Summary
The glycine riboswitch regulates gene expression through the cooperative recognition of its amino acid ligand by a tandem pair of aptamers. A 3.6Å crystal structure of the tandem riboswitch from the glycine permease operon of Fusobacterium nucleatum reveals the glycine binding sites and an extensive network of interactions, largely mediated by asymmetric A-minor contacts, that serve to communicate ligand binding status between the aptamers. These interactions provide a structural basis for how the glycine riboswitch cooperatively regulates gene expression.
Introduction
Cooperative ligand binding, as exemplified by oxygen binding to hemoglobin, is typically thought to be the exclusive domain of proteins, but the discovery of the glycine riboswitch demonstrated that cooperative ligand binding also plays a role in RNA based systems (Barrick et al., 2004; Mandal et al., 2004). Riboswitches effect genetic control through the recognition of a specific small molecule ligand, and are typically found in the untranslated regions of mRNAs (Nudler and Mironov, 2004). Although some riboswitches make use of multiple domains to attenuate gene expression in response to changes in the concentration of one or more ligands, the tandem-aptamer glycine riboswitch is unique in that its two aptamers, or ligand binding domains, each recognize glycine and binding is cooperative (Figure 1A) (Lee et al., 2010; Mandal et al., 2004; Sudarsan et al., 2006; Welz and Breaker, 2007). The RNA acts as a transcriptional “on” switch to increase the expression of glycine transport and cleavage genes as concentrations of glycine increase, allowing greater flexibility for carbon metabolism in the host organism (Mandal et al., 2004). The ability to bind cooperatively allows the glycine riboswitch to transition from a fully off-state to a fully on-state across a narrow concentration gradient – improving the sensitivity of gene regulation (Kwon and Strobel, 2008; Mandal et al., 2004).
Figure 1.
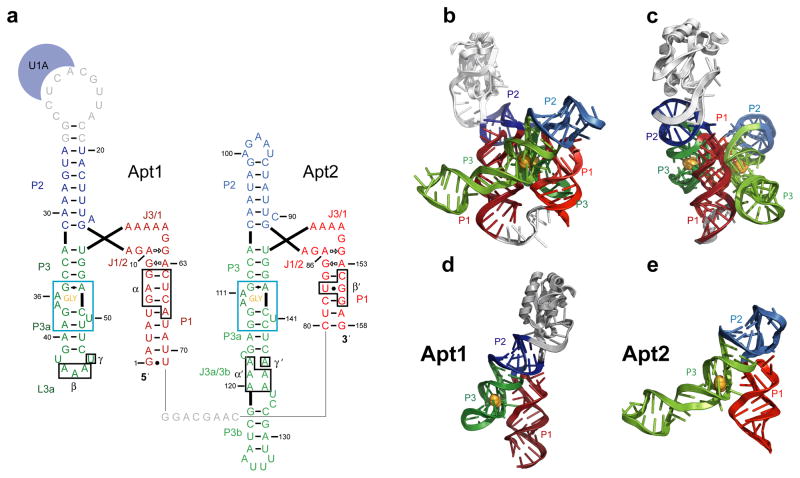
Structure of the glycine riboswitch tandem aptamer domain. (A) Secondary structure representation of the crystallized glycine aptamers. Helices P1, P2, and P3 are colored red, blue, and green respectively. Glycine is shown in orange and residues that form the glycine binding pocket are boxed. The U1A protein, U1A binding loop, and linker region are shown in gray. Numbering is the same as was previously used for this sequence (Kwon and Strobel, 2008). Watson-Crick base pairs are denoted by dashes. Other base pairs are indicated using standard nomenclature(Leontis and Westhof, 2001). Tertiary contacts between the aptamers are boxed and labeled α/α′, β/β′, andγ/γ′. (B) Crystal structure of the F. nucleatum glycine riboswitch aptamer domain. Coloring is the same as used in (A). (C) 90° rotation of (B). (D & E) Structures of the single aptamer domains, Apt1 (D) and Apt2 (E) See also Figure S1. Coloring is the same as in (A).
