Abstract
Fibroblast growth factor (FGF) 19 is an enterokine synthesized and released when bile acids are taken up into the ileum. We show that FGF19 stimulates hepatic protein and glycogen synthesis but does not induce lipogenesis. The effects of FGF19 are independent of the activity of either insulin or the protein kinase Akt and, instead, are mediated through a mitogen-activated protein kinase signaling pathway that activates components of the protein translation machinery and stimulates glycogen synthase activity. Mice lacking FGF15 (the mouse FGF19 ortholog) fail to properly maintain blood concentrations of glucose and normal postprandial amounts of liver glycogen. FGF19 treatment restored the loss of glycogen in diabetic animals lacking insulin. Thus, FGF19 activates a physiologically important, insulin-independent endocrine pathway that regulates hepatic protein and glycogen metabolism.
Fibroblast growth factor 19 (FGF19, also called FGF15 in rodents) is a member of a subfamily of fibroblast growth factors that govern nutrient metabolism (1). FGF19 is expressed in the distal small intestine, where its synthesis is regulated by the nuclear bile acid receptor, FXR, after the postprandial uptake of bile acids (2, 3). Thus, in response to feeding, the concentration of circulating FGF19 increases (4). FGF19 binds to a receptor complex composed of the FGF receptor 4 (FGFR4) and a coreceptor called β-Klotho, which are both highly expressed in liver. Binding of FGF19 to the FGFR4–β-Klotho complex results in activation of the small guanosine triphosphatase Ras and extracellular signal–regulated protein kinase (ERK) signaling pathway (5, 6).
FGF19 plays an important role in hepatic bile acid homeostasis by inhibiting expression of CYP7A1, the first and rate-limiting enzyme in the major bile acid synthesis pathway (2, 3). FGF19 also promotes relaxation and refilling of the gall-bladder after a meal (7). In addition to its roles in bile acid metabolism, FGF19 lowers serum glucose and triglycerides in diabetic mice through an unclear mechanism (8).
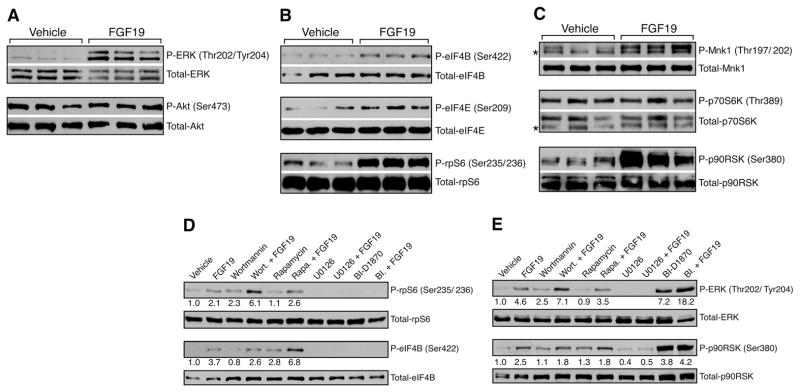
To elucidate whether FGF19 may have other effects on metabolism, we investigated FGF19-induced signaling in liver in normoglycemic wild-type animals (9). Because recombinant mouse FGF15 is unstable and has variable bioactivity, we used human FGF19 for these studies. FGF19 increased the phosphorylation of liver ERK1 and ERK2 of mice fasted overnight (Fig. 1A and fig. S1). In contrast, insulin, but not FGF19, induced phosphorylation of the protein kinase Akt (fig. S1), which demonstrated that FGF19 and insulin likely work through independent kinase signaling pathways. However, both FGF19 and insulin stimulated the phosphorylation of the eukaryotic initiation factors eIF4B on Ser422 and eIF4E on Ser209 in liver (Fig. 1B and fig. S1). These proteins are components of the eIF4F complex that mediates binding of mRNA to the ribosome, and their phosphorylation promotes the initiation of translation (10). Treatment of animals with insulin or FGF19 produced similar increases in phosphorylation of Ser235 and Ser236 of ribosomal protein S6 (rpS6) (Fig. 1B and fig. S1). Phosphorylation of rpS6 improves the efficiency of global protein synthesis by inducing cap-dependent translation (11).
