Abstract
Cardiolipin (CL), a major phospholipid in bacterial cell walls, is sequestered from the immune system in mammalian mitochondria and is, therefore, a potential “danger signal”. Based on growing evidence that phospholipids constitute natural ligands for CD1 and that CD1d-restricted T cells recognize phospholipids, we hypothesized that CD1d binds and presents CL, and that T cells in the normal immune repertoire respond to CL in a CD1d-restricted manner. We determined the murine CD1d-CL crystal structure at 2.3 Å resolution and established through additional lipid loading experiments that CL, a tetra-acylated phospholipid, binds to murine CD1d with two alkyl chains buried inside the CD1d binding groove and the remaining two exposed into the solvent. We furthermore demonstrate the functional stimulatory activity of CL, showing that splenic and hepatic γδ T cells from healthy mice proliferate in vitro in response to mammalian or bacterial CL in a dose-dependent and CD1d-restricted manner, rapidly secreting the cytokines interferon-γ and RANTES. Finally, we show that hepatic γδ T cells are activated in vivo by CD1d-bearing dendritic cells that have been pulsed with CL, but not, phosphatidylcholine. Together, these findings demonstrate that CD1d is able to bind and present CL to a subset of CL-responsive γδ T cells that exist in the spleen and liver of healthy mice, and suggest that these cells could play a role in host responses to bacterial lipids and, potentially, self-CL. We propose that CL-responsive γδ T cells play a role in immune surveillance during infection and tissue injury.
Keywords: CD1d, cardiolipin, phospholipid, γδ T cells, CD1-restricted, liver
Introduction
MHC and MHC-like proteins, including CD1, present distinct antigenic structures to T cells in response to microbial infections or cellular damage. While MHC molecules present peptides from protein Ags, CD1 molecules present lipid and glycolipid molecules (1–5). The Ags that bind naturally to CD1 remain poorly elucidated, but include both self and microbial ligands. Recognition of microbial lipid or glycolipid ligands underlies the innate-like antimicrobial functions mediated by activation of CD1-restricted T cells, which is characterized by the massive and rapid release of multiple cytokines and chemokines.
A growing body of evidence demonstrates that phospholipids constitute natural ligands for CD1. Glycerophospholipids, including phosphatidic acid, phosphatidylethanoloamine, phosphatidylcholine (PC), phosphatidylglycerol, phosphatidylserine, phosphatidylinositol (PI), and cardiolipin (CL), were found to be one of the two major categories of cellular lipids bound to human CD1d (6). Crystallographic studies of CD1d bound to PC (7) or phosphatidylinositol di-mannoside (PIM2) (8) demonstrate that such complexes can exist, while functional studies have shown that both αβ and γδ T cells can recognize phospholipids in a CD1d-restricted manner (9–11). Both natural and synthetic phospholipids are recognized by CD1-restricted T cells: phosphatidylethanolamine and PI are stimulatory to some murine CD1d (mCD1d)-restricted natural killer T (NKT) cell hybridomas (12,13); pollen phospholipids (including phosphatidylethanolamine and PC) are strong activators of human γδ CD1-restricted T cells, inducing the production of both IFN-γ and IL-4 (10,14); endogenous and exogenous phospholipids activate duodenal γδ T cells in a CD1-restricted manner, inducing the production of IL-4 and TGF-β (11); and lyso-PC stimulates GM-CSF and IFN-γ responses by human NKT cell clones and freshly isolated peripheral blood lymphocytes, respectively (15). Finally, CL has been shown to stimulate a murine γδ T cell hybridoma (16,17), but neither CD1 restriction nor generalizability to ex vivo T cells was assessed. Synthetic CL was not stimulatory to human NKT cell clones, but it efficiently blocked CD1d presentation of an antigenic glycolipid (15). Taken together, these findings suggest that phospholipids can serve as natural ligands for CD1d and induce strong CD1d-restricted T cell responses.
CL is a unique phospholipid that can be of both mammalian and bacterial origin. In prokaryotic cells, particularly gram negative bacteria, CL is a major phospholipid component of the cell membrane. In contrast, in eukaryotic cells, CL is absent from the cell membrane, but accounts for as much as 20% of mitochondrial lipid (because of its bacterial symbiotic origin). Thus, in healthy mammalian cells, CL is normally sequestered from the immune system.
The presence of CL in prokaryotic cell membranes, along with its absence in healthy eukaryotic cell membranes, suggests that prokaryotic CL may serve as a “danger signal” to the mammalian immune system. It is not clear, however, whether eukaryotic mitochondrial CL is normally presented to the immune system in the context of CD1, similar to MHC class I presentation of peptides from mitochondrial self-proteins (18). We hypothesize that by binding to CD1d, CL constitutes an important signal that induces a CD1d-restricted innate-like immune response to pathogens. Here, we demonstrate that CL is a natural ligand for CD1d and that the normal immune repertoire contains CD1d-restricted T cells that are responsive to this phospholipid Ag.
Materials and methods
Phospholipid preparation
Phospholipids and di-sulfatide (3,6-di-O-sulfo-β-D-C12-galactosylceramide) were obtained from Avanti Polar Lipids, Inc. (Alabaster, AL) and used without further purification. For lipid binding and crystallography studies, phospholipids were dissolved in either chloroform (CHCl3) at 7.5 mg/ml or ethanol at 4 mg/ml, while di-sulfatide was dissolved in DMSO at 5 mg/ml. For cellular studies, lyophilized phospholipids dissolved in CHCl3, or an equivalent volume of CHCl3 (vehicle control), were dried down in a round-bottomed glass tube under nitrogen gas. The lipid was resuspended in complete RPMI 1640 medium (10% fetal bovine serum, 2 mM L glutamine, 100 U/ml penicillin streptomycin, 10 mM Hepes, and 0.1% 2-ME), vortexed vigorously, and sonicated for 20 min at 37°C.
Protein expression, purification, and crystallization
Soluble and fully glycosylated mCD1d, a heterodimeric protein comprised of CD1d and β2-microglobulin, was expressed and purified as described (19). Synthetic TM-CL was loaded by incubating the protein with a 4-fold molar excess of lipid (4 mg/ml in ethanol) in 100 mM Tris buffer, pH 7.0, for 16 h at room temperature with gentle agitation. Synthetic TM-CL was used as a model for natural CL, as it captures most structural aspects of natural CL while possessing alkyl chains of uniform and short length, which are preferred for crystallization. Heavy precipitation was removed by centrifugation at 50,000 × g for 10 min at 4°C. The mCD1d-lipid solution was buffer-exchanged by ultrafiltration (Millipore Ultrafree, 30-kDa nominal molecular weight cut-off) against 10 mM Tris/HCl buffer, pH 8.0. Subsequent anion-exchange chromatography, using MonoQ (GE Healthcare), separated unloaded mCD1d from CD1d-TM-CL complexes and free lipid. Fractions containing CD1d-TM-CL complexes were pooled and buffer-exchanged to obtain a final buffer concentration of 10 mM Hepes and 30 mM NaCl, pH 7.5, and concentrated to 6 mg/ml using centrifugal filter devices (Millipore Ultrafree, 30-kDa molecular weight cut-off) for crystallization. Crystallization experiments were carried out using the sitting drop vapor diffusion method. The best crystals were obtained by mixing 0.5 μl of protein with 0.5 μl of precipitant (20% polyethylene glycol 4000, 0.3 M calcium acetate, 6% (v/v) ethylene glycol) and grown for several weeks at 22°C for data collection.
Structure determination and presentation
Crystals were flash-cooled at of 100oK in mother liquor containing 20% glycerol. Diffraction data from a single crystal were collected at Beamline 7.1 of the Stanford Synchrotron Radiation Laboratory (SSRL) and processed to 2.3 Å with the Denzo-Scalepack suite (20) in spacegroup P1 (unit cell dimensions: a=46.8 Å; b=50.9 Å; c=103.2 Å; α=105.1; β=92.2; γ=106.2). Two CD1-lipid complexes occupy the asymmetric unit with an estimated solvent content of 48.6%, based on a Matthews’ coefficient (Vm) of 2.39 Å3/Dalton. Molecular replacement in P1 was carried out in CCP4 (CCP4, 1994 #40) using the program MOLREP (21) and the CD1d-sulfatide structure (2AKR) as the search model, with the ligand removed, and resulted in a crystallographic R factor (Rcryst) of 38.0% and a correlation coefficient of 0.6. The initial refinement included several rounds of restrained refinement against the maximum likelihood target in REFMAC 5.2. At a later stage of refinement, carbohydrates were built in at all three N-linked glycosylation sites of CD1d. Refinement progress was judged by monitoring the free R factor (Rfree) for cross-validation (22). The model was rebuilt into SigmaA (σA)-weighted 2Fo−Fc and Fo−Fc difference electron density maps using the program COOT (23). Water molecules were assigned in COOT for >3σ peaks in an Fo−Fc map, and retained if they satisfied hydrogen-bonding criteria and returned 2Fo−Fc density >1σ after refinement in REFMAC. Starting coordinates for the TM-CL ligand were obtained from the Hetero-compound Information Centre Uppsala (HIC-Up) (compound CDL) and truncated at all four fatty acyl chains accordingly. The TM-CL library for REFMAC (24) was created using the Dundee PRODRG2 server (25). Final refinement steps were performed using the translation, libration, and screw-axis rotation (TLS) displacement procedure in REFMAC (26), with a total of three anisotropic domains (α1-α2 domain, α3-domain, and β2M), and resulted in improved electron density maps for the glycolipid ligand and a further drop in Rfree. The CD1d-TM-CL structure has a final Rcryst=23.2% and Rfree=27.3%, and the quality of the model (Table 1 (27)) was excellent, as assessed with the program Molprobity (28). The software program Pymol (DeLano Scientific, Palo Alto) was used to prepare Figure 2. The PDB2PQR server (29) and the program APBS (30) were used to calculate the electrostatic surface potentials in Figure 2.
Table 1.