The glycine riboswitch is composed of two similar but non-identical aptamers connected by a flexible tether. Each aptamer contains three helical segments (P1, P2 and P3) that converge around a central loop (Figure 1A). Helices P1 and P3 and the joining segments J1/2 and J3/1 show high levels of sequence conservation, while the sequence and helix length of P2 are not conserved (Barrick et al., 2004; Mandal et al., 2004). Residues within P3, particularly the adenosines in the P3a bulge, and the purines in the central loop (J1/2 and J3/1) become more structured upon glycine binding (Kwon and Strobel, 2008; Mandal et al., 2004). Biochemical studies suggested that a tertiary contact necessary for cooperativity between the aptamers occurs between P3 of the second aptamer (Apt2) and P1 of the first (Apt1) (Erion and Strobel, 2011; Kwon and Strobel, 2008). These tertiary contacts form as a part of large conformational rearrangements that cause compaction in the RNA (Erion and Strobel, 2011; Kwon and Strobel, 2008; Lipfert et al., 2007). In order to understand the structural basis of cooperative ligand binding in an RNA system, we determined the crystal structure (3.6Å resolution) of the tandem-aptamer glycine riboswitch from the glycine permease operon (FN0328) of Fusobacterium nucleatum (Figure 1B; Figure 1C, Table 1, Figure S1).
Table 1.
Data collection and refinement statistics
| Data collection | native | 0.5mM iridium hexamine | ||
|---|---|---|---|---|
| inflection | remote | peak | ||
| Space group | P212121 | P212121 | P212121 | P212121 |
| Cell dimensions | ||||
| a, b, c (Å) | 54.6, 104.6, 156.5 | 53.5, 98.7, 154.5 | 53.4, 98.4, 154.1 | 52.7, 97.5, 154.4 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Wavelength | 1.079 | 1.1054 | 1.08 | 1.1050 |
| Resolution (Å) | 3.6 (3.73–3.60)* | 3.85 (3.99–3.85) | 3.6 (3.73–3.6) | 4.1 (4.25–4.1) |
| Rmerge | 0.108 (>1.0) | 0.06 (0.847) | 0.055 (0.745) | 0.108 (0.906) |
| I/σI | 19.5 (1.76) | 16.2 (1.45) | 17.0 (0.98) | 12.1 (0.93) |
| Completeness (%) | 99.8 (99.9) | 94.5 (83.3) | 95.3 (78.3) | 99.0 (96.7) |
| Redundancy | 13.5 (11.9) | 2.4 (2.8) | 2.4 (3.0) | 3.7 (2.8) |
| Phasing | ||||
| Figure of Merit | 0.35 (MAD); 0.57 (after density modification) | |||
| Refinement | ||||
| Resolution (Å) | 25–3.6 | |||
| No. reflections | 10796 | |||
| Rwork/Rfree | 28.2/31.0 | |||
| No. atoms | ||||
| Protein | 749 | |||
| RNA | 3624 | |||
| Ligand/ion | 23 | |||
| Water | 20 | |||
| B-factors | ||||
| Protein | 169 | |||
| RNA | 200 | |||
| Ligand/ion | 154 | |||
| Water | 116 | |||
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.013 | |||
| Bond angles (°) | 1.32 | |||
Values in parenthesis are for highest-resolution shell.
Results and Discussion
Global description of the riboswitch structure
Based on the crystal structure, both aptamers adopt a similar y-shaped structure and interact to create a pseudosymmetric dimer (Figure 1). Within an individual aptamer the three helices are co-planar (Figure 1D; Figure 1E). Helices P2 and P3 stack to form an extended helix, and the third helix, P1, branches away from P2–P3 at an angle of 50° relative to helix P3. Several of the purine nucleotides in the central loop are unpaired but they form a continuous chain of stacked residues with P1. The two aptamers “dimerize” by packing the P1 helix of one aptamer against the P3 helix of the other resulting in a tertiary structural fold that is three helices thick. The P1 helices from both aptamers are adjacent and parallel to each other in the middle of the fold, sandwiched on both sides by a P2–P3 helix (Figure 1B; Figure 1C).
