Abstract
β-Catenin is a multifunctional protein, stabilization of which is a critical step in its functional activity. Stabilization can be conferred through several means, including mutations in β-catenin, common in many carcinomas. In this article, we explore the effect of viral infection on the β-catenin signaling pathway in B lymphocytes, including cell lines derived from lymphomas. Infection by the human tumor virus Epstein–Barr virus (EBV) generates several types of latency with different spectra of latent gene expression that are associated with different malignancies. In type III latency, exemplified by EBV lymphoproliferative diseases, the full range of these viral proteins is expressed, whereas in type I latency, only the EBNA1 protein is expressed and functional. We show that β-catenin is rapidly degraded in type I B lymphocytic lines but it is stabilized in type III B cell lines, and that β-catenin/T cell factor transcriptional activity is significantly higher in type III cells. Also we show the association of free cytoplasmic β-catenin with deubiquitinating enzymes, which may play a role in β-catenin stabilization. Activation of the β-catenin/T cell factor pathway by EBV may contribute to the lymphoproliferation characteristic of type III latency.
The multifunctional protein β-catenin plays a dual role in cells as a major structural component of cell–cell adherens junctions and as a signaling molecule in the Wnt pathway. The nonjunctional pool of β-catenin is normally degraded by the ubiquitin-proteasome system or, under certain conditions such as the activation of Wnt signaling, β-catenin is not degraded but stabilized, and it then activates transcription by providing a transactivation domain in a heterodimeric complex with lymphoid enhancer factor (Lef)/T cell factor (Tcf) transcription factors (1).
The amount of uncomplexed cytoplasmic β-catenin is generally regulated by a multiprotein complex including glycogen synthase kinase 3β (GSK3β), the adenomatous polyposis coli (APC) tumor suppressor protein, and Axin (2, 3). This complex targets β-catenin for phosphorylation, which triggers its ubiquitination and degradation.
Epstein–Barr virus (EBV) infects and immortalizes B lymphocytes, and the viral genome is regularly detected in a number of lymphoid malignancies (4, 5). EBV can establish different latency states in B lymphocytes. In type I latency, found in Burkitt's lymphoma, EBV nuclear antigen-1 (EBNA-1) is the chief gene product to be expressed. In type III latency, exemplified by EBV lymphoproliferative diseases, all of the viral latency proteins (six EBNAs and three lymphocyte membrane proteins) are expressed (6). It has been shown that EBNA2, which is expressed only in type III latency, induces c-Myc and cyclin D expression (7). β-Catenin also is involved in activation of these genes (8, 9).
Studies in recent years have shown that β-catenin is a potential oncogene, and its accumulation has been implicated in a variety of carcinomas (1, 10). The β-catenin/Tcf pathway also is significant in lymphocyte proliferation and differentiation during embryonic development in mice (11), and Wnt signaling plays an important role in self-renewal of hematopoietic stem cells (12). However, the possible effect of virus infection on the pathway has received little attention, and the role of the β-catenin/Tcf pathway in B cell lymphomas is largely unknown.
Ubiquitination is an essential step for proteasomal degradation. The identification and characterization of enzymes that recognize and ubiquitinate the substrates has been a major focus in the ubiquitin-proteasome field (13). Recently, interest in their enzymatic opposites, the deubiquitinating enzymes (DUBs), has rapidly emerged (14, 15). The presence of a large number of tissue-specific DUBs that share little sequence similarity suggests an as yet largely unexplored substrate specificity of this class of proteins (16).
It was shown recently that the herpesvirus-associated ubiquitin-specific protease can remove ubiquitin from the p53 tumor suppressor protein and rescue it from degradation, leading to p53-mediated cell-growth repression (17), and the tumor suppressor CYLD [which is mutated in familial cylindromatosis (18)], turns out to be a DUB that negatively regulates NF-κB signaling (19, 20). Yet knowledge about substrates of DUBs and their biological role in cells is otherwise very limited. Recently a new method for labeling active DUBs was reported (21) that will be helpful for study of the biological role of this family of proteins.
In this article, we show that type III latent EBV infection leads to β-catenin stabilization and activation in EBV-infected lymphoid cell lines, whereas type I latency does not. We found further that free cytoplasmic β-catenin is associated with active DUBs.