Data collection and refinement statistics for the CD1d-CL complex
| Data collection | |
| Resolution range (Å)1 | 50.0–2.3 (2.38–2.30) |
| Completeness (%)1 | 95.4 (88.4) |
| Number of unique reflections | 37,815 |
| Redundancy | 1.9 |
| Rsym1,2 (%) | 9.1 (38.4) |
| I/σ1 | 12.9 (1.9) |
| Refinement statistics | |
| Number of reflections (f>0) | 36,507 |
| Maximum resolution (Å) | 2.3 |
| Rcryst3 (%) | 23.2 (31.2) |
| Rfree4 (%) | 27.3 (32.7) |
| Number of atoms | 6,282 |
| Protein | 5,899 |
| Glycolipid ligand/spacer lipid | 92/36 |
| N-linked carbohydrate | 98 |
| Water | 157 |
| Ramachandran statistics (%) | |
| Favored | 96.8 |
| Disallowed | 0 |
| R.m.s. deviation from ideal geometry | |
| Bond length (Å) | 0.013 |
| Bond angles (°) | 1.5 |
| Average B values (Å2)5 | |
| Protein | 46.9 |
| Glycolipid/spacer lipid | 66.7/48.3 |
| Water molecules | 26.0 |
| Carbohydrates | 57.3 |
| Disordered residues | C89–C92, C110–111, C201–202 |
Number in parentheses refer to the highest resolution shell.
Rsym=(ΣhΣi|Ii(h)-<I(h)>I|(ΣhΣIIi(h))×100, where <I(h)> is the average intensity of i symmetry-related observations for reflections with Bragg index h.
Rcryst=(Σhkl|Fo−Fc|/Σhkl|Fo|)×100, where Fo and Fc are the observed and calculated structure factors, respectively, for all data.
Rfree was calculated as for Rcryst, but on 3% of data excluded from refinement.
B values were calculated with the CCP4 program TLSANL (27).
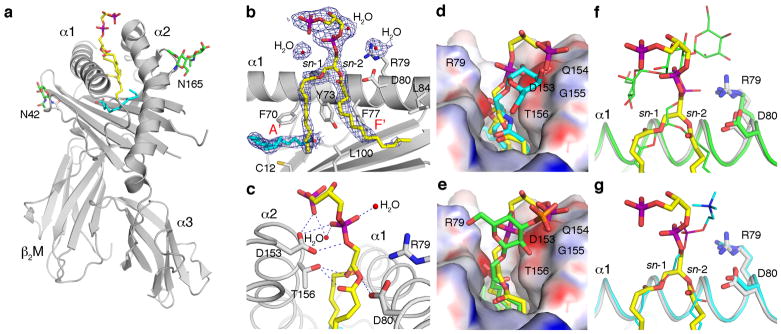
Figure 2. Schematic representation of the mCD1d-CL complex.
(a) Tetra-myristoyl CL (TM-CL) (yellow) is bound in the hydrophobic binding groove between the α1 and α2 helices of the mCD1d heavy chain (grey), which non-covalently associates with β2-microblobulin (β2M, grey). Two N-linked glycosylation sites (N42 and N165) carry well-ordered carbohydrates (green sticks). A spacer lipid (C16, cyan) is present in the binding groove to complement the shorter C14-alkyl chains of the synthetic CL. (b) The 2F0−Fc electron density map is contoured at 1σ and shown as a blue mesh around the CL ligand. The third and fourth acyl chains, as well as the connecting glycerol, are not ordered in the crystal structure, and only 11 carbons of the myristoyl chain that binds in the A′ pocket are ordered. (c) Hydrogen-bond interactions between CD1d residues (grey) and the polar moieties of CL (yellow) are represented. In addition, two water molecules also hydrogen-bond with the two phosphate groups of CL. (d) Comparison between the presentation of TM-CL (yellow) and α-galactosylceramide (α-GalCer, cyan) by mCD1d. (e) Comparison between the presentation of CL (yellow) and sulfatide (green) by mCD1d. The central phosphate-glycerol-phosphate moiety of CL is located farther above the binding groove as compared to the more intimate binding of α-GalCer (the predominant invariant NKT cell Ag). However, the polar groups of both CL and sulfatide occupy similar positions. Notably, the terminal phosphate of CL is in a similar position as the sulfate of sulfatide. (f) Binding comparison between TM-CL (yellow sticks) and PIM2 (thin green sticks), and (g) between TM-CL and PC (thin cyan sticks). Protein backbone is colored accordingly and side chains are shown for R79 and Asp80 for orientation. Surface representation, with electrostatic potential (red, electronegative, and blue, electropositive, contoured from −30 to +30 kT/e), is shown in panels d and e. Several aa residues, which are involved either in the shaping of the mCD1d-binding grooves (b, d, e) or in polar interactions with CL (c), are depicted using the single letter aa code with the relevant residue number. Individual atoms are colored as follows: carbon, yellow, green or cyan; oxygen, red; nitrogen, blue; phosphorus, purple; and sulfur, orange.
In vitro loading of lipid antigens
Aliquots of 10 μl of purified mCD1d at a concentration of 20 μM were loaded overnight at room temperature in the presence of 4–6 times molar excess of each ligand. PC and di-sulfatide were dissolved in DMSO (1–2 mg/ml), while all CL species were dissolved in ethanol (4 mg/ml). Final concentration of DMSO and ethanol in the loading experiment did not exceed 3–5%. Loading was performed in the presence of 100 mM Tris-HCl, pH 7. After lipid loading, samples were centrifuged (14,000 × g, 10 min) and 4 μl of the supernatant were used for isoelectric focusing (IEF) analysis. CD1d-lipid complexes were separated using precast gels (PhastGel 5–8 IEF) and the PhastSystem (GE Healthcare Bio-Sciences Corp., Piscataway, NJ). Staining of mCD1d was performed with Coomassie blue, and successful lipid loading is identified by a gel shift of the mCD1d band in relation to the control lanes.
Murine T cell preparation
All animal experiments were conducted according to the relevant Canadian and international guidelines, and were approved by the McGill University Animal Care Committee. Specific pathogen-free C57BL/6 mice were obtained from Harlan Sprague Dawley, and TCRβ-deficient mice (The Jackson Laboratory, Bar Harbor, ME) were bred in-house. CD1d−/− mice backcrossed for 12 generations (N12) in the C57BL/6J background, which were kindly provided by Drs. Peter van den Elzen and Rusung Tan (University of British Columbia), were originally derived by Drs. Steven Balk and Mark Exley (Harvard Medical School) (31). Anesthetized mice were perfused through the heart with PBS, pH 7.3, and the perfused spleen and liver were harvested post-mortem. T cells from spleen or liver were purified by negative selection using a murine T cell enrichment kit (EasySep, Stemcell Technologies, Vancouver, BC). Total splenic or hepatic T cells (~98% CD3+ cells) were isolated by negative selection (removal of cells expressing CD11b, CD19, CD45R, CD49b, and TER119) using a murine T cell enrichment kit (EasySep, Stemcell Technologies, Vancouver, BC). The isolated T cell population contained both αβ and γδ T cell subsets. In certain experiments (where specified), isolated T cells were labeled with CFSE using the CellTrace CFSE Cell Proliferation Kit (Invitrogen Corporation, Carlsbad, CA) according to the manufacturer’s instructions. In experiments using purified γδ T cells, the cells were isolated using a two step TCR γδ T cell isolation kit (Miltenyi Biotec, Auburn, CA). Briefly, a first step enrichment of T cells by negative selection (removal of CD11b- and CD45R-positive cells) was followed by positive selection of TCR γδ positive cells.
T cell stimulation with phospholipids
Purified total T cells (50 μl; 2.5 × 105/ml) or γδ T cells (50 μl; 1 × 105/ml) were stimulated with different final concentrations of phospholipids (30–60 μl of a 1 mg/ml suspension), including native bovine heart CL, hydrogenated bovine heart CL, or bovine heart PC, diluted in complete RPMI medium in presence of APCs (splenocytes treated with mitomycin C; (150 μl; 8 × 106/ml) in all experiments. The cells were cultured in a 96-well plate for 24 h or 48 h at 37°C (5% CO2). Blocking experiments were performed using anti-I-A (Y3P; generously provided by Dr. Philippa Marrack, Howard Hughes Medical Institute, National Jewish Medical and Research Center, Denver, CO); anti-CD1d (19G11) (32) or anti-H2Dd (34-2-12; BD Biosciences, Mississauga, ON). Cell proliferation was measured using a BrdU proliferation kit (Roche Diagnostics, Indianapolis, IN) or CFSE-labeled cells. T cells were characterized by flow cytometry using mAbs to murine CD3, IFN-γ, αβ TCR, and γδ TCR (BD Biosciences, Mississauga, ON). In certain studies (where specified), cells were stained with mAbs to murine CD25, CD69, Vγ2, Vγ3, Vγ4, or Vδ6.3/2; annexin V; and/or 7-AAD (BD Biosciences). To ensure inter-tube comparability, relative cell numbers were determined by adding 3000 TruCOUNT beads (BD Biosciences) to each tube prior to flow cytometry analysis and counting 500 True-Count bead events per sample during flow cytometry acquisition. These beads serve as an independent and absolute counting control, to ensure that equivalent volumes are counted for each tube (independent of cell composition or number).
In vivo γδ T cell response and flow cytometry
Mouse bone marrow derived DCs (BMDCs) were prepared by culturing bone marrow progenitors with 20 ng/ml GM-CSF (PeproTech) for 7 days following standard protocols. On day 7, BMDCs were washed, counted, and cultured with phospholipid or vehicle control (100 μg/ml) for 24 h. After being washed with PBS, phospholipid-pulsed BMDCs (5 × 105) were injected i.v. into mice. Concurrently, mice were injected i.p. with 1 mg BrdU (BD PharMingen). Liver mononuclear cells (LMNCs) positive for CD3 and γδ TCR (BD PharMingen) were analyzed directly ex vivo for BrdU incorporation and activation markers CD25 and CD69 (BD PharMingen) by flow cytometry. Cells were analyzed using an LSR II (BD Bioscience) and FlowJo software.
Cytokine detection
Supernatants of hepatic T cells stimulated with phospholipid (0.2 μM) or vehicle were harvested 48 h post stimulation, and a panel of 22 cytokines and chemokines was assayed using RayBio Mouse Cytokine Ab Array 1 (RayBiotech, Inc., Norcross, GA), according to manufacturer’s instructions. Furthermore, supernatants were assayed for IFN-γ (BD Biosciences) and RANTES (R&D Systems, Inc., Minneapolis, MN) by ELISA, according to manufacturer’s instructions.