Ligand recognition and binding pocket structure of the glycine riboswitch
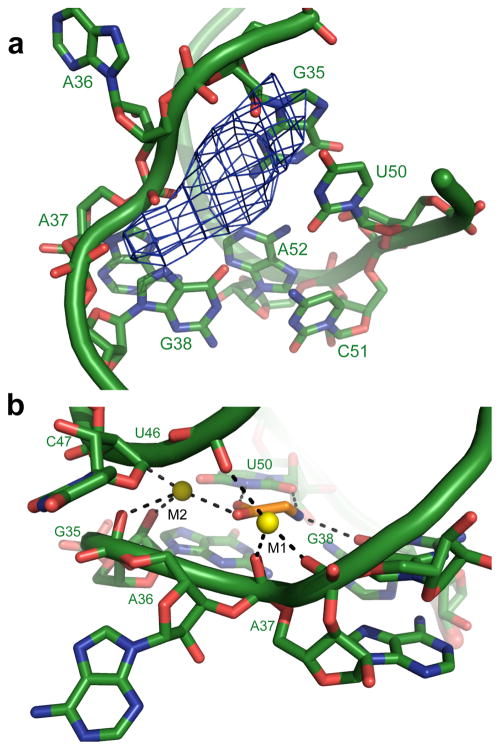
The riboswitch binds two molecules of glycine, one to each aptamer (Figure 2). In both cases the asymmetric A-rich bulge between helices P3 and P3a forms the amino acid binding pocket. Modification to nucleotides in P3a showed strong and symmetrical interference effects consistent with an important role in ligand binding (Kwon and Strobel, 2008). The two binding sites are close together in three dimensional space, with the glycines less than 25Å apart. A key feature of each binding site is U50 (Apt1 numbering), a universally conserved residue predicted to form a wobble pair with the opposing G (G38) in helix P3a. Instead, G38 pairs with C51, and U50 presents its Watson-Crick face to hydrogen bond with the amino acid (Figure 2A). The entrance to the glycine binding site is framed by the phosphate backbone of residues G35, A36, A37, G38, U46, and C47. The close approach of these phosphates is stabilized by a pair of metals (M1 and M2) that allow the base of A36 to flip out of the binding pocket and into the central loop where it mediates a cross-strand stack near the top of the P1 helix (Figure 2B; Figure S2A). Displacement of A36 creates a pocket into which glycine can bind and is likely to be a key element in cooperativity (see below).
Figure 2.
Glycine binding pocket. (A) Glycine binding pocket in Apt1. Coloring is the same as in (Figure 1A). Fo-Fc maps generated from a model lacking glycine and metals is shown in blue (contoured at 2.8σ). Universally conserved U50 presents its Watson-Crick face to the binding pocket, while A36 is flipped out to create a cavity for glycine binding. (B) Glycine binding pocket metals M1 and M2 (shown in yellow). See also Figure S2.
We observe clear density for glycine, but the symmetrical character of the ligand and relatively low resolution of the structure are insufficient to unambiguously orient the amino acid within the binding pocket. One chemically plausible orientation consistent with the electron density has the carbonyl of glycine coordinated to M2 and the N3 of U50, while the amine hydrogen bonds with the O2 carbonyl group of U50 and the O6 carbonyl of G38 (Figure 2B). In this orientation, the α-carbon of the amino acid makes Van der Waals contact with the major groove functional groups of the U49:A39 base pair, which may explain how the riboswitch is able to distinguish glycine from alanine and all other amino acids with a side chain. There is sufficient space near the carboxylic acid group to accommodate the glycine methyl ester, one of the few glycine derivatives that retain some affinity for the RNA (Mandal et al., 2004). Alternatively the glycine may occupy the reverse orientation with its carbonyl coordinated to M1 and the amino group hydrogen-bonded with the O2 of U50 (Figure S2B). The first model is a better fit to the binding geometry and electron density, but in both models U50 and metal ions play a prominent role in glycine recognition.