Fig. 1.
Stimulation by FGF19 of signaling pathways that regulate protein synthesis in liver. (A to C) Mice fasted overnight were injected intravenously (i.v.) with vehicle or 1 mg of FGF19 protein per kg of body weight. This concentration is based on the optimal systemic dose to observe the physiologic effects of FGF19 on bile acid metabolism. The animals were killed 1 hour after the injections. Proteins from liver homogenates were separated by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and identified by Western blotting with the indicated antibodies. Results represent triplicate experiments. (D and E) Overnight serum-starved HepG2 cells were starved for amino acids in Hank’s buffered salt solution medium for 1 hour. Vehicle, wortmannin (200 nM), rapamycin (20 nM), U0126 (10 μM), or BI-D1870 (10 μM) was added for a further 1-hour treatment. The cells were treated with vehicle or 250 ng/ml FGF19 and harvested after 30 min. Proteins were identified by Western blotting with the indicated antibodies. BI-D1870 treatment blocks the negative-feedback effect of p90RSK on ERK, which results in increased basal phosporylation of ERK and p90RSK. Numbers below blots represent fold-change relative to the vehicle group. Asterisk (*) marks nonspecific band (C).
We investigated the kinases that might mediate phosphorylation of eIF4 or rpS6 in response to FGF19. Ser209 of eIF4E is a target for the protein kinase Mnk1, which can be activated by phosphorylation at Thr197 and Thr202 by ERK (12). FGF19 induced phosphorylation of Mnk1, which implicated FGF19 as the upstream stimulus of a Ras-ERK-Mnk1 signaling cascade that activates eIF4E (Fig. 1C). rpS6 and eIF4B are well-known targets of p70 ribosomal S6 kinase (p70S6K), which is activated by insulin (fig. S1). However, FGF19 treatment did not induce the phosphorylation of p70S6K or Akt, which is known to activate mammalian target of rapamycin (mTOR) to stimulate p70S6K (Fig. 1, A and C, and fig. S1). Instead, FGF19 induced the phosphorylation of p90 ribosomal S6 kinase (p90RSK), which also is known to phosphorylate rpS6 and eIF4B (13, 14).
Because p90RSK is a downstream target of ERK, our results indicate that FGF19 utilizes a Ras-ERK-p90RSK pathway to induce phosphorylation of rpS6 and eIF4B. In human hepatocarcinoma HepG2 and rat hepatoma HII4E cells that express FGFR4 and β-Klotho, FGF19 induced the phosphorylation of fibroblast growth factor substrate 2α (FRS2α), ERK, and p90RSK in a dose-dependent manner (fig. S2A). Likewise, FGF19 treatment increased the phosphorylation of rpS6 and eIF4B in HepG2 cells (Fig. 1D). However, this effect was not inhibited by wortmannin, a potent phosphoinositide 3-kinase (PI3K) inhibitor, or rapamycin, an mTOR inhibitor, which suggested that FGF19 does not act through the Akt-mTOR-S6K pathway (Fig. 1D and fig. S2, B and C). Indeed, FGF19 treatment failed to induce the phosphorylation of Akt or p70S6K (Fig. 1E and fig. S2B). In contrast, the ERK pathway inhibitor U0126 and p90RSK inhibitor BI-D1870 (15) completely inhibited FGF19-induced phosphorylation of ERK and p90RSK (Fig. 1E), and both inhibitors blocked basal and FGF19-dependent phosphorylation of rpS6 and eIF4B (Fig. 1D).