Materials and Methods
Cell Lines and Reagents. All cell lines were cultured in RPMI medium 1640 (GIBCO) supplemented with 10% FBS (Sigma), penicillin, and streptomycin.
MG132 (benzyloxycarbonyl-leu-leucinyl-leucinal) and MG101 (N-acetyl-leu-leu-norleucinal) were obtained from Calbiochem. TCF reporter plasmids were obtained from Upstate Biotechnology (Lake Placid, NY). Ubiquitin aldehyde (Ubal) and protein A/G PLUS-Agarose were obtained from Santa Cruz Biotechnology.
Antibodies. β-Catenin (Transduction Laboratories, Lexington, KY), Axin (Zymed), GSK3β (Transduction Laboratories), APC (c-20; Santa Cruz Biotechnology), Tcf-4 (clone 6H5–3; Upstate Biotechnology), actin (Sigma), and hemagglutinin (HA) tag (F-7; Santa Cruz Biotechnology). Antibodies were used at the indicated dilutions.
Western Blotting. Cells were lysed in lysis buffer [50 mM Hepes, pH 7.4/150 mM NaCl/10% glycerol/1 mM EDTA/1 mM sodium orthovanadate/100 mM NaF/1% Triton X-100/protease inhibitor mixture (Roche Diagnostics)]. Protein concentration was determined by the Bradford assay (Bio-Rad). Total cell proteins were resolved on SDS/10% PAGE, transferred to nitrocellulose membrane (Osmonics, Minnetonka, MN), blocked in 5% milk/Tris-buffered saline, and incubated overnight with β-catenin (1:500), Tcf (1:250), and actin (1:10,000) antibodies.
Northern Blotting. Total RNAs of Sav I and Sav III cells were separated by electrophoresis on 1% agarose gels containing 3.7% formaldehyde; equal loading was assessed by analysis of the rRNA band on ethidium bromide-stained gels. Total RNA was transferred on nylon membrane and crosslinked by UV light. The Northern blots were probed with biotin-labeled cDNA specific for β-catenin according to the manufacturer's protocol (Ambion, Austin, TX). The blots then were washed twice with 2× SSPE (20× SSPE consists of 0.2 M sodium phosphate, pH 7.4, containing 25 mM EDTA and 3 M NaCl) at room temperature, twice with 2× SSPE containing 1% SDS at 55°C, and twice with 0.2× SSPE at room temperature. β-Catenin mRNA was detected with a nonisotopic detection kit (Ambion) according to the protocol.
Immunoprecipitation. Whole-cell lysates were precleared with A/G PLUS-Agarose beads for 1 h at 4°C and incubated with the indicated antibodies [Axin (1:500) or HA tag (1:100)] or an equal amount of control Ig overnight at 4°C and then with beads for an additional 1 h at 4°C. After intensive washing and centrifugation, immune complexes were separated by SDS/PAGE and probed with antibodies to the following proteins as indicated: GSK3β (1:1,000), APC (1:500), Axin (1:1,000), and β-catenin (1:500).
Reporter Assay. Approximately 107 Sav I or Sav III cells per transfection were electroporated in 0.5 ml of RPMI medium 1640 (10% FBS) with a Bio-Rad Gene-Pulser at 260 V and 975 μF. For reporter assays, each transfection contained 10 μgofthe TOPFLASH or FOPFLASH plasmids. After electroporation, cells were resuspended in 10 mM of complete medium and incubated at 37°C. A β-galactosidase-expressing vector was included as an internal control for transfection efficiency. Forty-eight hours later, cells were collected and luciferase reporter assays were performed according to standard procedures (Promega). Relative light units were measured in an Lmax luminometer (Autolumat LB953; Perkin–Elmer).
Labeling Cells with HAUbVME and Ubal. Sav III cells were lysed in labeling buffer (50 mM Tris, pH 7.4/5 mM MgCl2/250 mM sucrose/1 mM DTT/2 mM ATP). Nuclei and debris were removed by centrifugation, and cell extracts were incubated with HAUbVME probe (0.2 μg/μl) for 1 h at 37°C or with 1 μM Ubal for 2 h at 37°C.