Sites of interaction between aptamers
The global structure of the tandem aptamer reveals the inter-aptamer contacts that play a role in cooperative ligand binding. These are predominantly pseudo-dyad symmetric interactions between the P1 region of one aptamer and the P3 stem of the other. There are three sets of interactions that comprise the inter-aptamer contacts, which we have termed α, β, and γ (Figure 1A; Figure 3).
Figure 3.
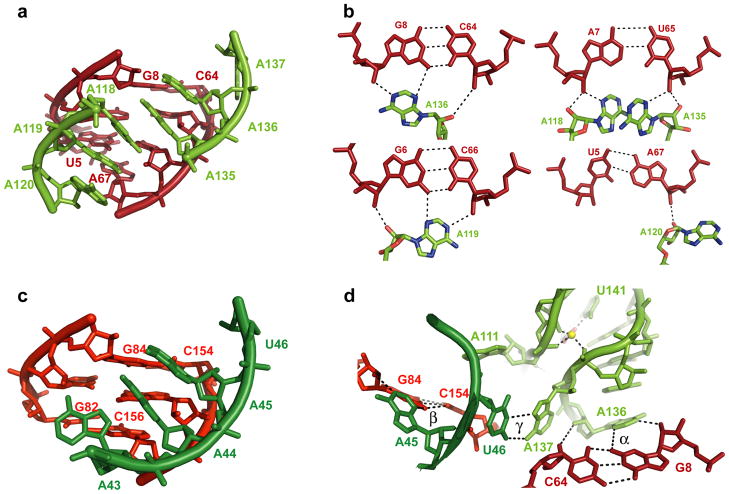
Inter-aptamer interactions leading to cooperative ligand binding. (A) Global view of cooperative interface α, Apt1 P1 shown in dark red and Apt2 J3a/3b shown in light green. 2Fo-Fc map contoured at 1σ displayed in light blue. (B) A-minor motif interactions that compose the α cooperative interface. See also Figure S3. (C) Global view of interface β, Apt1 L3a shown in dark green and Apt2 P1 shown in red. (D) Global view of interface γ. Coloring is the same as in (1A). Nucleotides in γ lie directly between cooperative interfaces α or β and the ligand binding pocket.
The α interaction is the most extensive of the interaptamer contacts. It forms between the top three base pairs of P1 in Apt1 and the Apt2 A-rich bulge between P3a and P3b (J3a/3b), just three base pairs from the glycine binding site. Five adenosines, two from one side (A135 and A136) and three from the other side (A118-A120) of the J3a/3b asymmetric loop, create a continuous base stack and form five consecutive A-minor interactions with the minor groove edge of P1 (Figure 3A; Figure 3B) (Nissen et al., 2001). The first and third base pairs of the P1 helix (G8-C64 and G6-C66) each form a type I A-minor interaction (with A136 and A119 respectively) (Figure 3B), while the second base pair (A7-U65) forms two type II A-minor contacts (A135 and A118), one on each strand of the helix (Figure 3B). Even the fourth base pair (U5-A67) is contacted, albeit less extensively by a type 0 A-minor interaction with A120 (Figure 3B). A number of these interactions explain previous biochemical observations, such as the critical role of the 2′-OHs of A7, A118, and A135 for riboswitch function (Kwon and Strobel, 2008). Consistent with the role of these interactions in cooperative ligand binding, mutation of the third base pair of P1 (a C-G base pair) to a U G in the Vibrio cholerae glycine riboswitch of the VC1422 gene resulted in loss of cooperativity, presumably by disrupting the formation of the type I A minor interaction (Kwon and Strobel, 2008).