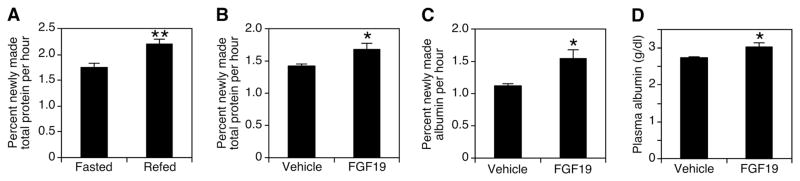
The above findings link FGF19 to stimulation of protein synthesis in liver. We analyzed protein synthesis in mouse liver using 2H2O labeling (16, 17). When injected into animals, 2H2O equilibrates with body water within 90 min, and 2H incorporates into amino acids. To determine the normal rate of protein synthesis after fasting and refeeding, mice were fasted overnight and then refed or continually fasted for another 6 hours. Refeeding caused a 25% increase in the rate of liver protein synthesis (Fig. 2A) (18). In comparison, injection of FGF19 significantly increased total protein synthesis by 18% (Fig. 2B). The de novo synthesis rate of albumin, the major protein product of liver, was increased 40% by FGF19 (Fig. 2C). Moreover, continuous treatment with FGF19 significantly increased plasma albumin levels by 10% (Fig. 2D). Thus, FGF19 is a positive regulator of hepatic protein synthesis.
Fig. 2.
Increased rates of global protein synthesis and albumin synthesis in mouse liver treated with FGF19. (A) Mice fasted overnight received 0.5 ml 2H2O intraperitoneally (i.p.); 90 min later, the animals were refed or kept fasted for 6 hours and killed (n = 10). Protein samples were hydrolyzed, and 2H labeling of alanine was determined by mass spectrometry. (B and C) Mice fed ad libitum received 0.5 ml 2H2O; 90 min later (at 6 p.m.), vehicle or 1 mg/kg FGF19 was injected subcutaneously. The next morning (8 a.m.), animals were injected again with the same dose and, 6 hours later, were killed (n = 10). Protein samples were hydrolyzed, and 2H labeling of alanine was determined by mass spectrometry. For albumin synthesis, 2H incorporation into plasma albumin was measured in the same way. (D) Over a 3-day period, mice (n = 6) received vehicle or 1 mg/kg FGF19 subcutaneously 3 times at 6 p.m. and once on the day they were killed at 8 a.m. Then, 6 hours after the last injection, the livers were harvested. Plasma albumin levels were determined with a Vitros 250 instrument. Values are means ± SEM. Statistics by two-tailed t test. *P < 0.05, **P < 0.005.
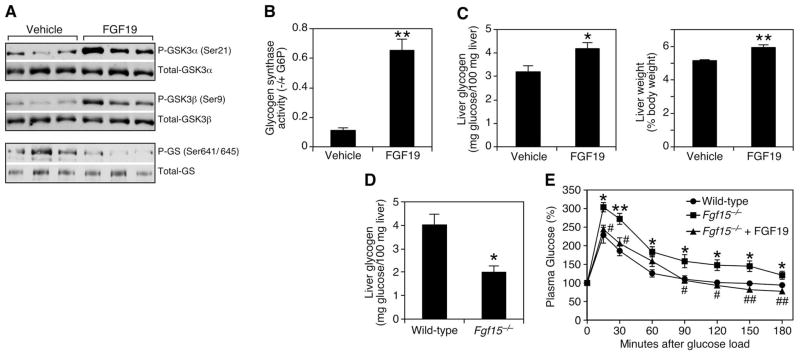
The effects of FGF19 on protein synthesis prompted us to investigate glycogen synthesis, another target of insulin action. Glycogen synthesis in liver is negatively regulated by glycogen synthase kinase (GSK) 3α and GSK3β, which phosphorylate and inhibit the enzyme glycogen synthase (GS). Phosphorylation also inactivates GSK3 kinases, which prevents inhibition of GS and thus increases glycogen synthesis (19). In animals fasted overnight, FGF19 induced phosphorylation of both GSK3α (Ser21) and GSK3β (Ser9), which correlated with decreased phosphorylation of Ser641 and Ser645 on GS (Fig. 3A) and increased GS activity (Fig. 3B). Concomitantly, there was a 30% increase in liver glycogen content that led to a small but significant increase in liver weight in FGF19-treated mice compared with that of control animals (Fig. 3C). FGF19 treatment had no effect on liver cholesterol or triglycerides (fig. S3, A and B), nor did it change plasma insulin or glucagon concentrations, which strengthens the idea that it acts directly on liver (fig. S3, C and D). We also analyzed hepatic glycogen concentration in mice lacking Fgf15 (the mouse ortholog of FGF19). Fed Fgf15−/− mice had >50% less hepatic glycogen than did wild-type animals (Fig. 3D), which demonstrated the physiologic requirement for FGF15 in maintaining normal glycogen metabolism. Moreover, Fgf15−/− mice showed impaired glucose uptake from the circulation. FGF19 administration completely rescued this phenotype (Fig. 3E).
