Abstract
The Dicistroviridae is a growing virus family characterized by a dicistronic genome, wherein each open reading frame (ORF) is translated from an independent internal ribosome entry site (IRES). The 5′ IRES that translates the first open reading frame (ORF1) is similar to the picornaviral IRESs. However the second IRES, referred to as the intergenic region (IGR) IRES, - translates ORF2 by and uses an unusual mechanism of initiating protein synthesis. It folds into a compact RNA structure that can bind directly to 40S ribosomal subunits and form 80S complexes to initiate translation in the absence of any initiation factors. Despite its unusual mechanism, the IGR IRES has proven to be an elegant model for elucidating initiation mechanisms employed by IRESs, as well as making it a powerful research tool with diverse applications.
Keywords: IRES, Dicistroviridae, Translation initiation, Ribosome, IGR IRES, Hepatitis C virus, RPS25
Introduction
The Dicistroviridae is an emerging family of positive sense RNA viruses of the order Picornavirales. They were originally referred to as either the picorna-like viruses or cricket paralysis virus-like viruses (Mayo, 2002). The 14 classified members of the Dicistroviridae family infect a diverse group of insects, except for the Taura syndrome virus (TSV), which infects shrimp from six orders of arthropoda (for a detailed review see Bonning and Miller, 2010). Natural infections of Dicistroviridae occur through the oral–fecal route and are usually asymptomatic or causes intestinal illness (Lautie-Harivel, 1992). However, infection can lead to paralysis or death. Several members of this virus family are pathogenic to commercially important organisms. For example, TSV has a large impact in the shrimp industry (Lightner and Redman, 1998), and the Israeli acute paralysis virus (IAPV) was correlated to colony collapse disorder (CCD) of honey bees (Cox-Foster et al., 2007), a major agricultural and global environmental concern. On the bright side, other Dicistroviridae members infect insect pests and have been evaluated as biological pesticides. For example, the Triatoma virus (TrV) infects the insect vector of the Trypanosoma cruzi parasite, which causes Chagas disease, leading to life threatening chronic heart or digestive disorders (Czibener et al., 2005). The Homalodisca coagulata virus-1 (HoCV-1), Rhopalosiphum padi virus (RhPV), and aphid lethal paralysis virus (ALPV) infect agricultural pests (Bonning and Miller, 2010). Metagenomic studies suggest that Dicistroviridae-like viruses are abundant and that the full scope and host range of this virus family remains undetermined. Understanding dicistroviruses’ mechanism of replication is important for developing strategies to combat infection of beneficial host organisms (like honeybees) and, conversely, to take advantage of the beneficial viruses as potential biological pesticides.
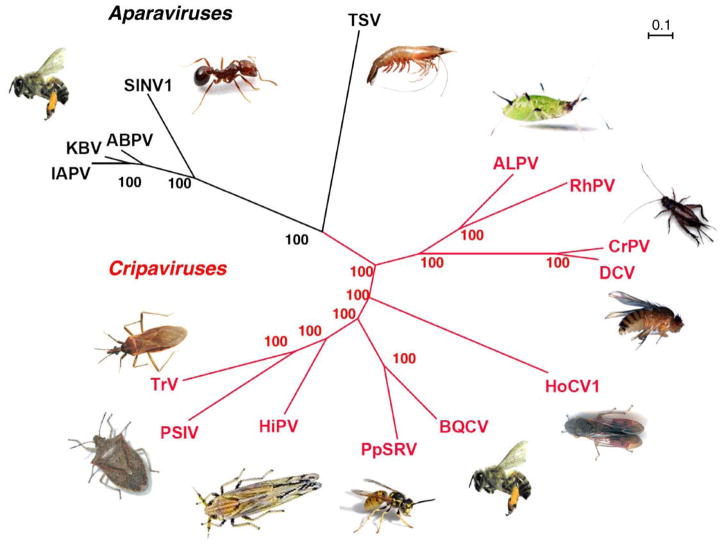
The Dicistroviridae family is characterized by an unusual bicistronic genome, wherein each ORF is translated by an independent IRES. The family is further divided into the genus Cripavirus (Fig. 1, red) and the proposed genus Aparavirus (Fig. 1, black). Phylogenetic analysis of the structural polyprotein (ORF2) (Fig. 1A) or IGR IRES that drives its translation (not shown) both divide the virus family cleanly into these two genera. However, the phylogenic tree for the 5′ non-structural polyprotein (ORF1) (data not shown) places TSV in Cripavirus instead of in Aparavirus. This is a possible evidence of recombination or different divergence rates between the non-structural proteins and the structural proteins. However, as TSV is the only virus that infects crustaceans and is very divergent from the insect infecting viruses it may be the first example of a new genus within this family. The future characterization of additional Dicistroviridae that infects marine organism hosts will establish whether or not TSV is an Aparavirus or a member of a novel genus in this family.
Fig. 1.
Phylogenetic tree for Dicistroviridae. The phylogenetic tree was derived from the protein sequence of the structural genes. The genera Cripavirus (red) and the Aparavirus (black) are shown with the host next to the virus. The predicted amino acid sequences were aligned using ClustalW2 (Chenna et al., 2003). Trees were predicted using MrBayes (Huelsenbeck and Ronquist, 2001) and formatted with Dendroscope (Huson et al., 2007). Bootstrap values are indicated at the nodes. The full virus names and GenBank accession numbers are as follows: IAPV = Israeli acute paralysis virus (NC009025), KBV = Kashmir bee virus (NC004807), ABPV = acute bee paralysis virus (AF150629), SINV-1 = Solenopsis invicta virus-1 (AY634314), TSV = Taura syndrome virus (AF277675), PSIV = Plautia stali intestine virus (AB006531), TrV = Triatoma virus (AF178440), HiPV = Himetobi P virus (AB017037), BQCV = black queen cell virus (AF183905), PpSRV = Pteromalus puparum small RNA virus (EU680971.1), HoCV-1 = Homalodisca coagulata virus-1 (NC008029), ALPV = aphid lethal paralysis virus (AF022937), RhPV = Rhopalosiphum padi virus (AF022937), CrPV = cricket paralysis virus (AF218039), and DCV = Drosophila C virus (AF014388).