Results
Crystal structure of cardiolipin bound to mouse CD1d
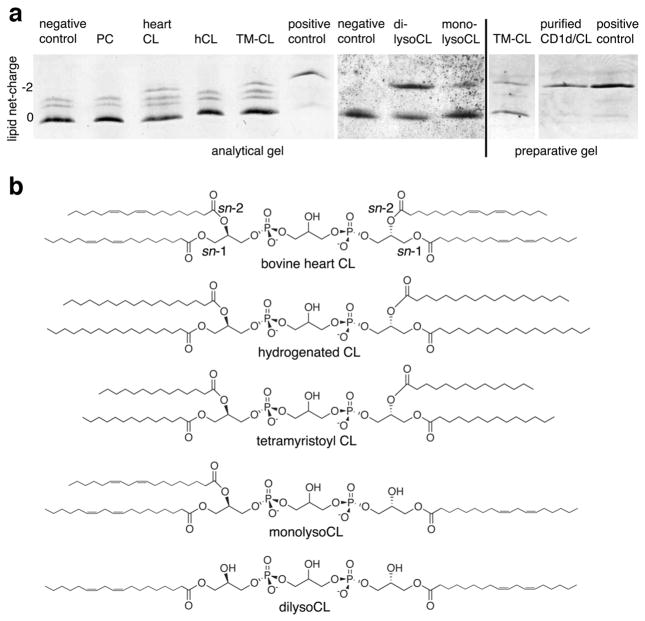
mCD1d is known to bind di-acylated glycolipids and phospholipids (33), but its ability to bind tetra-acylated phospholipids has not been studied. Recently, CL, among other lipids, has been identified by mass spectrometry as a natural human CD1d ligand (6). However, CL does not appear to be an antigen for human invariant NKT cells, as it failed to activate a panel of human iNKT cell clones (15). To determine whether mCD1d is able to bind to CL, a tetra-acylated phospholipid with two negatively charged phosphate groups, we performed ligand binding studies with recombinant mCD1d and native (bovine heart) and synthetic (tetramyristoyl (TM) [14:0]) forms of CL. We also evaluated hydrogenated bovine heart CL (hCL) to determine whether fatty acid chain saturation has an influence on CD1d binding. Bovine heart PC, which was derived from the same source as native CL and had similar fatty acid saturation, served as a control for mCD1d in the presence of a neutral phospholipid. Binding of CL to mCD1d was assessed by native IEF gel electrophoresis (Figure 1a). Upon incubation of mCD1d with native bovine heart CL or synthetic TM-CL, but not fully saturated bovine heart CL (hCL), two major bands were apparent (Figure 1a, left panel). The lower band corresponds to mCD1d containing an uncharged endogenous spacer lipid derived from its expression in insect cells (net charge 0), whereas the upper fainter band corresponds to mCD1d-CL complexes (net charge −2). In addition, purified mCD1d also contains a faint double band above the major species of mCD1d. This double band is likely a result of endogenous ligands with different charge values bound to mCD1d, as it disappears upon loading with di-sulfatide (Figure 1a, left panel, labeled positive control).
Figure 1. CL binds to mCD1d.
(a) Synthetic tetramyristoyl CL (TM-CL) (14:0) and native bovine heart CL can bind to mCD1d, as detected by native IEF gel electrophoresis. Analytical gel (left panel): mCD1d (negative control) was incubated with bovine heart phosphatidylcholine (PC) (net charge 0), bovine heart cardiolipin (CL) (net charge −2), hydrogenated bovine heart CL (hCL) (net charge −2), or TM-CL (net charge −2) for 16 hr. Removal of the third (monolysoCL) and fourth (dilysoCL) alkyl chain from CL increases loading efficiency (middle panel). Preparative gel (right panel): mCD1d-TM-CL complexes can be fully separated from mCD1d molecules by anion exchange chromatography (purified CD1d/CL). A positive control is shown, where mCD1d was loaded with di-sulfatide (a nonphospholipid ligand with a net negative charge of −2). The preparative gel depicts the large-scale TM-CL loading and purification of mCD1d-TM-CL used for crystallographic studies. The data in panel a are representative of two to four independent experiments. (b) Chemical structure of the variants of CL used in the binding studies. Interestingly, while bovine heart CL, TM-CL, monolysoCL, and dilysoCL bind to mCD1d, hCL fails to bind to mCD1d under these conditions. This is likely due to reduced solubility of hCL during loading, as a result of the long and fully saturated C18 acyl chains. Note the faint upper double bands in the negative control lane (left panel), which are likely the result of endogenously bound charged lipids.
The mCD1d preparation used for the lipid loading experiment is shown in the middle and right panels (Figure 1a), and is more homogeneous and does not contain endogenously bound charged lipids. These findings demonstrate that recombinant mCD1d protein binds native bovine heart CL and TM-CL (band at −2 net charge), but does not bind saturated bovine heart CL (hCL). mCD1d incubated with buffer or neutrally charged bovine heart PC showed a major band for the CD1d endogenous spacer lipid (net charge 0), but no band with a net charge of −2. Notably, binding of TM-CL and bovine heart CL occurred in the absence of lipid transfer proteins (none were present in the system), although binding was less efficient than that of the positive control (di-sulfatide with only 2 alkyl chains). We evaluated whether mCD1d could also bind bovine CL that had one (monolysoCL) or two (dilysoCL) fatty acyl chains removed (Figure 1). mCD1d bound both lyso forms of CL, and binding was maximal with dilyso-CL and appeared to decrease with the addition of fatty acyl chains. These data indicate that maximal binding of CL to mCD1d occurs with only two alkyl chains and that additional alkyl chains are not required for binding to mCD1d, although they could be required for recognition.
mCD1d-TM-CL complexes, purified using anion exchange (MonoQ) chromatography, showed a single band on IEF gel electrophoresis (Figure 1a, right panel), with the same migration as purified complexes of mCD1d and di-sulfatide (the control for mCD1d loaded with a non-phospholipid Ag having a net −2 charge), and were used for subsequent crystallization. These findings suggest that CL (bovine heart CL and TM-CL), despite the presence of four alkyl chains (Figure 1b), can still bind to mCD1d and form a stable mCD1d-CL complex. The chemical structure of TM-CL raises the question whether TM-CL can bind simultaneously to two mCD1d molecules by inserting 2 alkyl chains in each of the CD1d binding grooves. However, size exclusion chromatography revealed that one TM-CL molecule is bound by only one mCD1d protein molecule (data not shown).
To identify how CL binds to mCD1d, we crystallized mCD1d-CL complexes and determined the crystal structure to 2.3 Angstroms (Å) resolution (Figure 2 and Table 1). No gross structural differences in mCD1d are observed when comparing mCD1d-TM-CL to other mCD1d-glycolipid complexes (8,19,34–36). Two of the four alkyl chains are inserted into the A′ and F′ pockets of the CD1d binding groove (Figure 2a), while the other two alkyl chains are exposed to the solvent. The central phosphate-glycerol-phosphate moiety of TM-CL is located above the binding groove in a position similar to the head groups of other glycolipid ligands (8,19,34–36). No clear electron density is observed for the two solvent-exposed alkyl chains of TM-CL, indicating that these chains make no major contact with CD1d residues and are disordered. Therefore, these two solvent exposed alkyl chains were not built into the final structure and are not depicted in Figure 2. It is important to note that we observed a cavity within the crystal lattice. This cavity is big enough to accommodate the third and fourth alkyl chains. Moreover, it is clear from our ligand binding and size exclusion chromatography experiments (Figure 1a and data not shown) that one intact tetra-acylated TM-CL molecule binds to one CD1d molecule, and that fully loaded mCD1d-TM-CL complexes have been purified and crystallized. Interestingly, there are no CD1d residues capable of neutralizing the negative charge of both phosphate groups of TM-CL. Despite this, mCD1d residues Asp80, Asp153, and Thr156 interact with the polar center of TM-CL through hydrogen-bonding (Figure 2c), thereby stabilizing the polar core of TM-CL and giving rise to very well-ordered electron density (Figure 2b). In addition, two water molecules provide additional hydrogen-bond interactions with both phosphate groups of TM-CL.
We next compared the mCD1d binding of TM-CL to that of other mCD1d ligands, including α-galactosyl ceramide (α-GalCer), which stimulates Vα14-invariant NKT cells (34,37); sulfatide, which stimulates a minor subset of non-Vα14 NKT cells (19,38,39); and two phosphoglycerolipids, PIM2 and PC (7,8). The comparative structures (Figures 2d–g) reveal that the lipid backbone of the various Ags binds slightly differently inside mCD1d and, therefore, leads to a slightly different presentation of the polar epitopes. Although binding of α-GalCer to mCD1d shows greater differences because of its α-linked galactose, binding of TM-CL and sulfatide to mCD1d follows a similar pattern, in which the terminal phosphate of TM-CL, or sulfate of sulfatide, is located just above a slightly electro-positive patch formed by the Glu154 and Gly155 backbones of the α2-helix. When TM-CL binding to mCD1d is compared to either PIM2 or PC (Figures 2f and 2g), the binding of all three phosphoglycerolipids appear quite similar. However, the binding of the diacylglycerolipid backbones, in general, appear less conserved than the binding of the glycosphingolipid backbones, which all superimpose very closely when bound by mCD1d (33,40). The position of the proximal phosphate is slightly different in all three structures, and this orientation affects the presentation of the subsequent headgroup (Figures 2, f and g). In addition, the phosphoglycerolipids appear to bind in opposite orientations in the CD1d binding groove, leading to subtle differences in the binding orientation of the diacylglycerolipid backbone, similar to the binding of two α-galactosyl diacylglycerolipids from Borrelia burgdorferi to mCD1d (41). While both TM-CL and PIM2 bind with the sn-1 linked fatty acid in the A′ pocket and the sn-2 linked acyl chain in the F′ pocket, the binding orientation of PC is reversed (Figures 2f and 2g). In summary, mCD1d binds and presents structurally diverse glycolipids and complex phospholipids in an orientation that reflects the differences in their lipid backbones, while presenting the polar moieties for recognition by specific T cells.