Results and Discussion
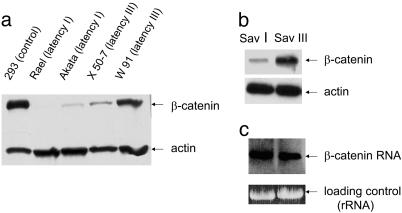
To investigate whether there are differences in β-catenin protein levels in either type I or type III latency, we examined several B cell lines with different latency types by using a human kidney epithelial cell line (293) that expresses high levels of β-catenin as a positive control. Western analysis shows a greater accumulation of β-catenin in type III latency than in type I (Fig. 1a). We then looked at β-catenin levels in the B cell lines Sav I and Sav III. These cell lines are genetically identical, differing in their EBV latency status, type I and type III, respectively (22). The level of β-catenin in Sav III is significantly higher than in Sav I, implying that a type III latency product(s) influences β-catenin accumulation (Fig. 1b). We also tested for β-catenin RNA levels by Northern analysis and detected no significant difference in type I and type III cells (Fig. 1c).
Fig. 1.
β-Catenin expression in different types of latency in EBV-infected B cell lines. (a) Total lysates of lines with type I latency (Akata and Rael) and type III latency (X 50-7 and W 91) were resolved in 10% PAGE and probed with β-catenin and actin antibodies. Total extract from the 293 cell line was used as the positive control for β-catenin expression. (b) Total protein extracts of genetically identical B cell lines with type I and type III latency (Sav I and Sav III) were probed with β-catenin and actin antibodies. (c) Northern blot of total RNA extracts of Sav I and Sav III cells was probed with biotin-labeled β-catenin DNA probe.
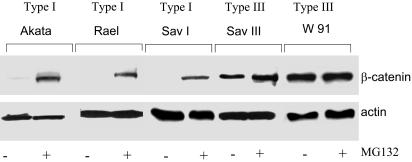
To determine whether the low level of β-catenin detected in type I cells was caused by proteasomal degradation, we incubated cells with MG132, a potent proteasome inhibitor, and then performed Western blot analysis. In all three latency type I cell lines tested, significant accumulation of β-catenin was detected in the presence of the inhibitor. In contrast, in type III cells β-catenin was not further stabilized by MG132 (Fig. 2). Therefore, the difference in β-catenin levels between type I and type III cell lines is attributable not to differential expression but to differential degradation.
Fig. 2.
Proteasome inhibition causes β-catenin accumulation in type I latency. Cells were treated with 10 μM MG132 for 1 h at 37°C before collection. Western blots of total protein extracts from B cell lines were probed with β-catenin and actin antibodies.
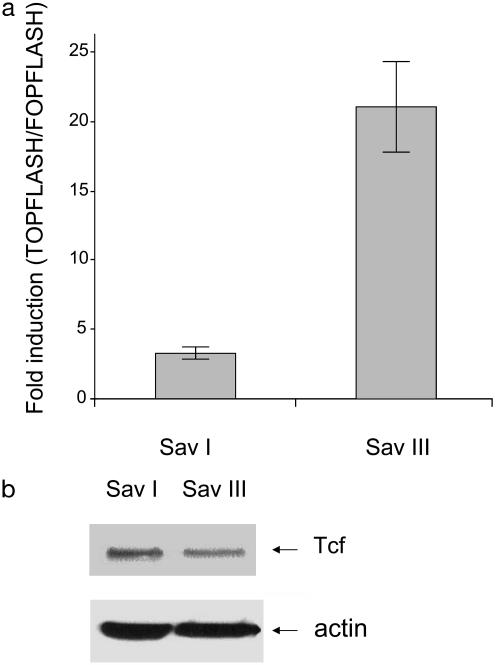
It has been shown that stabilized β-catenin forms a complex with Tcf/lymphoid enhancer factor transcriptional factors and that this complex transactivates downstream targets such as c-myc and cyclin D1. These genes have been implicated in cell transformation and tumor development (8, 9). To investigate whether stabilization of β-catenin in latency III results in transcriptional activation of Tcf, we transfected Sav I and Sav III cell lines with Tcf reporter plasmids containing three copies of WT Tcf-binding site (TOPFLASH) and three copies of mutated site as a negative control (FOPFLASH). The results show that stabilization of β-catenin in type III latency leads to constitutive transcriptional activation of β-catenin/Tcf complex, as determined by the luciferase assay (Fig. 3a), and that transcriptional activity of β-catenin does not depend on the Tcf-expression level in type I and type III latency (Fig. 3b).