An active searching for viruses has revealed four members that infect bees and a recently proposed member, Pteromalus puparum small RNA virus (PpSRV), that infects wasps (Zhu et al., 2008). The wide diversity in bees suggests that these viruses may be found in more insect genera. Indeed, metagenomic sampling has found many Dicistroviridae-like viruses in diverse environments. The bat guano virome contained viruses that share a high degree of homology with the acute bee paralysis virus and Kashmir bee virus (Li et al., 2010). A study of stool from children with acute flaccid paralysis identified a virus, Ervivirus, which shares 35% amino acid similarity with Dicistroviridae (Victoria et al., 2009). The similarity between Ervivirus and known Dicistroviridae is comparable with current diversity within the virus family. Due to their relatively error prone polymerase, the ALPV and CrPV, DCV or RhPV has 28 to 38% similarity between their coding regions (Van Munster et al., 2002). It is unlikely that these viruses isolated from bats or humans are pathogenic in those hosts, as there are no known cases of Dicistroviridae infecting vertebrates and efforts thus far to propagate these viruses in mammalian cell lines have failed (Pantoja et al., 2004). It is more likely that they were exposed to the viruses through their diet.
Culley et al. (2006) examined the viral diversity in seawater and discovered a large number of positive-stranded RNA viruses, which challenged the dogma that the majority of oceanic viruses were DNA bacteriophages. They were able to reconstruct the complete genome of two Dicistroviridae-like viruses named JP-A and JP-B (Culley et al., 2007). A separate study found freshwater lake sequences that represented all 14 classified Dicistroviridae (Djikeng et al., 2009).
Due to the high mutation rate and diversity of this family, efforts to discover new Dicistroviridae using degenerate primers would present a challenge and would bias results towards discovering only viruses similar to the known Dicistroviridae. The most impartial approach, although not the most direct, would be through deep sequencing of environmental samples (Djikeng and Spiro, 2009).
Genome organization
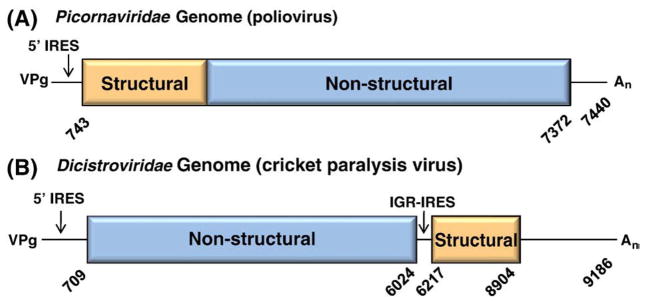
Dicistroviridae are distinguished from the other virus families in the order Picornavirales by a unique genomic structure. Picornaviruses have one long open reading frame (ORF) with the structural genes upstream of the non-structural genes (Fig. 2A). The Dicistroviridae genome is similar in length, averaging around 8–10 kb, but has two ORFs, which gave rise to its name (Fig. 2B). The first ORF encodes the non-structural genes: helicase, protease, VPg, and RNA-dependent RNA polymerase. The second ORF encodes the structural genes, VP1–4. Each ORF is translated by independent IRESs, with ORF2 being produced in super molar excess over ORF1 (Wilson et al., 2000a, 2000b). This independent regulation allows the virus to produce enzymes early in infection and then mass-produce the structural proteins for virions late in infection.
Fig. 2.
Genomic organization of (A) Picornaviridae and (B) Dicistroviridae genomes drawn to scale. Numbering is based on the nucleotide sequence from the reference sequences, PV (V01149.1) and CrPV (NC003924). The structural genes (orange) and the non-structural genes (blue) comprise one ORF in picornaviruses but are two independently translated ORFs in dicistroviruses. Both genomes have a VPg protein at the 5′ end and a 3′ poly-A tail (An).
The IGR IRES, which controls the expression of the structural genes, is approximately 200 nucleotides, folds into a compact structure that is largely conserved throughout the virus family. In contrast, the 5′ IRES is highly structured but forms an extended RNA element bearing more resemblance to picornaviral IRESs (Czibener et al., 2005; Shibuya and Nakashima, 2006; Wilson et al., 2000b; Woolaway et al., 2001) and shows very low sequence and structural homology across Dicistroviridae. In fact, each 5′ IRES has distinct cell and translational lysate preferences. For example, the RhPV 5′ IRES requires several canonical initiation factors for activity (Groppelli et al., 2007) and is functional in both rabbit reticulocyte lysate and wheat germ extract while the 5′ IRES from PSIV is unable to initiate translation in these cell-free translation systems (Shibuya and Nakashima, 2006). Overall this suggests that the 5′ IRESs from Dicistroviridae are not a closely related IRES family.
Additional features of the Dicistroviridae genome include a hypothetical overlapping gene, pog (predicted overlapping gene) or ORFX located in the +1 reading frame within ORF2 of the bee Aparaviruses. However, a gene product has not been identified (Sabath et al., 2009; Firth et al., 2009). There is also a predicted stem–loop at the 3′ end of ORF1 upstream of the IGR IRES in some members that is proposed to aid IGR IRES activity (Firth et al., 2009).
Mechanism of translation initiation by the IGR IRES
mRNAs are translated by either cap-dependent or IRES-dependent (cap-independent) mechanisms. The majority of mRNAs are translated through a cap-dependent mechanism that requires 10 to 13 translation initiation factors working together to recruit the 40S ribosomal subunit to the cap structure at the 5′ end of the mRNA. The 5′ cap is recognized by eIF4E, in a complex with eIF4G and eIF4A. Then, a 43S pre-initiation complex (PIC) is recruited to the 5′ end of the mRNA, which contains: eIF3, the ternary complex ( ), eIF1, eIF1A, eIF5, and a 40S ribosomal subunit. Once the ribosome binds to the mRNA, it scans down the mRNA in a 5′ to 3′ direction for an AUG start codon and positions the AUG in the P-site (peptidyl site) of the ribosome. When the is correctly positioned into the P-site of the ribosome, the eIFs are released as the 60S subunit joins in a reaction facilitated by eIF5B and GTP. Then translation elongation ensues. However, under adverse conditions, the cell shuts down cap-dependent translation and relies on IRES-dependent translation to synthesize proteins required to cope with the stress. Some viruses also shut down cap-dependent translation in order to usurp host ribosomes for the translation of viral proteins via the IRES-mediated mechanism. IRES-mediated translation requires an IRES sequence, which is generally located in the 5′ untranslated region (UTR) of the mRNA, is highly structured, and is able to directly recruit the 40S subunit internally to the message. An IRES is defined by a functional assay performed either by using a dicistronic reporter or a circular mRNA to demonstrate that the ribosome is recruited internally to the mRNA independent of the 5′ end. While the mechanism for this recruitment is not well understood, our best insights have come from studies of viral IRESs, and particularly of the Dicistroviridae IGR IRESs.