Murine T cells proliferate in response to CL in a dose-dependent manner and produce IFN-γ
To determine whether CL-reactive T cells are found in the immune repertoire of healthy, untreated mice, we studied total spleen- and liver-derived T cells. Both splenic and hepatic T cells from C57BL/6 mice proliferated in response to the maximal concentration of native bovine heart CL (0.2 μM), but only hepatic T cells showed a dose-dependent response at lower concentrations of CL (i.e., ≤0.1 μM) (Figure 3a). Moreover, the maximal proliferative response of hepatic T cells was greater than three-fold higher than that of splenic T cells (OD450 = 1.086 ± 0.110 and 0.333 ± 0.100, respectively). This is consistent with the fact that the liver contains a higher percentage of CD1-reactive T cells, such as NKT cells, than the spleen (42,43). In contrast, we observed no significant proliferative response to hCL, a completely saturated form of bovine heart CL (Figure 3a), in agreement with the finding that hCL did not bind to mCD1d (Figure 1a). Similarly, PC, from the same source (bovine heart) as CL and with a high degree of fatty acid unsaturation, also failed to stimulate T cell proliferation (Figure 3a), consistent with its inability to bind to mCD1d (Figure 1a). We also evaluated whether monolysoCL or dilysoCL, potential breakdown products of bovine heart CL that lack one or two fatty acyl chains, respectively, could stimulate hepatic T cells. T cell proliferation was maximal with native CL and appeared to decrease with removal of fatty acyl chains (Supplementary Figure 1a). These data indicate that maximal T cell proliferation occurs with the native form of the phospholipid and not with its breakdown products. To determine whether the T cell response was limited to bovine heart CL, we evaluated two other forms of CL: TM-CL (synthetic CL) and E. coli CL (bacterial CL) (Supplemental Figure 1b). TM-CL induced a low level of T cell proliferation, similar to that observed with dilysoCL. In contrast, E. coli CL induced marked levels of T cell proliferation that were statistically indistinguishable from those seen with bovine CL (p < 0.8). These results suggest that murine hepatic T cells can differentiate between different forms of CL, but recognize both eukaryotic (bovine heart) and prokaryotic (E. coli) CL.
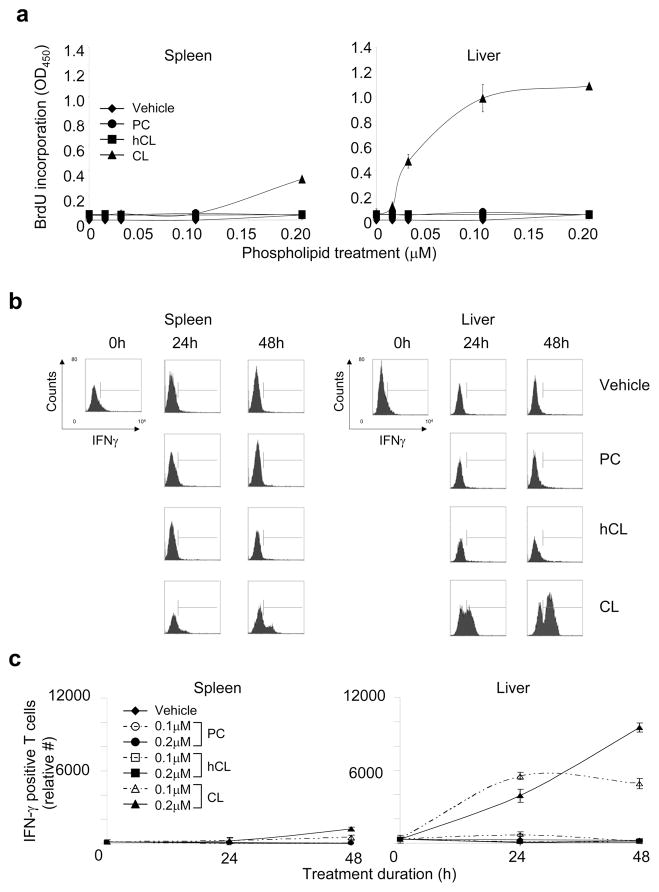
Figure 3. Murine splenic and hepatic T cells proliferate in response to CL in a dose-dependent manner and express IFN-γ.
Purified T cells from spleen (left panels) or liver (right panels) of C57BL/6 mice were incubated with vehicle or phospholipid (cardiolipin [CL], hydrogenated CL [hCL], or phosphatidylcholine [PC]) in presence of APCs in vitro. Phospholipids were used at the following final concentrations: 0.015–0.2 μM (panel a); 0.2 μM (panel b); and 0.1 or 0.2 μM (panel c)]. (a) Proliferation was evaluated by BrdU incorporation during the last 24 h of treatment. The data show the mean values (OD450) ±SD of triplicate samples from one representative experiment of three independent experiments. Other controls (not shown) included con A and cell medium, which had OD450 (± SD) values of 1.194 (0.037) and 0.063 (0.004), respectively. (b and c) IFN-γ production by CD3+ T cells was evaluated by flow cytometry in treated T cells at 0 h, 24 h, or 48 h. Panel b shows the raw flow cytometry data from a representative experiment, while panel c shows the mean IFN-γ-positive T cells ±SD of three independent flow cytometry experiments (including the one in panel b).
We next studied the time course of the T cell response to CL by monitoring IFN-γ expression of CD3+ T cells by flow cytometry. Purified splenic and hepatic T cells were stimulated for different periods of time with CL or other phospholipids (0.1 and 0.2 μM). Approximately 35.4% of splenic T cells treated with CL produced IFN-γ at 48 h, but far fewer cells (14.3%) produced IFN-γ at 24 h (Figure 3b). In contrast, little or no IFN-γ production was detected in T cells treated with hCL or PC. Hepatic T cells were particularly responsive to CL treatment, and >5-fold more IFN-γ-producing T cells were found in hepatic, compared to splenic, T cell preparations after treatment with 0.2 μM CL for 48 h (Figure 3c). Maximal stimulation was observed with 0.2 μM CL at 48 h, with approximately half-maximal stimulation observed after 24 h. At 72 h, <25% of the CL-responsive T cells observed at 48 h were still viable (data not shown). These findings demonstrate that T cells proliferate and produce IFN-γ rapidly in a dose-dependent manner to CL. CL-responsive T cells are found predominantly in the liver, but are also detected in the spleen of healthy C57BL/6 mice.
CL-responsive T cells express γδ TCR
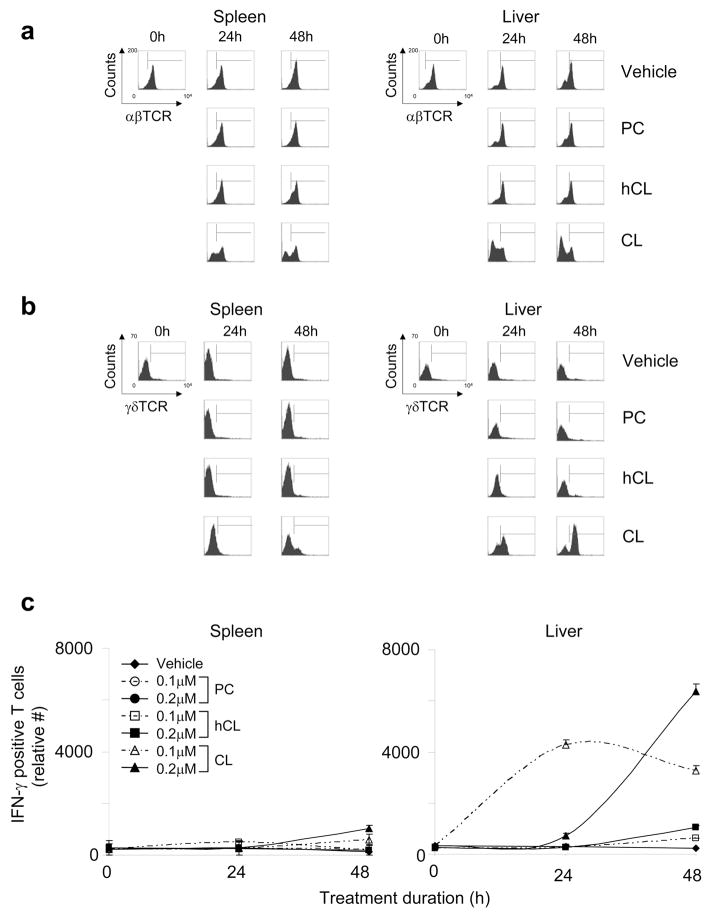
To determine the TCRs expressed by CL-responsive T cells, we monitored αβ and γδ TCR positivity of phospholipid-stimulated hepatic and splenic CD3+ T cells by flow cytometry. Vehicle-treated splenic and hepatic T cells were largely αβ TCR-positive and γδ TCR negative (96.4% and 97.1%, respectively; Figures 4a and 4b). In contrast, CL stimulation resulted in a reduction in αβ TCR+ CD3+ T cells and an increase in γδ TCR+ CD3+ T cells. Reduction in αβ TCR+ cells was due not to increased apoptosis (as detected by annexin V and 7-AAD staining), but instead to a specific proliferation of γδ TCR+ CFSE-labeled T cells (20.2% for CL versus 2.0% for vehicle) (Supplementary Figure 2). These changes were specific to native CL and were not observed with hCL or PC. Stimulation of γδ TCR-bearing T cells was maximal in both hepatic and splenic T cells at 48 h, but the relative cell numbers (as defined in Methods) differed substantially (6,367 [hepatic] versus 1,029 [splenic]) (Figure 4c), and suggest that there are approximately six-fold more CL-responsive γδ T cells in the liver than spleen. When hepatic CL-responsive γδ T cells were evaluated for Vγ2, Vγ3, Vγ4, and Vδ6.3/2 expression, only Vγ3 expression was found to be significantly increased (2.4%, compared to 0.3% in vehicle-stimulated cells) (data not shown). Taken together, our data indicate that CL-responsive cells are γδ T cells located more predominantly in the liver than in the spleen, and suggest that Vγ2, Vγ3, Vγ4, and Vδ6.3/2 are not expressed on the predominant γδ T cell subset that responds to CL.
Figure 4. CL-responsive T cells express γδ TCR.