Fig. 3.
(a) β-Catenin/Tcf transcriptional activity is higher in latency III. Sav I and Sav III cells were transfected with luciferase reporter plasmids containing WT (TOPFLASH) or mutated (FOPFLASH) Tcf promoter. After 48 h cells were collected, and Tcf activity was determined by a luciferase assay. The data represent the average fold induction of TOPFLASH/FOPFLASH for each cell line from two independent experiments prepared in duplicate. (b) Tcf protein levels are similar in type I and type III latency. Total protein extracts of Sav I and Sav III cells were probed with Tcf and actin antibodies.
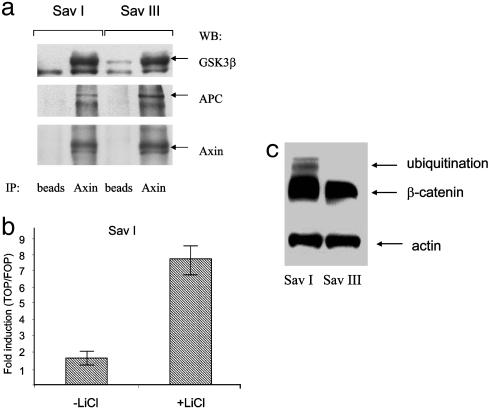
The β-catenin phosphorylation complex, the action of which targets β-catenin for degradation, has at least three essential components: Axin, APC, and GSK3β (10). Recently it was shown that the Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen can dysregulate this complex by binding to GSK3β (23). To test whether there are any differences in the phosphorylation complex in type I and type III latency, we isolated by immunoprecipitation the complex components with Axin antibody. All three expected components were detected, and there was no difference between the complexes detected in the Sav I and Sav III cell lines (Fig. 4a). Therefore the destruction complex is structurally intact in lines of either type, so that the difference in total β-catenin levels in type III compared with type I cells (Fig. 1b) was not attributable to failure to form this characteristic complex. To determine whether it is possible to restore Tcf transcriptional activity in type I latency, we inhibited GSK3β activity with LiCl, a well known activator of the Wnt pathway (3). In the presence of LiCl, Tcf transcriptional activity is significantly higher in type I latency (Fig. 4b), which indicates that the destruction complex is functional in these cells.
Fig. 4.
(a) Destruction complex of β-catenin exists in both type I and type III latency. Immunoprecipitation from cell extracts of Sav I and Sav III cell lines was performed with Axin antibody. The components of the complex were detected with Axin, GSK3β, and APC antibodies. (b) Destruction complex of β-catenin is functional in type I latency. Sav I cells were transfected with luciferase reporter plasmids containing WT (TOPFLASH) or mutated (FOP-FLASH) Tcf promoter after treatment with 20 mM LiCl. After 48 h cells were collected, and Tcf activity was determined by luciferase assay. (C) Ubiquitinated β-catenin is detected in latency I but not in latency III. Sav I and Sav III cells were incubated with 25 μM MG101 for4hat37°C. Western blot of total lysates was probed with β-catenin antibody.
Our experiments suggest that in B cells infected with EBV and with established type I latency, practically all expressed β-catenin is rapidly degraded. The observation that in type III latent EBV infection β-catenin is stabilized (Figs. 1 and 2) suggests that a tumor virus is able to influence the β-catenin pathway by inhibiting β-catenin degradation. On the other hand, virus products in type III latency do not influence the expression of components of the β-catenin phosphorylation complex nor do they prevent complex assembly (Fig. 4a). We thus examined other steps in the destruction process.
Ubiquitination is an essential step in proteasomal degradation (13). To determine whether there is a difference in β-catenin ubiquitination in latency I and III, we treated Sav I and Sav III cells with the proteasomal inhibitor MG101 (ALLN), which normally results in the accumulation of ubiquitinated β-catenin (24, 25). In the presence of MG101, type I latency cells accumulate ubiquitinated β-catenin, whereas the same cells with latency III phenotype do not (Fig. 4c). The interest in biological antagonists of ubiquitinating enzymes, DUBs, has been growing recently. Herpesvirus-associated ubiquitin-specific protease can remove ubiquitin from p53 (17), whereas the DUB FAM (fat facets in mouse) can stabilize β-catenin (25). However, we could find no evidence of FAM protein expression in human B cells (data not shown). It is possible that DUB expression is tissue- and cell-specific.