IGR IRES-mediated initiation
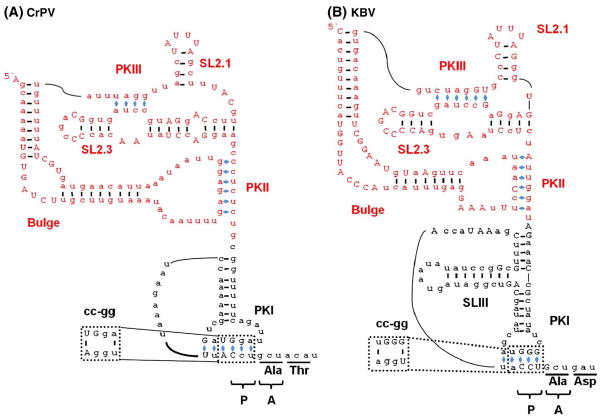
During canonical translation initiation, the ribosomal peptidyl-site (P-site) is occupied by an AUG codon base-paired to the anticodon of the initiator . The IGR IRES deviates from this by initiating protein synthesis from the acceptor site (A-site) of the ribosome and therefore requires neither the initiator nor an AUG codon (Kamoshita et al., 2009; Sasaki and Nakashima, 1999; Wilson et al., 2000a). Instead, the P-site is occupied by a CCU (proline) codon that base-pairs to IRES sequence located upstream, forming a pseudoknot structure (Wilson et al., 2000a). Thus, pseudoknot I (PKI; see Fig. 3) of the IGR IRES occupies the P-site and structurally mimics a tRNA anticodon–codon interaction (Costantino et al., 2008; Kieft, 2009). The disruption of PKI destroys IRES activity (<1%); however, making the compensatory mutations to restore base-pairing restores IGR IRES activity to 50% of wild type (Fig. 3) (Wilson et al., 2000a). This finding indicates that the important interaction is the pseudoknot structure and not the codon sequences. In fact, a stop codon can be substituted for the CCU codon and the IRES remains active, as long as the compensatory mutation is made to maintain base pairing (Sasaki and Nakashima, 1999; Wilson et al., 2000a).
Fig. 3.
Secondary structure for the Class I and II IGR IRESs (A) Class I CrPV IGR IRES. (B) Class II KBV IGR IRES. The RNA binding domain (red) and PKI (black) are indicated. Bases in uppercase denote conserved sequences within the IGR IRES subclass. Base pairing of PKs are indicated with blue diamonds (◆). The codon triplet that occupies the P- and A-sites of the ribosome before elongation is indicated by P and A respectively. Disruption of PKI by a two nucleotide mutation (cc to gg; shown in the dotted-lines) inhibits PKI formation leading to a complete loss of IRES activity.
Translation initiation by the IGR IRES is unique because the first codon translated is in the A-site of the ribosome. Upon the addition of an aminoacyl-tRNA (aa-tRNA) to the A-site by elongation factor 1A (eEF1A) the ribosome moves one codon in the 3′ direction and protein synthesis ensues (Wilson et al., 2000a). In canonical initiation, the movement of the peptidyl-tRNA/mRNA from the A-site to the P-site is a process referred to as translocation. Generally, translocation requires GTP hydrolysis, elongation factor 2 (eEF2) and peptide bond formation. However, since the PKI of the IGR IRES occupies the P-site, no GTP hydrolysis occurs, and no peptide bond is formed. Thus, this movement is referred to as pseudotranslocation. Therefore, the initiation of the IGR IRES is more reminiscent of elongation rather than initiation.
Interestingly, a recent study has shown that under certain conditions the IGR IRES can initiate translation from the P-site of the ribosome. If PKI is disrupted or melted this may provide a condition in which the P-site codon can be used to initiate translation if there is either an AUG codon in the P-site or a suppressor Met–tRNAi that recognizes the P-site codon (Kamoshita et al., 2009). None of the known dicistroviruses contains an AUG codon that would be positioned in the P-site, thus, making it unlikely that this mechanism of initiation would be used during viral infection for the known IGR IRESs (Nakashima and Uchiumi, 2009). However, this does suggest that initiation from the IGR IRES can occur from the P-site or the A-site once the RNA binding domain has bound to the ribosome. It is notable that the IGR IRESs, IAPV, ABPV, SINV-1, and TSV, contain upstream in-frame AUG codons (Nakashima and Uchiumi, 2009). Perhaps if the initiation from the P-site can occur at any of these upstream AUGs then this may provide an alternate mechanism of initiation that may be evolutionarily advantageous since the PKI structure has been found to be less stable than the RNA binding domain of the IRES (Costantino et al., 2008). Further experiments will be required to determine if any of these upstream AUGs can be used for the P-site initiation and if they are eIF2-dependent.
Initiation factor free translation initiation of the IGR IRES
Biochemical and genetic experiments have demonstrated that the CrPV IGR IRES is capable of binding 40S subunits in the absence of any translation initiation factors, GTP, and (Deniz et al., 2009; Jan and Sarnow, 2002; Pestova and Hellen, 2003; Wilson et al., 2000a). Upon addition of purified 60S ribosomal subunits to the IGR IRES/40S complex, 80S complexes are formed (Jan and Sarnow, 2002; Pestova and Hellen, 2003). Subsequent studies using mutant or deleted initiation factors demonstrated that the IGR IRES does not require initiation factors in vivo (Deniz et al., 2009). The binding of the IGR IRES to the 40S subunit results in a conformational change in the 40S (Schuler et al., 2006; Spahn et al., 2004) that resembles the change observed when the initiation factors eIF1 and eIF1A bind to the 40S resulting in an opening of the mRNA binding channel to allow the mRNA to bind (Passmore et al., 2007). This conformational change upon the IGR IRES binding to the ribosome demonstrates that the IRES is capable of manipulating the ribosome and suggests that the IGR IRES functions as an RNA-based translation factor (Kieft, 2008).