Purified T cells from spleen (left panels) or liver (right panels) of C57BL/6 mice were incubated with vehicle or phospholipid (CL, hCL, or PC) in presence of APCs in vitro. Phospholipids were used at the following final concentrations: 0.2 μM (panel a); and 0.1 or 0.2 μM (panel b). αβ (a) or γδ (b) TCR expression by CD3+ T cells was evaluated by flow cytometry in treated T cells at 24 h or 48 h, and cell numbers were established using True-Count beads. (c) IFN-γ production by γδ-expressing splenic and hepatic T cells was evaluated by flow cytometry in treated T cells at 0 h, 24 h, or 48 h. Panels a and b show the raw flow cytometry data from a representative experiment of three independent experiments, while panel c shows the mean IFN-γ-positive T cells ±SD of three independent flow cytometry experiments (including the one in panel b).
CL-responsive γδ T cell expansion is independent of αβ T cells
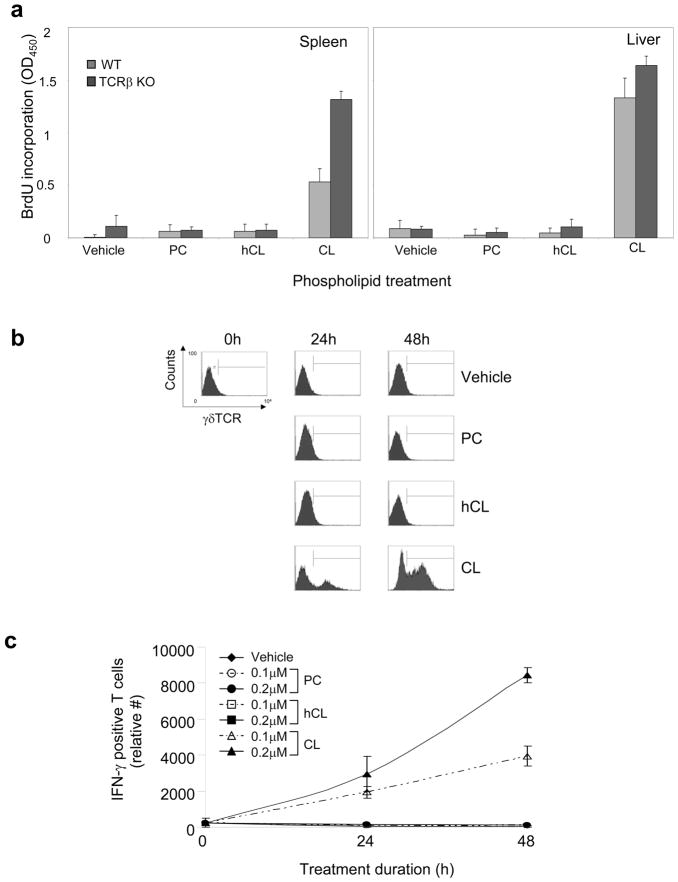
To exclude the possibility that αβ TCR-bearing cells (e.g., NKT cells) provide a first signal that induces a secondary γδ T cell response, we compared the responses of T cells from TCRβ-deficient and wild type (WT) (C57BL/6) mice. As shown in Figure 5a, splenic and hepatic T cells from TCRβ-deficient mice demonstrated strong proliferative responses to CL, similar to those of T cells from WT mice. The relative number of CL-responsive, IFN-γ-producing T cells found in the liver of TCRβ-deficient mice was also similar to that observed in WT mice (Figures 5b and 5c). These results demonstrate that the proliferation of γδ T cells in response to CL is independent of the αβ T cell population and not secondary to activation of an αβ T cell subset. Furthermore, we have observed CL-specific stimulation of γδ T cells purified from spleen and liver of WT mice in the absence of any other cells except APCs, and these data recapitulate our findings using total T cells (Supplementary Figure 3). Timing of the IFN-γ response in purified γδ T cells was similar to that of purified T cells. Furthermore, use of purified γδ T cells enabled a more accurate estimate of CL-induced proliferation, and suggested a 3-fold increase in liver γδ T cells at 24 hours and a 12-fold increase at 48 hours (data not shown).
Figure 5. CL-responsive γδ T cell expansion is independent of αβ T cells.
Purified T cells from spleen (left panels) or liver (right panels) of C57BL/6 (wild type) or TCRβ-deficient (TCRβ KO) mice were treated with vehicle or phospholipid (CL, hCL, or PC) in presence of APCs in vitro. Phospholipids were used at the following final concentrations: 0.2 μM (panels a and b); and 0.1 or 0.2 μM (panel c). (a) Proliferation was evaluated by BrdU incorporation during the last 24 h of treatment. The data show the mean values (OD450) ± SD of triplicate samples from one representative experiment of two independent experiments. (b) γδ TCR expression by CD3+ T cells was evaluated by flow cytometry in treated T cells at 24 h or 48 h, and cell numbers were established using True-Count beads. (c) IFN-γ production by γδ-expressing splenic and hepatic T cells was evaluated by flow cytometry in treated T cells at 0 h, 24 h, or 48 h. Panel b shows the raw flow cytometry data from a representative experiment of two independent experiments, while panel c shows the mean IFN-γ-positive T cells ±SD of two independent flow cytometry experiments (including the one in panel b).
γδ T cells respond to CL in a CD1d-restricted manner
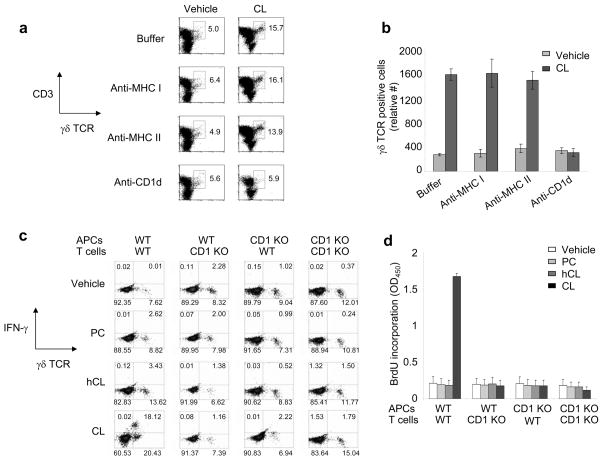
Given the binding of CL to CD1d (Figure 1a), we investigated whether CL-induced expansion of γδ T cells is CD1d-restricted by performing blocking studies with mAbs to either CD1d, MHC I, or MHC II. γδ T cell expansion in response to CL was unchanged by the addition of anti-MHC I or anti-MHC II Ab, but was completely abolished by the addition of anti-CD1d blocking Ab (Figures 6a and 6b). To confirm the CD1d restriction indicated by the Ab blocking experiments, we investigated whether CL-induced expansion of γδ T cells occurs in the presence of APCs derived from C57BL/6 CD1d−/−(CD1d KO) mice. γδ T cells derived from WT mice were unresponsive to CL presented by APCs from CD1d KO mice, but responded normally to CL presented by WT APCs (Figures 6c and 6d). Furthermore, CL-responsive γδ T cells were undetectable in CD1d KO mice, whether APCs from CD1d KO or WT mice were used (Figures 6c and 6d). The findings were the same whether detected by flow cytometry (γδ TCR, IFN-γ positivity) (Figure 6c) or BrdU proliferation assay (Figure 6d). Together, these data demonstrate that CL-responsive γδ T cells are restricted by CD1d, and that CD1d is required for presentation of CL.
Figure 6. γδ T cells respond to CL in a CD1d-restricted manner.
(a and b) Purified hepatic T cells from C57BL/6 mice were incubated in vitro with 0.1 μM CL, or vehicle, and APCs for 48 h in the presence of different blocking Abs (anti-MHC I, anti-MHC II, or anti-CD1d). Relative expansion of the γδ TCR-expressing CD3+ cell population was evaluated by flow cytometry using True-Count beads. Raw flow cytometry data, shown in panel a, are represented graphically in panel b. (c and d) Purified hepatic T cells from either C57BL/6 (WT) or C57BL/6 CD1d−/− (CD1d KO) mice were incubated in vitro with 0.2 μM phospholipid (PC, hCL, or CL), or vehicle, and APCs (from either WT or CD1d KO mice) for 48 h. Relative expansion of the CD3+, γδ TCR+, IFN-γ+ cell population was evaluated by flow cytometry using TruCOUNT beads (panel c) or BrdU proliferation assay (panel d). Panels a and b show data from a representative experiment of three independent experiments, while panels c and d show data from a representative experiment of two independent experiments. Error bars in panels b and d indicate SD of the means of duplicate (panel b) or triplicate (panel d) samples.
CD1-restricted γδ T cells express the activation marker CD25, and produce RANTES and IFN-γ in response to CL
To characterize the activation state of CL-responsive γδ T cells, we determined their expression of two activation markers (CD69 and CD25) 48 h after stimulation with CL (Supplementary Figure 4). Stimulation of hepatic T cells with CL resulted in γδ TCR+ cells that were >98% negative for the expression of the early activation marker CD69. In contrast, a proportion (17.4%) of these same γδ TCR+ cells expressed CD25. Notably, IFN-γ expression was observed in 37.2% of CD25+γδ TCR+ cells, compared to 5.2% of CD25−γδ TCR+ cells. These data indicate that CL-responsive IFN-γ-producing γδ T cells are clearly activated by CL, and express the later activation marker, CD25, after 48 h of stimulation.