Fig. 3.
FGF19 inhibits GSK3 signaling to increase liver glycogen in mice. (A) Mice fasted overnight were treated i.v. with vehicle or 1 mg/kg FGF19 and killed 10 min later. Proteins from liver homogenates were separated by SDS-PAGE and identified by Western blotting with the indicated antibodies. Results represent triplicate experiments. (B) The ability of glycogen synthase in the homogenates of the same livers to incorporate radiolabeled uridine 5′-diphosphate–glucose into glycogen in the absence and presence of glucose-6-phosphate was measured, and the ratio was shown as glycogen synthase activity (n = 3). (C) Mice fed ad libitum were injected subcutaneously with vehicle or 1 mg/kg FGF19 at 6 p.m. and the next morning at 8 a.m. Then, 6 hours after the last injection, the animals were killed, and liver weight and glycogen content were determined (n = 6). (D) Liver glycogen content was determined in wild-type and Fgf15−/− mice fed ad libitum (n = 5). (E) Oral glucose tolerance test in wild-type and Fgf15−/− mice (n = 6). Values are means ± SEM. Asterisks (*) refer to differences between wild-type and Fgf15−/− groups; number signs (#) refer to differences between Fgf15−/− and Fgf15−/− plus FGF19 groups. Statistics by two-tailed t test. *P < 0.05, **P < 0.005, #P < 0.05, ##P < 0.005.
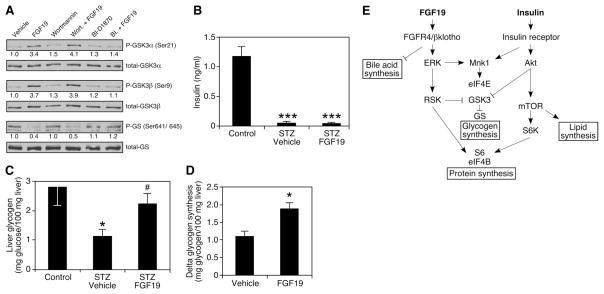
Akt and p90RSK phosphorylate the same residues of GSK3α and β (19–22). To test whether p90RSK might mediate phosphorylation of GSK3 kinases for FGF19, we treated HepG2 cells with FGF19 and either the PI3K or p90RSK inhibitor. FGF19-induced phosphorylation of GSK3 kinases in HepG2 cells was compromised when cells were treated with BI-D1870, but not when treated with wortmannin (Fig. 4A). These data further support the idea that FGF19 acts through an insulin-independent Ras-ERK-p90RSK pathway to regulate glycogen synthesis.
Fig. 4.
