surface when it is coexpressed with a CD63-YEVI mutant that can assemble with AP-3 but not with AP-2. These data indicate that an interaction between CD63 and AP-2 is required to initiate the redistribution of cell-surface HKβ to intracellular compartments. Because the HKβ is transported with CD63 into CD63-containing late endosomal–lysosmal structures, we conclude that CD63 acts as an adaptor not only between the HKβ and the AP-2 complex but also between the HKβ and the AP-3 complex. Consequently, CD63 may act as an adaptor protein that facilitates the extended trafficking of associated proteins from the cell surface all of the way to the late endosome–lysosomal compartment (Fig. 7).
CD63 has additional interaction partners that may participate also in the internalization of this tetraspanin and its associated proteins. Previous work has demonstrated that CD63 interacts with type II phosphatidylinositol 4-kinase (10). Phosphatidylinositol 4-kinase is essential for the formation of phosphatidylinositol 4,5-bisphosphate, which is a necessary participant in both endocytosis and secretion. Phosphatidylinositol 4,5-bisphosphate appears to facilitate the interaction between the AP-2 complex and membranes, concentrating the adaptin in endocytic “hot spots” (32). Thus, CD63 may help to localize phosphatidylinositol 4-kinase near the AP-2 complex, bringing phosphatidylinositol 4,5-bisphosphate and AP-2 into close proximity and stimulating endocytosis.
The tetraspanin CD63 is resident in many intracellular structures, including endothelial cell Weibel–Palade bodies, B cell MHC class II compartments, and, as we have demonstrated, parietal cell TVEs. Proteins in each of these types of compartments cycle from intracellular vesicles to the cell surface on physiologic stimulation and return to intracellular vesicles when stimulation ceases. Interactions with CD63 may be involved in directing the trafficking of these proteins. Thus, the interaction between CD63 and the HKβ may play a role in inactivating gastric acid secretion by helping to the return the pump to the parietal cell TVEs. Interestingly, Franic et al. (33) have shown that mice that do not express the HKβ lack TVEs. It is tempting to suggest that the endocytically active complex created by the interaction between CD63 and the HKβ may play a crucial role in the formation of these subcellular compartments. Confirmation of this hypothesis must await the creation of animal model systems in which the functional activity of CD63 has been altered genetically.
Previous work in this laboratory has shown that a tyrosine-based motif in the intracellular tail of the HKβ facilitates efficient internalization of the pump in gastric parietal cells of mice (18). We find that the tyrosine-based motif in the HKβ tail is not required for its interaction with CD63 and that the association is likely to be mediated by the transmembrane or extracellular domains of these proteins (data not shown). Furthermore, intracellular accumulation of a mutant β-subunit lacking the tyrosine motif is observed also when it is coexpressed with CD63 in COS cells (data not shown), although we have not quantified the rate and extent of the mutant HKβ internalization. It is possible that the tyrosine-based motif of the HKβ plays a more prominent role in protein internalization in parietal cells than it does in COS cells because parietal cells are highly specialized to undergo massive exocytic and endocytic transformations routinely. Alternatively, these findings suggest that the internalization signals of the β-subunit and CD63, as well as other associated tetraspanins, may act synergistically. The HKβ contains an internalization signal, and associated tetraspanins could provide additional AP-2 binding sites, facilitating the putative clathrin-mediated internalization of the β-subunit from the cell surface (34).
In summary, we have demonstrated that the association of the HKβ with CD63 on the cell surface enhances the internalization of the β-subunit. This internalization appears to be mediated at least in part by μ2 and μ3, the association partners of CD63. The ability of CD63 to interact with both AP-2 and AP-3 makes it suited uniquely to the tasks of both enhancing the endocytosis of associated proteins and delivering these proteins to intracellular compartments. Our findings suggest that CD63 may be involved in the process of returning membrane proteins that are resident in secretory vesicles to their original compartments after their exocytotic delivery to the cell membrane.