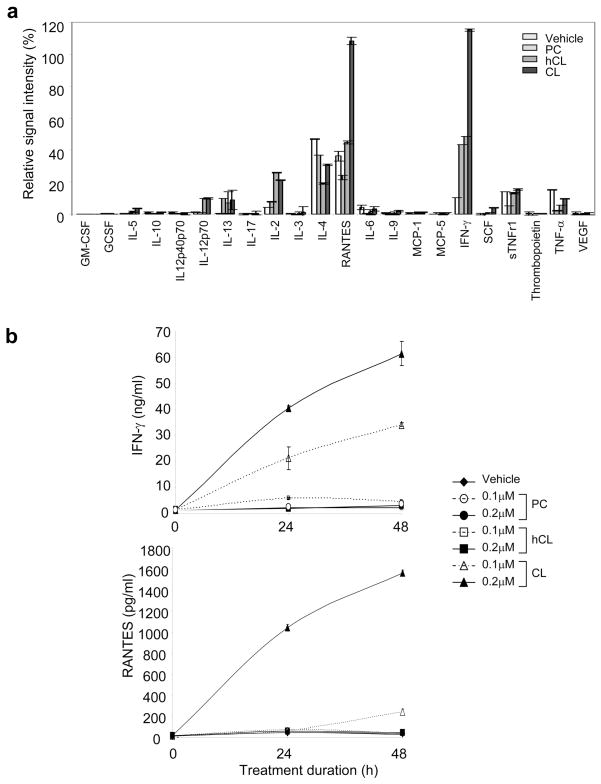
To investigate the potential biological effects of CL-responsive γδ T cells, we used an Ab array to examine the production of 22 cytokines and chemokines by hepatic T cells stimulated with CL, vehicle, or other phospholipids (Figure 7a). Two cytokines, RANTES and IFN-γ, were significantly elevated (>2-fold) in supernatants from CL-treated T cells, compared to control cells (vehicle-, hCL-, or PC-treated). IL-4 was also detected in supernatants of vehicle-treated cells, but its presence was decreased by phospholipid treatment. Minimal levels of IL-13, IL-2, IL-12, sTNFR1, and TNF-α were detected, but did not correlate with CL treatment. IFN-γ and RANTES levels in the supernatants of treated cells were 61.4 ± 0.7 ng/ml and 1,563.8 ± 25.1 pg/ml, respectively, after treatment with 0.2 μM CL. Lower, but significantly elevated, levels of both cytokines (34.0 ± 4.7 ng/ml for IFN-γ and 244.6 ± 25.9 pg/ml for RANTES) were also observed with 0.1 μM CL (Figure 7b). In contrast, only background levels (4.6 ± 0.1 ng/ml and 45.0 ± 4.4 pg/ml, respectively) of these cytokines were detected in supernatants of cells treated with hCL or PC (0.1 or 0.2 μM) (Figure 7b). In parallel studies, 17.4% of CL-responsive γδ T cells were shown to express the activation marker CD25 (data not shown). Taken together, our results show that hepatic γδ T cells are activated and secrete significant amounts of the cytokines, IFN-γ and RANTES, when stimulated by CL.
Figure 7. CD1d-restricted γδ T cells produce RANTES and IFN-γ in response to CL.
Purified hepatic T cells from C57BL/6 mice were incubated for 48 h in vitro with vehicle or 0.2 μM phospholipid (CL, hCL, or PC) in presence of APCs. (a) Supernatants of treated cells were tested for the presence of 22 cytokines and chemokines using an Ab array quantitated by densitometry. Values indicate the signal intensity (%) relative to the signal for the positive control (set at 100%) for each cell treatment. (b) IFN-γ or RANTES in supernatants of treated cells was quantified by ELISA. The data in panels a and b show the mean ± SD of two independent experiments. Panels a and b show data from a representative experiment of two independent experiments. Error bars in panels a and b indicate SD of the means of duplicate samples.
CL activates γδ T cells in vivo
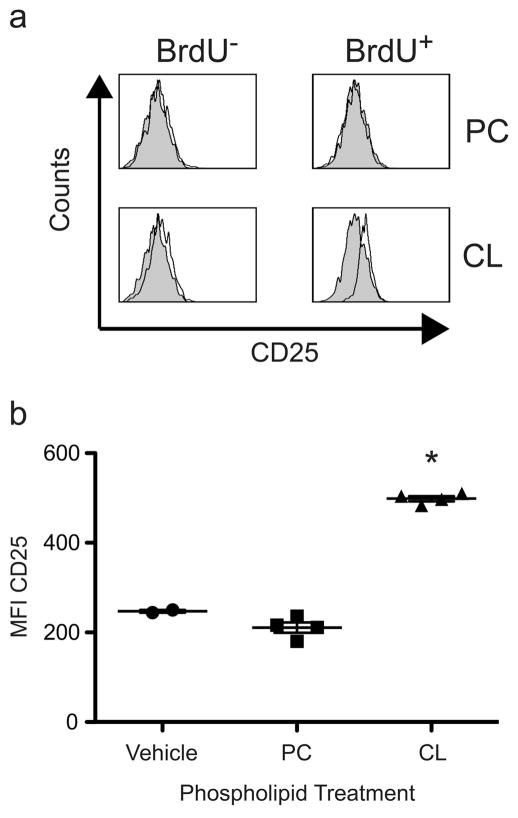
To determine whether CL also stimulates γδ T cell in vivo, we injected mice that had received BrdU with BMDCs pulsed with CL or PC. 24 h later, we analyzed the expression of activation markers (CD25 and CD69) on CD3+, γδ TCR+ T cells in the liver. Injection of CL-pulsed BMDCs resulted in increased expression of CD25 on the proliferating BrdU+ γδ T cells in the liver (MFI = 500, compared to 220 and 200 for vehicle and PC, respectively), while CD25 expression remained unaffected in the majority of the BrdU− cells. (Figure 8). In contrast, PC-pulsed BMDCs did not induce the expression of activation markers on BrdU+ cells. Interestingly, this up-regulation was observed in the liver and not the spleen, and up-regulation of CD69 was not observed (data not shown). Importantly, the percentage of γδ T cells proliferating remained unaltered regardless of the phospholipid (CL or PC) used to pulse BMDCs (data not shown). Furthermore, when Cd1d−/− BMDCs were pulsed with CL and injected into mice, we did not observe induction of CD25 on BrdU+ γδ T cells (data not shown).
Figure 8. γδ T cells are activated by CL in vivo.
C57BL/6 mice, which had received 1 mg BrdU i.p., were injected i.v. with 5 × 105 100 μg/ml phospholipid (CL or PC) or vehicle treated BMDCs. (a) BrdU− (left) and BrdU+ (right) populations of CD3+, γδ TCR+ liver mononuclear cells were analyzed for CD25 expression (open histograms) compared to vehicle treated controls (shaded histograms) at 24 h. Histograms are representative plots of 2 independent experiments of 2–4 mice per group. (b) Scatter plot of CD25 MFI of 2–4 mice per group 24 h post injection with phospholipid pulsed BMDCs from a minimum of 2 independent experiments. A statistically significant difference (p < 0.001 indicated with *) was found for CL, compared to PC pulsed BMDCs, using the equal variance Student t test.
Discussion
There is a growing body of evidence showing that CD1-restricted T cells recognize di-acylated phospholipids (10,11,13). We demonstrate here that CD1d interacts specifically with CL, a tetra-acylated phospholipid. Moreover, we show that γδ T cells from the normal immune repertoire (particularly, the liver) of healthy mice are stimulated by CL in a CD1d-dependent manner, proliferate rapidly, and secrete IFN-γ and RANTES. Our data suggest that CL-reactive γδ T cells could play a role in host responses to bacteria and, potentially, to self-CL by promoting IFN-γ and RANTES-mediated responses.
We present here the structure of mCD1d complexed with CL, the first structure of CD1d with a tetra-acylated phospholipid. While the structures of several CD1d-glycolipid complexes have been determined, the structures of only two CD1d-phospholipid complexes have been elucidated: endogenously bound PC (7) and exogenously added phosphatidylinositol-di-mannoside (PIM-2) (8). PC and PIM-2 bind in opposite orientations in the lipid binding groove, indicating a less stringent binding orientation for glycerolipids in general, as compared with the binding of ceramide-based ligands, such as sulfatide and α-GalCer. CL, like PIM-2, binds with the sn-1 fatty acid inserted into the A′ pocket, and the sn-2 fatty acid in the F′ pocket. In line with previous data on glycerolipid antigens from B. burgdorferi (41), we postulate that variations in the fatty acid composition of naturally occurring phosphoglycerolipids, such as length and saturation of the fatty acid (C14:0, C16:0, C18:0, C18:1, or C18:2), could influence the overall binding orientation of the glycerolipid in the binding groove of mCD1d, leading to subtle structural changes in the exposed T cell epitope and, possibly, altered T cell recognition. While both bovine heart CL and TM-CL interacted with mCD1d, hCL did not. Chemically, hCL differs from native bovine heart CL only in the saturation of its fatty acid chains. Intriguingly, the role of the third and fourth alkyl chain of CL in T cell activation is currently unclear. On a molecular level, both exposed alkyl chains are flexible and can be accommodated between the CD1-TCR interface, similar to the “squashed” model that has been proposed for the highly exposed iGb3 ligand (36). MonolysoCL and dilysoCL (bovine heart CL with one and two fatty acids, respectively, removed) bound to mCD1d in our binding studies. However, lipid loading into CD1d is directly linked to the solubility of the lipid in question. Our findings for CD1d binding are consistent with this, in that binding/loading of dilysoCL > monolysoCL > TM-CL. In contrast, in T cell proliferation studies, dilysoCL was only minimally stimulatory and monolysoCL was not stimulatory (and appeared to be toxic to the cells). These data suggest that the solvent-exposed third and fourth alkyl chains of CL may be important for T cell recognition and activation.
CL was recognized predominantly by hepatic, but also by splenic, γδ T cells. T cell stimulation occurred in a dose-dependent and CL-specific manner. Indeed, treatment with neither hCL nor PC induced similar T cell stimulation. In contrast, CL from both prokaryotic and eukaryotic cells was able to induce hepatic T cell proliferation to the same degree. The only difference between CL from different species is the fatty acid composition. Both E. coli CL and bovine heart CL are of natural origin and, as such, contain fatty acids that are both common (palmitic acid and stearic acid) and unique to prokaryotes (cyclopropyl fatty acids). Our limited data suggest that CL-reactive hepatic γδ T cells are not sensitive to the differences in fatty acid composition between eukaryotic and prokaryotic forms of CL, unlike invariant Vα14 NKT cells that have been reported to be sensitive to the fatty acid composition of Borrelia burgdorferi α-galactosyl diacylglycerolipids (44) at least in part as a result of the two opposite binding orientations of the lipids observed inside the CD1d groove (41). To fully address the question of fatty acid specificity of murine hepatic γδ T cells, a panel of synthetic versions of prokaryotic and eukaryotic CL is required. In contrast to similar recognition of E. coli and mammalian CL, murine hepatic γδ T cells clearly differentiated between saturated (hCL) and unsaturated (native CL) forms of mammalian CL. CL from different organisms are not characterized by unique degrees of fatty acyl chain length or unsaturation (dominant fatty acids may vary in length between 14 and 22 carbon atoms, and contain between 0 and 6 double bonds) (45). In light of these data, unsaturation is unlikely to be essential for the biological function of CL. However, as unsaturation tends to lower the transition temperature of a phospholipid, it can affect lipid solubility and fluidity. We have previously shown that CD1d-restricted recognition of phosphatidylethanolamine by a NKT cell hybridoma (24.8.A) correlated with the degree of acyl chain unsaturation (13). While it is tempting to speculate that CL induces γδ T cell activation because it is able to load more efficiently into the mCD1d molecule than hCL, it is equally possible that acyl chain unsaturation fine tunes the binding orientation of the phospholipid in the CD1d binding groove, leading to slight structural changes in the exposed T cell epitope which, possibly, affects T cell recognition.