FGF19-induced glycogen synthesis is independent of insulin. (A) Overnight serum-starved HepG2 cells were pretreated with vehicle, wortmannin (200 nM), or BI-D1870 (10 μM) for 1 hour. The cells were lysed 30 min after vehicle or FGF19 (250 ng/ml) treatment. Proteins were identified by Western blotting with the indicated antibodies. Numbers below blots represent fold-change relative to the vehicle group. (B and C) Mice were treated i.p. with STZ (175 mg/kg). Eight days later, diabetic animals were chosen and treated with vehicle or 1 mg/kg FGF19 i.p. at 6 p.m. for seven consecutive days, and killed 6 hours after the last injection at 8 a.m.(n = 5 to 9). Liver glycogen content and plasma insulin levels were determined. *P < 0.05 is between control and STZ-vehicle groups; #P < 0.05 is between STZ-vehicle and STZ-FGF19 groups. (D) Three-hour hyperglycemic clamp study was performed on rats fasted overnight (n = 5 to 7). Animals were continuously infused with insulin and somatostatin to maintain low levels of insulin and glucagon and variably infused with glucose to maintain hyperglycemia. Net glycogen synthesis was determined by assessing the glycogen content in the clamped animals subtracted by the glycogen content of unclamped animals that were killed after the same duration of fasting. (E) Insulin and FGF19 act through different signaling pathways to coordinate overlapping but distinct postprandial responses in liver. (B, C, and D) Values are means ± SEM. Statistics by two-tailed t test. *P < 0.05, ***P < 0.0005.
Streptozotocin (STZ)–treated mice are severely diabetic and have almost no detectable insulin in the blood (Fig. 4B). STZ treatment reduced liver glycogen content to 50% of that of control animals. FGF19 treatment restored hepatic glycogen amounts and had a significant effect on glucose disposal (Fig. 4C and fig. S4). Insulin-independent effects of FGF19 on the rate of net hepatic glycogen synthesis were also investigated in rats fasted overnight. A hyperglycemic clamp was used in combination with somatostatin to inhibit endogenous insulin and glucagon secretion. Under matched conditions of plasma glucose, insulin, and glucagon concentrations (fig. S5), FGF19 increased net hepatic glycogen synthesis by 70% compared with that in control rats (Fig. 4D).
Taken together, these studies suggest that FGF19 acts in parallel to and independent from insulin to govern postprandial metabolism in liver. Like insulin, pharmacologic administration of FGF19 can induce protein and glycogen synthesis, whereas loss of the physiologic hormone in Fgf15−/− mice results in glucose intolerance and reduced hepatic glycogen. However, whereas insulin reaches its maximum serum concentration within 1 hour of a meal in humans, peak FGF19 levels are achieved ~3 hours after a meal (4) just before glycogen accumulation peaks in the liver (23, 24). Thus, we propose insulin and FGF19 work in a coordinated temporal fashion to facilitate the proper postprandial storage of nutrients. Of the anabolic enterokines (e.g., the incretins, GLP-1 and GIP), FGF19 is unusual in that it mimics insulin action rather than stimulating its release.
The different signaling pathways used by FGF19 and insulin permit overlapping but distinct biological effects for the two hormones (Fig. 4E). For example, unlike insulin, FGF19 did not increase hepatic triglycerides (fig. S3B) or induce sterol regulatory element–binding protein, isoform 1c (SREBP-1c)–dependent lipogenic gene expression (fig. S6), which requires the PI3K-Akt-mTOR signaling pathway (25, 26). Indeed, FGF19 appears to be unique in its ability to differentially govern glycogen synthesis and lipogenesis. FGF15 (or FGF19) is required to maintain normal glycogen levels in fed mice, and it uses the alternative Ras-ERK-p90RSK pathway; taken together, these findings may explain the puzzling observation that liver-specific loss of insulin signaling in IRS1-IRS2 null mice does not fully block glycogen storage in response to feeding (27, 28). These results may also help explain the glucose- and insulin-lowering actions of FGF19 in diabetic rodents (8). Thus, pharmacologically targeting the FGF19 pathway might be an attractive alternative to using insulin to increase glycogen storage without affecting lipogenesis.
Supplementary Material
Acknowledgments
We thank T. Inagaki, X. Ding, and A. Bookout for their help with animal experiments and M. Cobb and P. Scherer (University of Texas Southwestern Medical Center) for discussion and comments. This research was supported by the Howard Hughes Medical Institute (G.I.S., D.J.M.), the NIH (DK67158 and DK62434 to S.A.K. and D.J.M., DK40936 and U24 DK076169 to G.I.S.), the Robert A. Welch Foundation (I-1275 to D.J.M. and I-558 to S.A.K.), and the Yale and Case Western Reserve University Mouse Metabolic Phenotyping Centers. A Materials Transfer Agreement is required for sharing materials from the Van Andel Institute.