Acknowledgments
We thank Drs. P. Cresswell, P. Deen, J. Forte, and C. T. Okamoto for generous gifts of reagents; Drs. P. Cresswell, P. De Camilli, and I. Mellman for critical readings of the manuscript; S. Mentone for performing immunohistochemical experiments; and members of the Caplan laboratory for helpful discussions. This work was supported in part by National Institutes of Health Grants GM 07205 (to A.D.), GM 42136, and DK 17433 (to M.J.C.), and European Molecular Biology Organization Long-Term Fellowship ALTF-155–2001 (to E.-J.K.).
Abbreviations: AP-2 and AP-3, adaptor protein complexes 2 and 3; AQP4, aquaporin-4; CD63-FL, FLAG-tagged CD63; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate; HKα and HKβ, H,K-ATPase α- and β-subunits; pAb, polyclonal antibody; MesNa, 2-mercaptoethanesulfonic acid, sodium salt; TVE, tubulovesicular element.
References
- 1.Hemler, M. E. (2001) J. Cell Biol. 155, 1103–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boucheix, C. & Rubinstein, E. (2001) Cell. Mol. Life Sci. 58, 1189–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maecker, H. T., Todd, S. C. & Levy, S. (1997) FASEB J. 11, 428–442. [PubMed] [Google Scholar]
- 4.Sincock, P. M., Mayrhofer, G. & Ashman, L. K. (1997) J. Histochem. Cytochem. 45, 515–525. [DOI] [PubMed] [Google Scholar]
- 5.Escola, J. M., Kleijmeer, M. J., Stoorvogel, W., Griffith, J. M., Yoshie, O. & Geuze, H. J. (1998) J. Biol. Chem. 273, 20121–20127. [DOI] [PubMed] [Google Scholar]
- 6.Kobayashi, T., Vischer, U. M., Rosnoblet, C., Lebrand, C., Lindsay, M., Parton, R. G., Kruithof, E. K. & Gruenberg, J. (2000) Mol. Biol. Cell 11, 1829–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arribas, M. & Cutler, D. F. (2000) Traffic 1, 783–793. [DOI] [PubMed] [Google Scholar]
- 8.Hammond, C., Denzin, L. K., Pan, M., Griffith, J. M., Geuze, H. J. & Cresswell, P. (1998) J. Immunol. 161, 3282–3291. [PubMed] [Google Scholar]
- 9.Berditchevski, F. (2001) J. Cell Sci. 114, 4143–4151. [DOI] [PubMed] [Google Scholar]
- 10.Berditchevski, F., Tolias, K. F., Wong, K., Carpenter, C. L. & Hemler, M. E. (1997) J. Biol. Chem. 272, 2595–2598. [DOI] [PubMed] [Google Scholar]
- 11.Yauch, R. L. & Hemler, M. E. (2000) Biochem. J. 351, 629–637. [PMC free article] [PubMed] [Google Scholar]
- 12.Robinson, M. S. & Bonifacino, J. S. (2001) Curr. Opin. Cell Biol. 13, 444–453. [DOI] [PubMed] [Google Scholar]
- 13.Rous, B. A., Reaves, B. J., Ihrke, G., Briggs, J. A., Gray, S. R., Stephens, D. J., Banting, G. & Luzio, J. P. (2002) Mol. Biol. Cell 13, 1071–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boehm, M. & Bonifacino, J. S. (2002) Gene 286, 175–186. [DOI] [PubMed] [Google Scholar]
- 15.Caplan, M. J. (1997) Am. J. Physiol. 272, G1304–G1313. [DOI] [PubMed] [Google Scholar]