Our finding that CL is able to trigger the proliferation and activation of γδ T cells in the context of CD1d reveals a new subset of γδ T cells. γδ T cells have a relatively limited diversity of TCRs and respond to Ags that include: (1) several conserved unconventional microbial Ags (e.g., phosphoantigens) (46), and (2) inducible self-Ags that reflect the status of a cell or tissue (e.g., heat shock proteins) (17). It is noteworthy that CL, a phospholipid that can be either bacterial or self in origin, falls into both of these γδ T cell Ag groups.
We speculate that CL-responsive γδ T cells may play a biological role in several settings, especially in host responses to infection in the liver. The liver is an important site of visceral infection and mucosal immunity, and the low hepatic blood pressure and high surface area in contact between blood and parenchymal cells provide pathogens with an easy route of access. Immune surveillance for pathogens in the liver is an important, but poorly understood, process. CL-responsive CD1d-restricted T cells are particularly well-suited to hepatic immune surveillance since CD1d is expressed on hepatocytes, Kupffer cells, hepatic DCs, and endothelial cells of the sinusoid lining (47), all of which have significant Ag uptake capabilities (48). CL is a major prokaryotic phospholipid expressed by many genera of bacteria (45). Thus, bacterial infections may result in circulating CL that could act as a danger signal and trigger protective γδ T cell responses. Indeed, transient increases of circulating human γδ T cells are observed during the early acute stage of several bacterial, parasitic, and viral infections (47,49–52). In mice, the H3 variant of Coxsackievirus B3 (CVB3) activates splenic γδ T cells that recognize CD1d expressed by CVB3-infected myocytes, resulting in severe myocarditis (53).
Our results show that upon exposure to CL, γδ T cells are activated and secrete significant amounts of IFN-γ and RANTES. Both cytokines are important in amplifying the innate immune response and bridging the transition from innate to adaptive immunity. IFN-γ, a pleiotropic cytokine known to orchestrate many distinct cellular programs such as macrophage-mediated killing of intracellular pathogens and antiviral immunity, has wide-reaching effects that extend beyond conventional immune cells to nonprofessional host defense cells (e.g., endothelial cells, fibroblasts and hepatocytes). In contrast, RANTES (or CCL5) is a chemokine that plays an active role in recruiting leukocytes to sites of inflammation. In the presence of cytokines such as IFN-γ, RANTES also induces the activation and proliferation of natural-killer cells (54). Furthermore, RANTES is known to be secreted by certain T effector cells (55). We propose that exposure of γδ T cells to CL may create a pro-inflammatory hepatic microenvironment, resulting in potent antimicrobial responses implicated in immune surveillance.
CL-responsive T cells could also be expanded in the context of CL presentation as a self-Ag. Normally, CL is present in the inner mitochondrial membrane of eukaryotic cells (45). However, cell death or injury may result in CL being released or expressed on the cell surface (45). Exposure of endogenous CL, in a pathologic or nonphysiologic context, may constitute a danger signal and trigger a CL-specific γδ T cell response. Autoimmune responses to CL and other phospholipids are known to occur in certain pathological contexts, such as systemic lupus erythematosus and anti-phospholipid syndrome (56), and it is possible that CL-responsive γδ T cells may be activated in these disease states. To date, there is no published data on human CL-reactive γδ T cells. However, peripheral blood-derived CD1-restricted human γδ T cells recognize pollen phospholipids (phosphatidylethanolamine and PC), produce both IFN-γ and IL-4, and provide help for IgE production (10,14). Phosphatidylethanolamine and PC also activate human CD1-restricted duodenal γδ T cells, but the cytokines (IL-4 and TGF-β) they produce appear to be regulatory in nature (11). CL was not evaluated in the latter studies. Furthermore, little is known about the recognition of phospholipid by human splenic or hepatic γδ T cells.
In summary, we demonstrate that CL, a tetra-acylated phospholipid, is a natural ligand for mCD1d. Furthermore, we show that a subset of γδ T cells, found in the normal immune repertoire, is activated both in vitro and in vivo by CL in a CD1d-restricted manner and secretes IFN-γ and RANTES. We propose that prokaryotic, and possibly also eukaryotic, CL may serve as a danger signal for mammalian T cells. We further propose that CD1d-bound CL induces a potent innate-like immune response, which, through the release of IFN-γ and RANTES, promotes the transition to an adaptive immune response. In this manner, CL-reactive γδ T cells may play a key role in immune surveillance for CL expression during infection or tissue injury.
Supplementary Material
Acknowledgments
The authors are grateful to Drs. Peter van den Elzen and Rusung Tan for providing the CD1d-deficient mice (originally derived by Drs. Steven Balk and Mark Exley), and to Drs. Michel Lafleur and Gerhard Wingender for their careful review and comments about the manuscript. We also thank Rebecca Subang and David Salem for technical assistance; and Maria Da Silva Martins and Ekaterina Yurchenko for expert advice, assistance, and discussion in the later phase of the project. This paper is dedicated by Dr. Mélanie Dieudé to the memory of Rita Larochelle Boucher.
Footnotes
This work was supported by Canadian Institutes of Health Research operating grants (MOP-42391 [JR], MOP-67101 [JR], and IMHA Priority Announcement MUS-67101 [JR]); National Institutes of Health grants (AI 074952 [DMZ] and HL 080330 [SMB]): a Genzyme Renal Innovations Program Award (Genzyme Corporation) (JSL); an investigator award from the Cancer Research Institute (DMZ); and post-doctoral fellowships from the Research Institute of the McGill University Health Centre (MD), the Department of Medicine of McGill University (MD), and the National Institutes of Health (F32 AI 80087 to AJT).
Structure factors and coordinates are deposited into the PDB database with code 3MA7.
References
- 1.Beckman EM, Porcelli SA, Morita CT, Behar SM, Furlong ST, Brenner MB. Recognition of a lipid antigen by CD1-restricted αβ+ T cells. Nature. 1994;372:691–694. doi: 10.1038/372691a0. [DOI] [PubMed] [Google Scholar]
- 2.Sieling PA, Chatterjee D, Porcelli SA, Prigozy TI, Mazzaccaro RJ, Soriano T, Bloom BR, Brenner MB, Kronenberg M, Brennan PJ, Modlin RL. CD1-restricted T cell recognition of microbial lipoglycan antigens. Science. 1995;269:227–230. doi: 10.1126/science.7542404. [DOI] [PubMed] [Google Scholar]
- 3.Moody DB, Reinhold BB, Guy MR, Beckman EM, Frederique DE, Furlong ST, Ye S, Reinhold VN, Sieling PA, Modlin RL, Besra GS, Porcelli SA. Structural requirements for glycolipid antigen recognition by CD1b-restricted T cells. Science. 1997;278:283–286. doi: 10.1126/science.278.5336.283. [DOI] [PubMed] [Google Scholar]
- 4.Kawano T, Cui J, Koezuka Y, Toura I, Kaneko Y, Motoki K, Ueno H, Nakagawa R, Sato H, Kondo E, Koseki H, Taniguchi M. CD1d-restricted and TCR-mediated activation of vα14 NKT cells by glycosylceramides. Science. 1997;278:1626–1629. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 5.Tsuji M. Glycolipids and phospholipids as natural CD1d-binding NKT cell ligands. Cell Mol Life Sci. 2006;63:1889–1898. doi: 10.1007/s00018-006-6073-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox D, Fox L, Tian R, Bardet W, Skaley M, Mojsilovic D, Gumperz J, Hildebrand W. Determination of cellular lipids bound to human CD1d molecules. PLoS One. 2009;4:e5325. doi: 10.1371/journal.pone.0005325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giabbai B, Sidobre S, Crispin MD, Sanchez-Ruiz Y, Bachi A, Kronenberg M, Wilson IA, Degano M. Crystal structure of mouse CD1d bound to the self ligand phosphatidylcholine: a molecular basis for NKT cell activation. J Immunol. 2005;175:977–984. doi: 10.4049/jimmunol.175.2.977. [DOI] [PubMed] [Google Scholar]
- 8.Zajonc DM, Ainge GD, Painter GF, Severn WB, Wilson IA. Structural characterization of mycobacterial phosphatidylinositol mannoside binding to mouse CD1d. J Immunol. 2006;177:4577–4583. doi: 10.4049/jimmunol.177.7.4577. [DOI] [PubMed] [Google Scholar]
- 9.Chang DH, Deng H, Matthews P, Krasovsky J, Ragupathi G, Spisek R, Mazumder A, Vesole DH, Jagannath S, Dhodapkar MV. Inflammation-associated lysophospholipids as ligands for CD1d-restricted T cells in human cancer. Blood. 2008;112:1308–1316. doi: 10.1182/blood-2008-04-149831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Russano AM, Agea E, Corazzi L, Postle AD, De LG, Porcelli S, de Benedictis FM, Spinozzi F. Recognition of pollen-derived phosphatidyl-ethanolamine by human CD1d-restricted γδ T cells. J Allergy Clin Immunol. 2006;117:1178–1184. doi: 10.1016/j.jaci.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 11.Russano AM, Bassotti G, Agea E, Bistoni O, Mazzocchi A, Morelli A, Porcelli SA, Spinozzi F. CD1-restricted recognition of exogenous and self-lipid antigens by duodenal γδ+ T lymphocytes. J Immunol. 2007;178:3620–3626. doi: 10.4049/jimmunol.178.6.3620. [DOI] [PubMed] [Google Scholar]