Footnotes
References and Notes
- 1.Beenken A, Mohammadi M. Nat Rev Drug Discov. 2009;8:235. doi: 10.1038/nrd2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holt JA, et al. Genes Dev. 2003;17:1581. doi: 10.1101/gad.1083503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Inagaki T, et al. Cell Metab. 2005;2:217. doi: 10.1016/j.cmet.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Lundåsen T, Gälman C, Angelin B, Rudling M. J Intern Med. 2006;260:530. doi: 10.1111/j.1365-2796.2006.01731.x. [DOI] [PubMed] [Google Scholar]
- 5.Kurosu H, et al. J Biol Chem. 2007;282:26687. doi: 10.1074/jbc.M704165200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin BC, Wang M, Blackmore C, Desnoyers LR. J Biol Chem. 2007;282:27277. doi: 10.1074/jbc.M704244200. [DOI] [PubMed] [Google Scholar]
- 7.Choi M, et al. Nat Med. 2006;12:1253. doi: 10.1038/nm1501. [DOI] [PubMed] [Google Scholar]
- 8.Fu L, et al. Endocrinology. 2004;145:2594. doi: 10.1210/en.2003-1671. [DOI] [PubMed] [Google Scholar]
- 9.Materials and methods are available as supporting material on Science Online.
- 10.Gingras AC, Raught B, Sonenberg N. Annu Rev Biochem. 1999;68:913. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 11.Fumagalli S, Thomas G. In: Translational Control of Gene Expression. Sonenberg N, et al., editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. pp. 695–717. [Google Scholar]
- 12.Ueda T, Watanabe-Fukunaga R, Fukuyama H, Nagata S, Fukunaga R. Mol Cell Biol. 2004;24:6539. doi: 10.1128/MCB.24.15.6539-6549.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roux PP, et al. J Biol Chem. 2007;282:14056. doi: 10.1074/jbc.M700906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shahbazian D, et al. EMBO J. 2006;25:2781. doi: 10.1038/sj.emboj.7601166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sapkota GP, et al. Biochem J. 2007;401:29. doi: 10.1042/BJ20061088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dufner DA, et al. Am J Physiol Endocrinol Metab. 2005;288:E1277. doi: 10.1152/ajpendo.00580.2004. [DOI] [PubMed] [Google Scholar]
- 17.Rachdaoui N, et al. Mol Cell Proteomics. 2009;8:2653. doi: 10.1074/mcp.M900026-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson SR, Gilge DA, Steiber AL, Previs SF. Metabolism. 2008;57:347. doi: 10.1016/j.metabol.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen P, Frame S. Nat Rev Mol Cell Biol. 2001;2:769. doi: 10.1038/35096075. [DOI] [PubMed] [Google Scholar]
- 20.Sutherland C, Leighton IA, Cohen P. Biochem J. 1993;296:15. doi: 10.1042/bj2960015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stambolic V, Woodgett JR. Biochem J. 1994;303:701. doi: 10.1042/bj3030701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding Q, et al. Mol Cell. 2005;19:159. doi: 10.1016/j.molcel.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 23.Krssak M, et al. Diabetes. 2004;53:3048. doi: 10.2337/diabetes.53.12.3048. [DOI] [PubMed] [Google Scholar]
- 24.Taylor R, et al. J Clin Invest. 1996;97:126. doi: 10.1172/JCI118379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li S, Brown MS, Goldstein JL. Proc Natl Acad Sci USA. 2010;107:3441. doi: 10.1073/pnas.0914798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Porstmann T, et al. Cell Metab. 2008;8:224. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kubota N, et al. Cell Metab. 2008;8:49. doi: 10.1016/j.cmet.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 28.Dong X, et al. J Clin Invest. 2006;116:101. doi: 10.1172/JCI25735. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.