- 16.Okamoto, C. T. & Forte, J. G. (2001) J. Physiol. (London) 532, 287–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Urushidani, T. & Forte, J. G. (1997) J. Membr. Biol. 159, 99–111. [DOI] [PubMed] [Google Scholar]
- 18.Courtois-Coutry, N., Roush, D., Rajendran, V., McCarthy, J. B., Geibel, J., Kashgarian, M. & Caplan, M. J. (1997) Cell 90, 501–510. [DOI] [PubMed] [Google Scholar]
- 19.Biemesderfer, D., Dekan, G., Aronson, P. S. & Farquhar, M. G. (1992) Am. J. Physiol. 262, F55–F67. [DOI] [PubMed] [Google Scholar]
- 20.Gottardi, C. J. & Caplan, M. J. (1993) J. Biol. Chem. 268, 14342–14347. [PubMed] [Google Scholar]
- 21.Gottardi, C. J., Dunbar, L. A. & Caplan, M. J. (1995) Am. J. Physiol. 268, F285–F295. [DOI] [PubMed] [Google Scholar]
- 22.Kennel, S. J., Lankford, P. K., Foote, L. J. & Davis, I. A. (1998) Hybridoma 17, 509–515. [DOI] [PubMed] [Google Scholar]
- 23.Rabon, E. C., Im, W. & Sachs, G. (1988) Methods Enzymol. 157, 649–654. [DOI] [PubMed] [Google Scholar]
- 24.Callaghan, J. M., Toh, B. H., Pettitt, J. M., Humphris, D. C. & Gleeson, P. A. (1990) J. Cell Sci. 95, 563–576. [DOI] [PubMed] [Google Scholar]
- 25.Toh, B. H., Gleeson, P. A., Simpson, R. J., Moritz, R. L., Callaghan, J. M., Goldkorn, I., Jones, C. M., Martinelli, T. M., Mu, F. T., Humphris, D. C., et al. (1990) Proc. Natl. Acad. Sci. USA 87, 6418–6422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okamoto, C. T., Karam, S. M., Jeng, Y. Y., Forte, J. G. & Goldenring, J. R. (1998) Am. J. Physiol. 274, C1017–C1029. [DOI] [PubMed] [Google Scholar]
- 27.Weiner, I. D. & Milton, A. E. (1996) Am. J. Physiol. 270, F518–F530. [DOI] [PubMed] [Google Scholar]
- 28.Claas, C., Stipp, C. S. & Hemler, M. E. (2001) J. Biol. Chem. 276, 7974–7984. [DOI] [PubMed] [Google Scholar]
- 29.Asano, S., Kawada, K., Kimura, T., Grishin, A. V., Caplan, M. J. & Takeguchi, N. (2000) J. Biol. Chem. 275, 8324–8330. [DOI] [PubMed] [Google Scholar]
- 30.Misaka, T., Abe, K., Iwabuchi, K., Kusakabe, Y., Ichinose, M., Miki, K., Emori, Y. & Arai, S. (1996) FEBS Lett. 381, 208–212. [DOI] [PubMed] [Google Scholar]
- 31.Carmosino, M., Procino, G., Nicchia, G. P., Mannucci, R., Verbavatz, J. M., Gobin, R., Svelto, M. & Valenti, G. (2001) J. Cell Biol. 154, 1235–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jost, M., Simpson, F., Kavran, J. M., Lemmon, M. A. & Schmid, S. L. (1998) Curr. Biol. 8, 1399–1402. [DOI] [PubMed] [Google Scholar]
- 33.Franic, T. V., Judd, L. M., Robinson, D., Barrett, S. P., Scarff, K. L., Gleeson, P. A., Samuelson, L. C. & Van Driel, I. R. (2001) Am. J. Physiol. 281, G1502–G1511. [DOI] [PubMed] [Google Scholar]
- 34.Yao, X. & Forte, J. G. (2003) Annu. Rev. Physiol. 65, 103–131. [DOI] [PubMed] [Google Scholar]
- 35.Wolosin, J. M. & Forte, J. G. (1981) J. Biol. Chem. 256, 3149–3152. [PubMed] [Google Scholar]