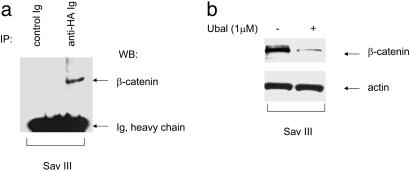
The activity of DUBs in complex samples such as mammalian cell extracts has been difficult to examine. Recently a new chemistry-based proteomics approach has allowed demonstration of DUB activity with specific active-site-directed probes against this group of enzymes (21). The active-site-directed probes, which contain an epitope-tagged ubiquitin (HAUb), can covalently modify only active DUBs. To determine whether cytoplasmic β-catenin can form a complex with DUBs we used the HAUbVME probe, which has broad reactivity (21), because we did not know which active DUB (or DUBs), if any, might be associated with β-catenin in our model system. Fig. 5a shows that stable cytoplasmic β-catenin is in the complex with active DUBs in cells with latency III. To investigate whether inhibition of DUB activity can influence β-catenin stabilization, we used a potential inhibitor of many DUBs, Ubal. Previously it has been shown that at concentrations <3 μM, Ubal increases ATP-dependent proteasomal degradation of different proteins (26). Fig. 5b shows that after incubation with 1 μM Ubal, the β-catenin level in Sav III cells is decreased, which indicates that inhibition of DUB activity leads to an increase in β-catenin degradation.
Fig. 5.
(a) β-Catenin is present in a complex with active DUBs. Sav III cell lysates were lysed in labeling buffer as described in Materials and Methods and incubated with HAUbVME active-site-directed probe for 1 h at 37°C. After immunoprecipitation (IP) with HA antibody, protein complexes were pulled down with A/G PLUS-Agarose, and Western blot was probed with β-catenin antibody. (b) Inhibition of DUB activity by Ubal leads to an increase of β-catenin degradation. Sav III cell lysates were incubated with Ubal probe for 2hat37°C. Western blots of total protein extracts were probed with β-catenin and actin antibodies.
Accumulation of β-catenin can be the result of different processes. For example, mutations in the APC tumor suppressor or in β-catenin itself trigger β-catenin accumulation in many human carcinomas, mostly those of colon and skin (10, 27, 28). Mutations in the Axin gene leading to β-catenin accumulation have been detected in hepatocellular carcinomas and cerebellar medulloblastomas (29, 30). All these mutations result in reduced degradation of β-catenin, which is believed to promote tumor formation by constitutive activation of β-catenin targets (2). Another pathway leading to β-catenin stabilization is the activation of Wnt signaling (31). Wnt genes are tumorigenic in mice (32) and also may be implicated in human cancer (2). Most results deal with carcinogenesis in epithelial cells, but almost nothing is known in lymphoid cells.
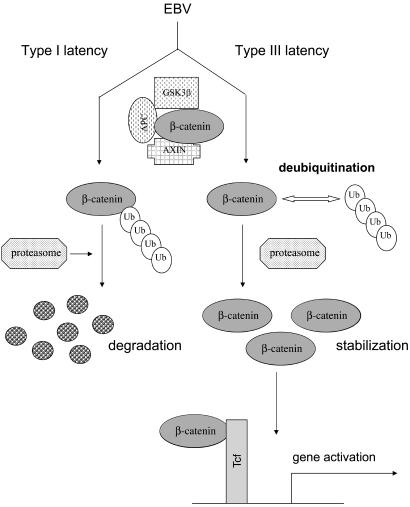
In this article we identify a means whereby β-catenin can be stabilized in cells, specifically lymphoid cells, namely by a tumor virus, EBV, through the inhibition of β-catenin degradation. Our results further suggest that deubiquitination may enable β-catenin stabilization and activation in type III latent infection (Fig. 6). In future experiments, we plan to identify and study the function and activity of the DUBs involved. It is possible that different EBV gene products and proteins encoded by other viruses also may produce such effects. We have preliminary evidence that the Epstein–Barr nuclear antigen 2 (EBNA2), which is essential for immortalization of cells by EBV, may be involved in β-catenin stabilization (data not shown).