Dicistroviridae modulation of host translation machinery
Dicistroviruses are positive stranded RNA viruses, thus the genome is used as an mRNA template for translation. The virus subverts the host’s translation by shutting down the cap-dependent translation and utilizing IRES-mediated translation to synthesize its proteins (Gebauer and Hentze, 2004; Thompson and Sarnow, 2000). Picorna-viruses force this switch by cleaving the host translation initiation factors with the virally encoded 2A and 3C proteases (Kuyumcu-Martinez et al., 2004; Lin et al., 2009; Perera et al., 2007; Ventoso et al., 1998). The dicistroviral non-structural genes, encoded by ORF1 are translated first, while IGR IRES-mediated translation peaks later in infection and produces high levels of the capsid proteins needed to assemble progeny (Garrey et al., 2010). The mechanism for Dicistroviridae inhibition of host protein synthesis is not understood. Nonetheless, the translation is shutdown and viral proteins are the predominate proteins synthesized during viral infection (Garrey et al., 2010). One cellular response to combat viral infection is the phosphorylation of the alpha subunit of eIF2 (eIF2α), which shuts down the recycling of the initiator methionine tRNA and thus rapidly shuts down the majority of the translation in the cell (for review see Gebauer and Hentze, 2004). However, the IGR IRES is enhanced when eIF2α is phosphorylated (Deniz et al., 2009; Thompson et al., 2001) since it does not use the canonical AUG initiation codon. Therefore, one would predict that its activity during infection would be stimulated by, or even dependent upon, the phosphorylation eIF2α. However, a time course experiment showed that translation by the IGR IRES peaks prior to the phosphorylation of eIF2α, indicating that the IRES does not depend on the phosphorylation of eIF2α (Garrey et al., 2010).
These viruses employ several methods to disable cellular antiviral responses. CrPV specifically shuts down the host translation by inhibiting the interaction of eIF4E, the cap binding protein, and eIF4G, the major scaffolding protein within the eIF4F mRNA binding complex (Garrey et al., 2010). The shutdown of the host protein synthesis is dependent on active infection, not just virus uptake, suggesting that the shutdown is caused by specific events during an infection and not just a general host stress response (Garrey et al., 2010). CrPV or DCV infection of the Drosophila S2 cell line down-regulates the formation of stress granules, which would normally sequester the cellular machinery required for the viral translation and replication, thus allowing the virus to effectively replicate (Khong and Jan, 2010).
Structural model of ribosome binding by the IGR IRES
A structure of the IGR IRES bound to human ribosomes resolved to 17.3 Å was determined using cryo-EM (Spahn et al., 2004). A higher resolution structure (7.3 Å) was later achieved with yeast ribosomes (Schuler et al., 2006). The IGR IRES occupies the inter-subunit region of the 80S ribosome, predominately binding to the E-site with PKI residing in the decoding center (Schuler et al., 2006; Spahn et al., 2004). The crystal structure of the IGR IRES has been solved independently for both the RNA binding domain and the PKI domain, then reconstructed to fit into the Cryo-EM density to generate a complete structural model of the IGR IRES (Costantino and Kieft, 2005; Costantino et al., 2008). The crystal structure the IGR IRES confirms its tight packing with the predicted PK structures Pfingsten (et al., 2006). The RNA binding domain (Fig. 3; red) of the IGR IRES forms a compact core that is sufficient for binding 40S subunits (Costantino and Kieft, 2005; Nishiyama et al., 2003). PKI folds (Fig. 3; blue) independently of the RNA binding domain (Pfingsten et al., 2007) and is positioned in the P-site of the ribosome, mimicking an initiator tRNA–mRNA interaction (Pfingsten et al., 2007; Costantino and Kieft, 2005; Costantino et al., 2008; Jan and Sarnow, 2002; Kanamori and Nakashima, 2001; Nishiyama et al., 2003; Pestova and Hellen, 2003; Pestova et al., 2004; Wilson et al., 2000a). While PKI is essential for translation initiation, it does not increase the affinity of the RNA binding domain for the 40S subunit (Costantino and Kieft, 2005; Nishiyama et al., 2003; Pfingsten et al., 2006).
The structural data support a model in which the 40S ribosomal subunit interacts with the stem loop (SL) 2.3 (Fig. 3), which bends upwards and is positioned close to SL2.1 within the E-site of the 40S subunit (Costantino and Kieft, 2005; Schuler et al., 2006; Spahn et al., 2004). Based on the proximity in the structural model, SL2.1 has been predicted to interact with the 40S ribosomal protein S5 (Rps5) and SL2.3 with an unknown 40S ribosomal protein (Pfingsten et al., 2006; Schuler et al., 2006; Spahn et al., 2004). However, cross-linking experiments using 4-thiouracil (a zero distance cross-linker) identified ribosomal protein S25 (Rps25p) as the ribosomal protein that interacts with SL2.1, and no cross-linking to Rps5p was detected (Nishiyama et al., 2007). Rps25p is next to Rps5p (Uchiumi et al., 1981). A recent high resolution cryo-EM structure of the eukaryotic ribosome has placed Rps25p in the E-site of the 40S ribosome next to Rps5p (Armache et al., 2010), which is consistent with Rps25p interacting with either SL2.1 or SL2.3. The structural data also predict that the bulge region (Fig. 3) interacts with the 60S ribosomal subunit, specifically with the ribosomal protein L1 (Rpl1) and Helix 76 and Helix 77. PKII is predicted to interact with ribosomal protein L11 (Rpl11) in the 60S subunit (Pfingsten et al., 2006; Schuler et al., 2006; Spahn et al., 2004). These ribosomal sites are part of the universally conserved tRNA binding sites. Structural models also predict that PKI interacts with Helix 34 and Helix 18 of the 60S subunit, which interact with the anticodon stem–loop of the A-site tRNA (Pfingsten et al., 2006; Schuler et al., 2006; Spahn et al., 2004). Taken together, these data suggest that the IGR IRES interacts with highly conserved regions of the ribosome, which may explain why it functions in yeast unlike other IRESs from higher eukaryotes (Deniz et al., 2009; Thompson et al., 2001).