- 12.Gumperz JE, Roy C, Makowska A, Lum D, Sugita M, Podrebarac T, Koezuka Y, Porcelli SA, Cardell S, Brenner MB, Behar SM. Murine CD1d-restricted T cell recognition of cellular lipids. Immunity. 2000;12:211–221. doi: 10.1016/s1074-7613(00)80174-0. [DOI] [PubMed] [Google Scholar]
- 13.Rauch J, Gumperz J, Robinson C, Skold M, Roy C, Young DC, Lafleur M, Moody DB, Brenner MB, Costello CE, Behar SM. Structural features of the acyl chain determine self-phospholipid antigen recognition by a CD1d-restricted invariant NKT (iNKT) cell. J Biol Chem. 2003;278:47508–47515. doi: 10.1074/jbc.M308089200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agea E, Russano A, Bistoni O, Mannucci R, Nicoletti I, Corazzi L, Postle AD, De LG, Porcelli SA, Spinozzi F. Human CD1-restricted T cell recognition of lipids from pollens. J Exp Med. 2005;202:295–308. doi: 10.1084/jem.20050773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fox LM, Cox DG, Lockridge JL, Wang X, Chen X, Scharf L, Trott DL, Ndonye RM, Veerapen N, Besra GS, Howell AR, Cook ME, Adams EJ, Hildebrand WH, Gumperz JE. Recognition of lyso-phospholipids by human natural killer T lymphocytes. PLoS Biol. 2009;7:e1000228. doi: 10.1371/journal.pbio.1000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Born WK, Vollmer M, Reardon C, Matsuura E, Voelker DR, Giclas PC, O’Brien RL. Hybridomas expressing γδ T-cell receptors respond to cardiolipin and β2-glycoprotein 1 (apolipoprotein H) Scand J Immunol. 2003;58:374–381. doi: 10.1046/j.1365-3083.2003.01315.x. [DOI] [PubMed] [Google Scholar]
- 17.Born WK, Jin N, Aydintug MK, Wands JM, French JD, Roark CL, O’Brien RL. γδ T lymphocytes-selectable cells within the innate system? J Clin Immunol. 2007;27:133–144. doi: 10.1007/s10875-007-9077-z. [DOI] [PubMed] [Google Scholar]
- 18.Hickman HD, Luis AD, Buchli R, Few SR, Sathiamurthy M, VanGundy RS, Giberson CF, Hildebrand WH. Toward a definition of self: proteomic evaluation of the class I peptide repertoire. J Immunol. 2004;172:2944–52. doi: 10.4049/jimmunol.172.5.2944. [DOI] [PubMed] [Google Scholar]
- 19.Zajonc DM, Maricic I, Wu D, Halder R, Roy K, Wong CH, Kumar V, Wilson IA. Structural basis for CD1d presentation of a sulfatide derived from myelin and its implications for autoimmunity. J Exp Med. 2005;202:1517–1526. doi: 10.1084/jem.20051625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Otwinowski Z, Minor W. HKL: Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 21.Vagin AA, Teplyakov A. MOLREP:an automated programm for molecular replacement. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 22.Brunger AT. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 23.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 24.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 25.Schuttelkopf AW, van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 26.Winn MD, supov MNI, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 27.Howlin B, Butler SA, Moss DS, Harris GW, Driessen HPC. Tlsanl - Tls Parameter-Analysis Program for Segmented Anisotropic Refinement of Macromolecular Structures. J Appl Cryst. 1993;26:622–624. [Google Scholar]
- 28.Lovell SC, I, Davis W, Arendall WB, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Ca geometry: phi,psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 29.Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Exley M, Bigley N, Cheng O, Tahir S, Carter Q, Garcia J, Patten K, Stills H, Alt F, Snapper S, Balk S. Innate immune response to EMCV infection mediated by CD1. Immunol. 2003;110:519–526. doi: 10.1111/j.1365-2567.2003.01779.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roark JH, Park SH, Jayawardena J, Kavita U, Shannon M, Bendelac A. CD1.1 expression by mouse antigen-presenting cells and marginal zone B cells. J Immunol. 1998;160:3121–3127. [PubMed] [Google Scholar]
- 33.Zajonc DM, Kronenberg M. CD1 mediated T cell recognition of glycolipids. Curr Opin Struct Biol. 2007;17:521–529. doi: 10.1016/j.sbi.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zajonc DM, Cantu C, III, Mattner J, Zhou D, Savage PB, Bendelac A, Wilson IA, Teyton L. Structure and function of a potent agonist for the semi-invariant natural killer T cell receptor. Nat Immunol. 2005;6:810–818. doi: 10.1038/ni1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu D, Zajonc DM, Fujio M, Sullivan BA, Kinjo Y, Kronenberg M, Wilson IA, Wong CH. Design of natural killer T cell activators: structure and function of a microbial glycosphingolipid bound to mouse CD1d. Proc Natl Acad Sci U S A. 2006;103:3972–3977. doi: 10.1073/pnas.0600285103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zajonc DM, Savage PB, Bendelac A, Wilson IA, Teyton L. Crystal structures of mouse CD1d-iGb3 complex and its cognate Valpha14 T cell receptor suggest a model for dual recognition of foreign and self glycolipids. J Mol Biol. 2008;377:1104–1116. doi: 10.1016/j.jmb.2008.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 38.Jahng A, Maricic I, Aguilera C, Cardell S, Halder RC, Kumar V. Prevention of autoimmunity by targeting a distinct, noninvariant CD1d-reactive T cell population reactive to sulfatide. J Exp Med. 2004;199:947–957. doi: 10.1084/jem.20031389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blomqvist M, Rhost S, Teneberg S, Löfbom L, Østerbye T, Brigl M, Månsson J-E, Cardell SL. Multiple tissue-specific isoforms of sulfatide activate CD1d-restricted type II NKT cells. Eur J Immunol. 2009;39:1726–1735. doi: 10.1002/eji.200839001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zajonc DM, I, Wilson A. Architecture of CD1 proteins. Curr Top Microbiol Immunol. 2007;314:27–50. doi: 10.1007/978-3-540-69511-0_2. [DOI] [PubMed] [Google Scholar]
- 41.Wang J, Li Y, Kinjo Y, Mac TT, Gibson D, Painter GF, Kronenberg M, Zajonc DM. Lipid binding orientation within CD1d affects recognition of Borrelia burgorferi antigens by NKT cells. Proc Natl Acad Sci U S A. 2010;107:1535–1540. doi: 10.1073/pnas.0909479107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matsuda JL, Naidenko OV, Gapin L, Nakayama T, Taniguchi M, Wang CR, Koezuka Y, Kronenberg M. Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J Exp Med. 2000;192:741–754. doi: 10.1084/jem.192.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eberl G, Lees R, Smiley ST, Taniguchi M, Grusby MJ, Macdonald HR. Tissue-specific segregation of CD1d-dependent and CD1d-independent NK T cells. J Immunol. 1999;162:6410–6419. [PubMed] [Google Scholar]
- 44.Kinjo Y, Tupin E, Wu D, Fujio M, Garcia-Navarro R, Benhnia MR, Zajonc DM, Ben-Menachem G, Ainge GD, Painter GF, Khurana A, Hoebe K, Behar SM, Beutler B, Wilson IA, Tsuji M, Sellati TJ, Wong CH, Kronenberg M. Natural killer T cells recognize diacylglycerol antigens from pathogenic bacteria. Nat Immunol. 2006;7:978–986. doi: 10.1038/ni1380. [DOI] [PubMed] [Google Scholar]
- 45.Schlame M. Cardiolipin synthesis for the assembly of bacterial and mitochondrial membranes. J Lipid Res. 2008;49:1607–1620. doi: 10.1194/jlr.R700018-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beetz S, Wesch D, Marischen L, Welte S, Oberg HH, Kabelitz D. Innate immune functions of human γδ T cells. Immunobiology. 2008;213:173–182. doi: 10.1016/j.imbio.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 47.Geissmann F, Cameron TO, Sidobre S, Manlongat N, Kronenberg M, Briskin MJ, Dustin ML, Littman DR. Intravascular immune surveillance by CXCR6+ NKT cells patrolling liver sinusoids. PLoS Biol. 2005;3:e113. doi: 10.1371/journal.pbio.0030113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adams DH, Eksteen B, Curbishley SM. Immunology of the gut and liver: a love/hate relationship. Gut. 2008;57:838–848. doi: 10.1136/gut.2007.122168. [DOI] [PubMed] [Google Scholar]
- 49.de PP, Basaglia G, Gennari D, Santini G. γδ T cells in infectious diseases. Allergol Immunopathol (Madr) 1991;19:123–127. [PubMed] [Google Scholar]
- 50.Jason J, Buchanan I, Archibald LK, Nwanyanwu OC, Bell M, Green TA, Eick A, Han A, Razsi D, Kazembe PN, Dobbie H, Midathada M, Jarvis WR. Natural T, γδ, and NK cells in mycobacterial, Salmonella, and human immunodeficiency virus infections. J Infect Dis. 2000;182:474–481. doi: 10.1086/315740. [DOI] [PubMed] [Google Scholar]
- 51.Perera MK, Carter R, Goonewardene R, Mendis KN. Transient increase in circulating γδ T cells during Plasmodium vivax malarial paroxysms. J Exp Med. 1994;179:311–315. doi: 10.1084/jem.179.1.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oliaro J, Dudal S, Liautard J, Andrault JB, Liautard JP, Lafont V. Vγ9Vδ2 T cells use a combination of mechanisms to limit the spread of the pathogenic bacteria Brucella. J Leukoc Biol. 2005;77:652–660. doi: 10.1189/jlb.0704433. [DOI] [PubMed] [Google Scholar]
- 53.Huber S, Sartini D, Exley M. Role of CD1d in coxsackievirus B3-induced myocarditis. J Immunol. 2003;170:3147–3153. doi: 10.4049/jimmunol.170.6.3147. [DOI] [PubMed] [Google Scholar]
- 54.Appay V, Rowland-Jones SL. RANTES: a versatile and controversial chemokine. Trends Immunol. 2001;22:83–87. doi: 10.1016/s1471-4906(00)01812-3. [DOI] [PubMed] [Google Scholar]
- 55.Tikhonov I, Deetz CO, Paca R, Berg S, Lukyanenko V, Lim JK, Pauza CD. Human Vγ2Vδ2 T cells contain cytoplasmic RANTES. Int Immunol. 2006;18:1243–1251. doi: 10.1093/intimm/dxl055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Levine JS, Branch DW, Rauch J. The antiphospholipid syndrome. N Engl J Med. 2002;346:752–763. doi: 10.1056/NEJMra002974. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.