Fig. 6.
Stabilization of β-catenin by EBV in type III latency. In type I EBV latency β-catenin is rapidly degraded. In type III latency EBV activates the β-catenin pathway by stabilizing β-catenin through inhibition of proteasomal degradation. Deubiquitination, at least in part, is involved in β-catenin stabilization and activation in type III latency EBV-infected cells.
From the EBV perspective, a scenario could be constructed as follows. In type I latency, β-catenin is degraded as expected and therefore cannot affect transforming potential by activating cell-proliferation genes. However, type I latency is invariably found only in Burkitt's lymphoma. In Burkitt's lymphoma, the c-myc oncogene is already dysregulated by characteristic chromosomal translocations that are the hallmark of this lymphoma, even in the absence of EBV (5, 6). Therefore, β-catenin stabilization would presumably contribute little. In contrast, in type III latency that characterizes lymphoproliferative diseases caused by EBV, mutations affecting c-myc expression are not found. Rather, presumably the virus exerts a stabilizing effect on β-catenin, thus making it available for activation of cell-proliferation genes such as c-myc or cyclin D and perhaps partly explaining the role of EBV in the genesis of lymphomatous conditions.
Acknowledgments
We are grateful to H. Ovaa (Harvard Medical School, Boston) and H. Ploegh (Harvard Medical School) for providing the HAUbVME probe and M. Peifer and R. Duronio for critical reading of the manuscript. This work was supported by National Cancer Institute Grant CA 19014.
Abbreviations: EBV, Epstein–Barr virus; Tcf, T cell factor; GSK, glycogen synthase kinase; APC, adenomatous polyposis coli; DUB, deubiquitinating enzyme; HA, hemagglutinin; Ubal, ubiquitin aldehyde.
Note Added in Proof. Since acceptance of this paper, Morrison et al. (33) have reported that EBV latent membrane protein 2A activates β-catenin in epithelial cells through the PI3/AKT pathway.
References
- 1.Korinek, V., Barker, N., Morin, P. J., van Wichen, D., de Weger, R., Kinzler, K. W., Vogelstein, B. & Clevers, H. (1997) Science 275, 1784–1787. [DOI] [PubMed] [Google Scholar]
- 2.Polakis, P. (2000) Genes Dev. 14, 1837–1851. [PubMed] [Google Scholar]
- 3.Barker, N. & Clevers, H. (2000) BioEssays 22, 961–965. [DOI] [PubMed] [Google Scholar]
- 4.Pagano, J. S. (1999) Proc. Assoc. Am. Physicians 111, 573–580. [DOI] [PubMed] [Google Scholar]
- 5.Katz, B. Z., Raab-Traub, N. & Miller, G. (1989) J. Infect. Dis. 160, 589–598. [DOI] [PubMed] [Google Scholar]
- 6.Kieff, E. & Rickinson, A. (2001) Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott, Philadelphia), 4th Ed., pp. 2511–2573.