The role of RPS25 in IGR IRES-mediated translation
While the mechanism of direct internal recruitment of ribosomes by IRESs to a mRNA is not understood, the fact that the IGR IRES functions in both yeast and mammals has allowed for both biochemical and genetic studies of the IGR IRES that have identified Rps25p as a key interaction partner that is critical for IGR IRES activity (Landry et al., 2009). In the absence of the non-essential ribosomal protein Rps25p, the IGR IRES does not bind to 40S ribosomal subunits (Landry et al., 2009). Structural data place both the IGR IRES ribosome binding domain (Schuler et al., 2006; Spahn et al., 2004) and Rps25p (Armache et al., 2010) in the E-site of the 40S ribosome. Taken together these data suggest an essential role for Rps25p in the IGR IRES binding that is consistent with all previous biochemical and structural data (Doring et al., 1994; Landry et al., 2009; Marion and Marion, 1988; Nishiyama et al., 2007; Schuler et al., 2006; Uchiumi et al., 1981; Wower et al., 1993; Yusupov et al., 2001). While RPS25 is nonessential, there is a slight (19%) decrease in protein synthesis rates when it is absent (Landry et al., 2009). While cap-dependent initiation is unaffected (Landry et al., 2009), the decrease in the rate of protein synthesis may be explained by a decrease in elongation, since the elongation factor 3 (eEF3) has been shown to interact with Rps25p in the E-site (Andersen et al., 2006; Armache et al., 2010). These studies on the IGR IRES have revealed differences in IRES-dependent versus cap-dependent translation that could be exploited to preferentially inhibit viral translation.
Significantly, the hepatitis C viral (HCV) IRES also requires Rps25p for activity (Landry et al., 2009). In contrast to the IGR IRES, the HCV IRES binds directly to the solvent side of the 40S ribosomal subunit and only contacts the E-site, where Rps25p is located, with a finger like structure (domain II) (Boehringer et al., 2005; Spahn et al., 2001). While the HCV IRES requires Rps25p for IRES activity, it is unlikely that the HCV IRES requires it to bind to the 40S subunit since the deletion of domain II does not disrupt HCV binding to the 40S ribosome (Kieft et al., 2001; Spahn et al., 2001). However, since both the HCV and IGR IRESs induce a similar conformational change in the 40S subunit upon binding, which is necessary for 60S joining (Schuler et al., 2006; Spahn et al., 2004, 2001), Rps25p could be required for the conformational change. Consistent with this model, Rps25p has an extension that reaches into the decoding and tRNA binding sites of the ribosome, thus bridging the E-site to the decoding center (Armache et al., 2010) and, when the HCV IRES domain II is deleted this conformational change is no longer observed (Spahn et al., 2001). Future experiments should address whether Rps25p is required for the IRES-induced 40S ribosomal subunit conformational change.
The fact that the HCV and the IGR IRESs bind to the 40S ribosomal subunit so differently, yet both require Rps25p for IRES activity strongly suggests that they share a common mechanism for IRES-mediated translation that is downstream of ribosome binding, such as the 40S conformational change. It is worth noting that an identical AGCC sequence is found both at the apical loop of domain II of the HCV IRES and in the loop region of SL2.3 of the IGR IRES. It has not been determined whether SL2.3 or SL2.1 of the IGR IRES, which are both found in the E-site, are interacting with Rps25p. Nonetheless, it is worth noting that the sequence AGCC is conserved in the apical loops of other viral IRESs (Honda et al., 1999). Future experiments should determine whether the AGCC motif interacts with Rps25p, which would demonstrate a similar mechanism of initiation, shared across structurally diverse IRESs.
The IGR IRES as a tool to understand translation and cellular stress
Thus far we have focused on how the IGR IRESs serve as an important tool to better understand the mechanism of IRES-dependent translation initiation. It serves as a powerful model IRES because the information that has been obtained from this simplified translation initiator has been successfully applied to other, more complex IRESs like the HCV IRES (Landry et al., 2009). Studies to determine the similarities between the IGR IRESs and cellular IRESs are also being pursued. One notable similarity between cellular IRES mechanisms and the IGR IRES was revealed in the studies on ribosomal RNA modifications. The human genetic disease, X-linked dyskeratosis, is caused by mutations in the DKC1 gene, a pseudouridine synthase that catalyzes the conversion of uridines to pseudouridines in the rRNA and small nucleolar RNAs (Ni et al., 1997). The study demonstrated that while no defect was observed in the global or cap-dependent translation in cells from X-DC patients, the translation of cellular IRESs and the CrPV IGR IRES were reduced to 50% of the level observed for wild-type cells (Yoon et al., 2006). This was the first reported evidence that the CrPV IGR IRES shares a common mechanism with cellular IRES-mediated translation.
The IGR IRES has also proven to be a powerful tool in the study of other aspects of translation, well beyond IRES-dependent initiation. Translation carried out in vitro using only purified factors allows for a very controlled system to study the different stages of translation. Unfortunately, it is currently a very laborious procedure requiring the purification of at least 9 translation initiation factors on top of the elongation and termination factors. However, cloning the CrPV IGR IRES into the 5′UTR of an expression construct circumvents the need to purify the initiation factors (Pestova and Hellen, 2005). This is a promising strategy for in vitro translation assays to study elongation or termination. The current drawback to this system is that the CrPV IGR IRES is much less efficient than the cap-dependent translation and the translation efficiency may not be high enough for some assays. This could be addressed by identifying or developing more active IGR IRESs.
The initiation factor-independent nature of the IGR IRES translation can be used to determine if a cellular process is initiation factor dependent. Petersen et al. used the IGR IRES to determine that miRNA inhibition of translation occurs post initiation (Petersen et al., 2006), since equivalent miRNA-mediated down-regulation was observed regardless of whether the translation initiation mechanism employed was cap-dependent, CrPV IGR IRES or HCV IRES driven. A similar strategy was used to determine that initiation factors were not required for recoding of a UGA codon into a selenocysteine (Sec). Equivalent incorporation of Sec was shown in both cap-dependent and CrPV IGR IRES driven translational reporters (Donovan and Copeland, 2010).