- 7.Kaiser, C., Laux, G., Eick, D., Jochner, N., Bornkamm, G. W. & Kempkes, B. (1999) J. Virol. 73, 4481–4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He, T. C., Sparks, A. B., Rago, C., Hermeking, H., Zawel, L., da Costa, L. T., Morin, P. J., Vogelstein, B. & Kinzler, K. W. (1998) Science 281, 1509–1512. [DOI] [PubMed] [Google Scholar]
- 9.Tetsu, O. & McCormick, F. (1999) Nature 398, 422–426. [DOI] [PubMed] [Google Scholar]
- 10.Morin, P. J. (1999) BioEssays 21, 1021–1030. [DOI] [PubMed] [Google Scholar]
- 11.Reya, T., O'Riordan, M., Okamura, R., Devaney, E., Willert, K., Nusse, R. & Grosschedl, R. (2000) Immunity 13, 15–24. [DOI] [PubMed] [Google Scholar]
- 12.Reya, T., Duncan, A. W., Ailles, L., Domen, J., Scherer, D. C., Willert, K., Hintz, L., Nusse, R. & Weissman, I. L. (2003) Nature 423, 409–414. [DOI] [PubMed] [Google Scholar]
- 13.Hershko, A. & Ciechanover, A. (1998) Annu. Rev. Biochem. 67, 425–479. [DOI] [PubMed] [Google Scholar]
- 14.Kim, J. H., Park, K. C., Chung, S. S., Bang, O. & Chung, C. H. (2003) J. Biochem. 134, 9–18. [DOI] [PubMed] [Google Scholar]
- 15.Wilkinson, K. D. (2000) Semin. Cell. Dev. Biol. 11, 141–148. [DOI] [PubMed] [Google Scholar]
- 16.Chung, C. H. & Baek, S. H. (1999) Biochem. Biophys. Res. Commun. 266, 633–640. [DOI] [PubMed] [Google Scholar]
- 17.Li, M., Chen, D., Shiloh, A., Luo, J., Nikolaev, A. Y., Qin, J. & Gu, W. (2002) Nature 416, 648–653. [DOI] [PubMed] [Google Scholar]
- 18.Bignell, G. R., Warren, W., Seal, S., Takahashi, M., Rapley, E., Barfoot, R., Green, H., Brown, C., Biggs, P. J., Lakhani, S. R., et al. (2000) Nat. Genet. 25, 160–165. [DOI] [PubMed] [Google Scholar]
- 19.Kovalenko, A., Chable-Bessia, C., Cantarella, G., Israel, A., Wallach, D. & Courtois, G. (2003) Nature 424, 801–805. [DOI] [PubMed] [Google Scholar]
- 20.Trompouki, E., Hatzivassiliou, E., Tsichritzis, T., Farmer, H., Ashworth, A. & Mosialos, G. (2003) Nature 424, 793–796. [DOI] [PubMed] [Google Scholar]
- 21.Borodovsky, A., Ovaa, H., Kolli, N., Gan-Erdene, T., Wilkinson, K. D., Ploegh, H. L. & Kessler, B. M. (2002) Chem. Biol. 9, 1149–1159. [DOI] [PubMed] [Google Scholar]
- 22.Nonkwelo, C., Skinner, J., Bell, A., Rickinson, A. & Sample, J. (1996) J. Virol. 70, 623–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujimuro, M., Wu, F. Y., ApRhys, C., Kajumbula, H., Young, D. B., Hayward, G. S. & Hayward, S. D. (2003) Nat. Med. 9, 300–306. [DOI] [PubMed] [Google Scholar]
- 24.Danilkovitch-Miagkova, A., Miagkov, A., Skeel, A., Nakaigawa, N., Zbar, B. & Leonard, E. J. (2001) Mol. Cell. Biol. 21, 5857–5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taya, S., Yamamoto, T., Kanai-Azuma, M., Wood, S. A. & Kaibuchi, K. (1999) Genes Cells 4, 757–767. [DOI] [PubMed] [Google Scholar]
- 26.Shaeffer, J. R. & Cohen, R. E. (1996) Biochemistry 35, 10886–10893. [DOI] [PubMed] [Google Scholar]
- 27.Chan, E. F., Gat, U., McNiff, J. M. & Fuchs, E. (1999) Nat. Genet. 21, 410–413. [DOI] [PubMed] [Google Scholar]
- 28.Oving, I. M. & Clevers, H. C. (2002) Eur. J. Clin. Invest. 32, 448–457. [DOI] [PubMed] [Google Scholar]
- 29.Clevers, H. (2000) Nat. Genet. 24, 206–208. [DOI] [PubMed] [Google Scholar]
- 30.Baeza, N., Masuoka, J., Kleihues, P. & Ohgaki, H. (2003) Oncogene 22, 632–636. [DOI] [PubMed] [Google Scholar]
- 31.Peifer, M. & Polakis, P. (2000) Science 287, 1606–1609. [DOI] [PubMed] [Google Scholar]
- 32.Nusse, R. & Varmus, H. E. (1992) Cell 69, 1073–1087. [DOI] [PubMed] [Google Scholar]
- 33.Morrison, J. A., Klingelhutz, A. J. & Raab-Traub, N. (2003) J. Virol. 77, 12276–12284. [DOI] [PMC free article] [PubMed] [Google Scholar]