An IGR IRES was similarly employed to determine that nonsense mediated decay (NMD) requires translation initiation by the canonical pathway. NMD is a process that rids cells of mRNAs containing premature stop codons. In mammals, this process is thought to rely on the recognition of the exon junction complexes (EJCs) which are normally deposited on mRNAs during splicing in the nucleus and removed during the pioneer round of translation (Isken and Maquat, 2007). If any EJCs remain on a message after termination, this signals to the NMD machinery that the termination was premature. It had been suggested that the eukaryotic initiation factor 3 (eIF3) was necessary for EJC recognition by the NMD machinery (Chiu et al., 2004). When a reporter with a premature stop codon was put under the translational regulation of the CrPV IGR IRES it was not recognized as an NMD substrate. However, reporters initiating with the encephalomyocarditis (EMCV) IRES, which does require eIF3, were recognized as a substrate for NMD. This supports the model that eIF3-dependent initiation is a prerequisite to NMD decay (Isken et al., 2008). It would be interesting to see if an HCV IRES driven NMD substrate message would also serve as a target for NMD, since the HCV IRES only requires eIF3 and eIF2 (Otto and Puglisi, 2004).
The EMCV IRES is often used in mammalian expression vectors. This allows for expression of a reporter gene and the gene of interest from a single transcript. Since IGR IRESs, unlike EMCV, can function when ternary complexes are low, the production of the protein will be maintained throughout the cell cycle and may even be up-regulated during stress. This would be advantageous to the study of proteins active during down-regulation of cap-dependent translation. The use of IGR IRESs in the expression vectors would allow for efficient translation of the protein of interest during stress. Future advances that increase the translation efficiency by IGR IRESs will improve both our understanding of IRES-mediated translation and increase the utility of the IRES as a tool.
Acknowledgments
We are grateful to Elliot J. Lefkowitz for help in generating the phylogenetic tree. We would like to thank Robert Curtis Hendrickson for critical reading of this manuscript. The research in the Thompson laboratory is supported by grants from the National Institutes of Health (R01GM084547; 3R01GM084547-01A1S1) and the National Cancer institute (CA13148-35) UAB Comprehensive Cancer Center Collaborative Programmatic Development Grant program.
References
- Andersen CBF, Becker T, Blau M, Anand M, Halic M, Balar B, Mielke T, Boesen T, Pedersen JS, Spahn CM, Kinzy TG, Andersen GR, Beckmann R. Structure of eEF3 and the mechanism of transfer RNA release from the E-site. Nature. 2006;443 (7112):663–668. doi: 10.1038/nature05126. [DOI] [PubMed] [Google Scholar]
- Armache JP, Jarasch A, Anger AM, Villa E, Becker T, Bhushan S, Jossinet F, Habeck M, Dindar G, Franckenberg S, Marquez V, Mielke T, Thomm M, Berninghausen O, Beatrix B, Soding J, Westhof E, Wilson DN, Beckmann R. Localization of eukaryote-specific ribosomal proteins in a 5.5-A cryo-EM map of the 80S eukaryotic ribosome. Proc Natl Acad Sci USA. 2010;107 (46):19754–19759. doi: 10.1073/pnas.1010005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehringer D, Thermann R, Ostareck-Lederer A, Lewis JD, Stark H. Structure of the hepatitis C virus IRES bound to the human 80S ribosome: remodeling of the HCV IRES. Structure. 2005;13 (11):1695–1706. doi: 10.1016/j.str.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Bonning BC, Miller WA. Dicistroviruses. Annu Rev Entomol. 2010;55:129–150. doi: 10.1146/annurev-ento-112408-085457. [DOI] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31 (13):3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu SY, Lejeune F, Ranganathan AC, Maquat LE. The pioneer translation initiation complex is functionally distinct from but structurally overlaps with the steady-state translation initiation complex. Genes Dev. 2004;18 (7):745–754. doi: 10.1101/gad.1170204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantino D, Kieft JS. A preformed compact ribosome-binding domain in the cricket paralysis-like virus IRES RNAs. RNA. 2005;11 (3):332–343. doi: 10.1261/rna.7184705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantino DA, Pfingsten JS, Rambo RP, Kieft JS. tRNA–mRNA mimicry drives translation initiation from a viral IRES. Nat Struct Mol Biol. 2008;15 (1):57–64. doi: 10.1038/nsmb1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox-Foster DL, Conlan S, Holmes EC, Palacios G, Evans JD, Moran NA, Quan PL, Briese T, Hornig M, Geiser DM, Martinson V, vanEngelsdorp D, Kalkstein AL, Drysdale A, Hui J, Zhai J, Cui L, Hutchison SK, Simons JF, Egholm M, Pettis JS, Lipkin WI. A metagenomic survey of microbes in honey bee colony collapse disorder. Science. 2007;318 (5848):283–287. doi: 10.1126/science.1146498. [DOI] [PubMed] [Google Scholar]
- Culley AI, Lang AS, Suttle CA. Metagenomic analysis of coastal RNA virus communities. Science. 2006;312 (5781):1795–1798. doi: 10.1126/science.1127404. [DOI] [PubMed] [Google Scholar]
- Culley AI, Lang AS, Suttle CA. The complete genomes of three viruses assembled from shotgun libraries of marine RNA virus communities. Virol J. 2007;4:69. doi: 10.1186/1743-422X-4-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czibener C, Alvarez D, Scodeller E, Gamarnik AV. Characterization of internal ribosomal entry sites of Triatoma virus. J Gen Virol. 2005;86 (Pt 8):2275–2280. doi: 10.1099/vir.0.80842-0. [DOI] [PubMed] [Google Scholar]
- Deniz N, Lenarcic EM, Landry DM, Thompson SR. Translation initiation factors are not required for Dicistroviridae IRES function in vivo. RNA. 2009;15 (5):932–946. doi: 10.1261/rna.1315109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djikeng A, Spiro D. Advancing full length genome sequencing for human RNA viral pathogens. Future Virol. 2009;4 (1):47–53. doi: 10.2217/17460794.4.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djikeng A, Kuzmickas R, Anderson NG, Spiro DJ. Metagenomic analysis of RNA viruses in a fresh water lake. PLoS ONE. 2009;4 (9):e7264. doi: 10.1371/journal.pone.0007264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan J, Copeland PR. The efficiency of selenocysteine incorporation is regulated by translation initiation factors. J Mol Biol. 2010;400 (4):659–664. doi: 10.1016/j.jmb.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doring T, Mitchell P, Osswald M, Bochkariov D, Brimacombe R. The decoding region of 16S RNA; a cross-linking study of the ribosomal A, P and E sites using tRNA derivatized at position 32 in the anticodon loop. EMBO J. 1994;13 (11):2677–2685. doi: 10.1002/j.1460-2075.1994.tb06558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth AE, Wang QS, Jan E, Atkins JF. Bioinformatic evidence for a stem–loop structure 5′-adjacent to the IGR-IRES and for an overlapping gene in the bee paralysis dicistroviruses. Virol J. 2009;6:193. doi: 10.1186/1743-422X-6-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrey JL, Lee YY, Au HH, Bushell M, Jan E. Host and viral translational mechanisms during cricket paralysis virus infection. J Virol. 2010;84 (2):1124–1138. doi: 10.1128/JVI.02006-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5 (10):827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groppelli E, Belsham GJ, Roberts LO. Identification of minimal sequences of the Rhopalosiphum padi virus 5′ untranslated region required for internal initiation of protein synthesis in mammalian, plant and insect translation systems. J Gen Virol. 2007;88 (Pt 5):1583–1588. doi: 10.1099/vir.0.82682-0. [DOI] [PubMed] [Google Scholar]
- Honda M, Beard MR, Ping LH, Lemon SM. A phylogenetically conserved stem–loop structure at the 5′ border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J Virol. 1999;73 (2):1165–1174. doi: 10.1128/jvi.73.2.1165-1174.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17 (8):754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Huson DH, Richter DC, Rausch C, Dezulian T, Franz M, Rupp R. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinform. 2007;8:460. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21 (15):1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- Isken O, Kim YK, Hosoda N, Mayeur GL, Hershey JW, Maquat LE. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell. 2008;133 (2):314–327. doi: 10.1016/j.cell.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan E, Sarnow P. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J Mol Biol. 2002;324 (5):889–902. doi: 10.1016/s0022-2836(02)01099-9. [DOI] [PubMed] [Google Scholar]
- Kamoshita N, Nomoto A, Rajbhandary UL. Translation Initiation from the ribosomal A Site or the P site, dependent on the conformation of RNA pseudoknot I in dicistrovirus RNAs. Mol Cell. 2009;35 (2):181–190. doi: 10.1016/j.molcel.2009.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamori Y, Nakashima N. A tertiary structure model of the internal ribosome entry site (IRES) for methionine-independent initiation of translation. RNA. 2001;7 (2):266–274. doi: 10.1017/s1355838201001741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khong A, Jan E. Modulation of stress granules and P bodies during dicistrovirus infection. J Virol. 2010 doi: 10.1128/JVI.02220-10. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft JS. Viral IRES RNA structures and ribosome interactions. Trends Biochem Sci. 2008;33 (6):274–283. doi: 10.1016/j.tibs.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft JS. Comparing the three-dimensional structures of Dicistroviridae IGR IRES RNAs with other viral RNA structures. Virus Res. 2009;139 (2):148–156. doi: 10.1016/j.virusres.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft JS, Zhou K, Jubin R, Doudna JA. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA. 2001;7 (2):194–206. doi: 10.1017/s1355838201001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuyumcu-Martinez NM, Van Eden ME, Younan P, Lloyd RE. Cleavage of poly (A)-binding protein by poliovirus 3C protease inhibits host cell translation: a novel mechanism for host translation shutoff. Mol Cell Biol. 2004;24 (4):1779–1790. doi: 10.1128/MCB.24.4.1779-1790.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry DM, Hertz MI, Thompson SR. RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes Dev. 2009;23 (23):2753–2764. doi: 10.1101/gad.1832209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lautie-Harivel N. Drosophila C virus cycle during the development of two Drosophila melanogaster strains (Charolles and Champetieres) after larval contamination by food. Biol Cell. 1992;76 (2):151–157. doi: 10.1016/0248-4900(92)90207-h. [DOI] [PubMed] [Google Scholar]
- Li L, Victoria JG, Wang C, Jones M, Fellers GM, Kunz TH, Delwart E. Bat guano virome: predominance of dietary viruses from insects and plants plus novel mammalian viruses. J Virol. 2010;84 (14):6955–6965. doi: 10.1128/JVI.00501-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightner DV, Redman RM. Strategies for the control of viral diseases of shrimp in the Americas. Fish Pathol. 1998;33:165–180. [Google Scholar]
- Lin JY, Chen TC, Weng KF, Chang SC, Chen LL, Shih SR. Viral and host proteins involved in picornavirus life cycle. J Biomed Sci. 2009;16:103. doi: 10.1186/1423-0127-16-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion MJ, Marion C. Ribosomal proteins S2, S6, S10, S14, S15 and S25 are localized on the surface of mammalian 40S subunits and stabilize their conformation. A study with immobilized trypsin. FEBS Lett. 1988;232 (2):281–285. doi: 10.1016/0014-5793(88)80753-1. [DOI] [PubMed] [Google Scholar]
- Mayo MA. A summary of taxonomic changes recently approved by ICTV. Arch Virol. 2002;147 (8):1655–1663. doi: 10.1007/s007050200039. [DOI] [PubMed] [Google Scholar]
- Nakashima N, Uchiumi T. Functional analysis of structural motifs in dicistroviruses. Virus Res. 2009;139 (2):137–147. doi: 10.1016/j.virusres.2008.06.006. [DOI] [PubMed] [Google Scholar]
- Ni J, Tien AL, Fournier MJ. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89 (4):565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- Nishiyama T, Yamamoto H, Shibuya N, Hatakeyama Y, Hachimori A, Uchiumi T, Nakashima N. Structural elements in the internal ribosome entry site of Plautia stali intestine virus responsible for binding with ribosomes. Nucleic Acids Res. 2003;31 (9):2434–2442. doi: 10.1093/nar/gkg336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama T, Yamamoto H, Uchiumi T, Nakashima N. Eukaryotic ribosomal protein RPS25 interacts with the conserved loop region in a dicistroviral intergenic internal ribosome entry site. Nucleic Acids Res. 2007;35 (5):1514–1521. doi: 10.1093/nar/gkl1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto GA, Puglisi JD. The pathway of HCV IRES-mediated translation initiation. Cell. 2004;119 (3):369–380. doi: 10.1016/j.cell.2004.09.038. [DOI] [PubMed] [Google Scholar]
- Pantoja CR, Navarro SA, Naranjo J, Lightner DV, Gerba CP. Nonsusceptibility of primate cells to Taura syndrome virus. Emerg Infect Dis. 2004;10 (12):2106–2112. doi: 10.3201/eid1012.040419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell. 2007;26 (1):41–50. doi: 10.1016/j.molcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- Perera R, Daijogo S, Walter BL, Nguyen JH, Semler BL. Cellular protein modification by poliovirus: the two faces of poly(rC)-binding protein. J Virol. 2007;81 (17):8919–8932. doi: 10.1128/JVI.01013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Hellen CU. Translation elongation after assembly of ribosomes on the cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes Dev. 2003;17 (2):181–186. doi: 10.1101/gad.1040803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Hellen CU. Reconstitution of eukaryotic translation elongation in vitro following initiation by internal ribosomal entry. Methods. 2005;36 (3):261–269. doi: 10.1016/j.ymeth.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Pestova TV, Lomakin IB, Hellen CU. Position of the CrPV IRES on the 40S subunit and factor dependence of IRES/80S ribosome assembly. EMBO Rep. 2004;5 (9):906–913. doi: 10.1038/sj.embor.7400240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen CP, Bordeleau ME, Pelletier J, Sharp PA. Short RNAs repress translation after initiation in mammalian cells. Mol Cell. 2006;21 (4):533–542. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Pfingsten JS, Costantino DA, Kieft JS. Structural basis for ribosome recruitment and manipulation by a viral IRES RNA. Science. 2006;314 (5804):1450–1454. doi: 10.1126/science.1133281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfingsten JS, Costantino DA, Kieft JS. Conservation and diversity among the three-dimensional folds of the Dicistroviridae intergenic region IRESes. J Mol Biol. 2007;370 (5):856–869. doi: 10.1016/j.jmb.2007.04.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabath N, Price N, Graur D. A potentially novel overlapping gene in the genomes of Israeli acute paralysis virus and its relatives. Virol J. 2009;6:144. doi: 10.1186/1743-422X-6-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki J, Nakashima N. Translation initiation at the CUU codon is mediated by the internal ribosome entry site of an insect picorna-like virus in vitro. J Virol. 1999;73 (2):1219–1226. doi: 10.1128/jvi.73.2.1219-1226.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler M, Connell SR, Lescoute A, Giesebrecht J, Dabrowski M, Schroeer B, Mielke T, Penczek PA, Westhof E, Spahn CM. Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat Struct Mol Biol. 2006;13 (12):1092–1096. doi: 10.1038/nsmb1177. [DOI] [PubMed] [Google Scholar]
- Shibuya N, Nakashima N. Characterization of the 5′ internal ribosome entry site of Plautia stali intestine virus. J Gen Virol. 2006;87 (Pt 12):3679–3686. doi: 10.1099/vir.0.82193-0. [DOI] [PubMed] [Google Scholar]
- Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science. 2001;291 (5510):1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- Spahn CM, Jan E, Mulder A, Grassucci RA, Sarnow P, Frank J. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: the IRES functions as an RNA-based translation factor. Cell. 2004;118 (4):465–475. doi: 10.1016/j.cell.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Thompson SR, Sarnow P. Regulation of host cell translation by viruses and effects on cell function. Curr Opin Microbiol. 2000;3 (4):366–370. doi: 10.1016/s1369-5274(00)00106-5. [DOI] [PubMed] [Google Scholar]
- Thompson SR, Gulyas KD, Sarnow P. Internal initiation in Saccharomyces cerevisiae mediated by an initiator tRNA/eIF2-independent internal ribosome entry site element. Proc Natl Acad Sci USA. 2001;98 (23):12972–12977. doi: 10.1073/pnas.241286698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiumi T, Terao K, Ogata K. Identification of neighboring protein pairs cross-linked with dimethyl 3, 3′-dithiobispropionimidate in rat liver 40S ribosomal subunits. J Biochem (Tokyo) 1981;90 (1):185–193. doi: 10.1093/oxfordjournals.jbchem.a133449. [DOI] [PubMed] [Google Scholar]
- Van Munster M, Dullemans AM, Verbeek M, Van Den Heuvel JF, Clerivet A, Van Der Wilk F. Sequence analysis and genomic organization of Aphid lethal paralysis virus: a new member of the family Dicistroviridae. J Gen Virol. 2002;83 (Pt 12):3131–3138. doi: 10.1099/0022-1317-83-12-3131. [DOI] [PubMed] [Google Scholar]
- Ventoso I, MacMillan SE, Hershey JW, Carrasco L. Poliovirus 2A proteinase cleaves directly the eIF-4G subunit of eIF-4F complex. FEBS Lett. 1998;435 (1):79–83. doi: 10.1016/s0014-5793(98)01027-8. [DOI] [PubMed] [Google Scholar]
- Victoria JG, Kapoor A, Li L, Blinkova O, Slikas B, Wang C, Naeem A, Zaidi S, Delwart E. Metagenomic analyses of viruses in stool samples from children with acute flaccid paralysis. J Virol. 2009;83 (9):4642–4651. doi: 10.1128/JVI.02301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JE, Pestova TV, Hellen CU, Sarnow P. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000a;102 (4):511–520. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
- Wilson JE, Powell MJ, Hoover SE, Sarnow P. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol Cell Biol. 2000b;20 (14):4990–4999. doi: 10.1128/mcb.20.14.4990-4999.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolaway KE, Lazaridis K, Belsham GJ, Carter MJ, Roberts LO. The 5′ untranslated region of Rhopalosiphum padi virus contains an internal ribosome entry site which functions efficiently in mammalian, plant, and insect translation systems. J Virol. 2001;75 (21):10244–10249. doi: 10.1128/JVI.75.21.10244-10249.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wower J, Scheffer P, Sylvers LA, Wintermeyer W, Zimmermann RA. Topography of the E site on the Escherichia coli ribosome. EMBO J. 1993;12 (2):617–623. doi: 10.1002/j.1460-2075.1993.tb05694.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon A, Peng G, Brandenburger Y, Zollo O, Xu W, Rego E, Ruggero D. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science. 2006;312 (5775):902–906. doi: 10.1126/science.1123835. [DOI] [PubMed] [Google Scholar]
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292 (5518):883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- Zhu JY, Ye GY, Fang Q, Wu ML, Hu C. A pathogenic picorna-like virus from the endoparasitoid wasp, Pteromalus puparum: initial discovery and partial genomic characterization. Virus Res. 2008;138 (1–2):144–149. doi: 10.1016/j.virusres.2008.09.001. [DOI] [PubMed] [Google Scholar]