The β interaction is thematically similar but less extensive than the contacts made in α. This tertiary contact involves the matching regions of the two aptamers, namely Apt1 adenosines in the L3a loop and the P1 stem of Apt2 (Figure 3C). Like α, the β interaction is within a few base pairs of a glycine binding site, but in this case only three base pairs of Apt2 P1 are contacted (Figure S3). As in α, the first base pair in P1 (G84-C154) is recognized by a type I A-minor interaction (with A45), but the second base pair (U83-G155) is contacted by just a single type II interaction (G155 with A44). The difference between the α and β contacts are even more evident at the third base pair (C82-G156) which instead of forming an extensive type I contact, makes a single hydrogen bond to A43, and no contacts are made to the fourth base pair. The extent of the β interaction appears to be limited by the geometry of the Apt1 adenosines, which are constrained by the sharp hairpin turn of L3a. The smaller interface of the β relative to the α interaction is consistent with NAIM and biochemical results which showed that the third base pair of P1 in Apt1 was critical for cooperativity, but minor groove chemical modifications and even a wobble pair were fully tolerated at the third base pair of Apt2 (Kwon and Strobel, 2008).
A third inter-aptamer contact (γ) is positioned such that it may play an important role in communicating the status of ligand binding between the two binding pockets. The γ interaction forms between U46 in Apt1 and A137 in Apt2 (Figure 3D). A137 flips out of the helical stack and adopts a syn conformation which allows it to make a Hoogsteen base pair with U46. These two nucleotides are flanked on the 5′ end by the adenosines that form the α and β interfaces and flanked on the 3′ end by nucleotides that form the glycine binding pocket. While the particular nucleotides are not highly conserved, multiple sequence alignments indicate that when one nucleotide is a U there is a bias towards the interaction partner occurring as an A. The sum of these tertiary interactions results in an RNA that can communicate the ligand binding status from one aptamer to the other (Erion and Strobel, 2011; Kwon and Strobel, 2008; Mandal et al., 2004).
Subsequent to the submission of this paper, the structure of a single aptamer glycine riboswitch was independently reported by Huang et al at 2.85Å resolution (Huang et al., 2010). It is noteworthy that the major features of an isolated aptamer, including helix orientation, nucleotide displacement from the binding pocket and glycine binding appear to be consistent with that reported for the single aptamer. The core region of each structure superposes well, with an average displacement of just 0.92Å and the glycine orientation depicted in Figure 2B matches that reported by Huang et al. Although their structure is of an isolated single aptamer, the RNA forms a homodimer within the crystal’s asymmetric unit. Based upon the interaptamer contacts made in the crystal, they proposed that A-minor interactions between P1 and P3 are responsible for cooperative ligand binding. Our structure confirms that the A minor interactions (α, β) present in their crystal contacts do occur in a natural tandem glycine riboswitch, and reveals the asymmetric character of these interactions. They also observe interaptamer pi-stacking interactions between the two A65 residues, which are structurally different, but thematically similar to the noncanonical γ interaction between U46 and A137 that is present in this dual aptamer structure.
Significance
This structure in concert with previous biochemical data suggest a mechanism for cooperative ligand binding. The question is how does glycine binding to one aptamer promote glycine binding to the second? In hemoglobin, the classic model for cooperative ligand binding, structural changes between subunits are triggered by ligand binding at one site resulting in a transition from the low affinity (T) to high affinity (R) state (Ackers et al., 1992; Royer et al., 2001; Turner et al., 1992). Many of the fundamental elements observed in hemoglobin cooperativity are consistent with structural features of cooperative glycine binding to this RNA (Perutz, 1970). The J1/3, J3/1, and P1 regions become more structured with the addition of glycine, likely due to a series of stacking interactions that can only form when A36 or A111 are displaced from the binding pocket (Kwon and Strobel, 2008; Mandal et al., 2004). Stabilization of the P1 region enhances formation of inter-aptamer contacts α or β, which have been shown to play a role in cooperativity (Erion and Strobel, 2011; Kwon and Strobel, 2008). Our structure reveals the close proximity of all the inter-aptamer contacts to the two binding pockets (α and β are within 11Å of glycine, γ is within 8Å), which suggests that structural rearrangements induced by glycine binding in one pocket are translated into conformational changes that enhance ligand binding in the other pocket through these interfaces. In the case of both hemoglobin and the glycine riboswitch, structural rearrangements induced when the first ligand is bound trigger changes in inter-domain contacts that stabilize a state more competent to bind a second ligand.
Experimental Procedures
Crystallization, Data Collection, and Refinement
The Fusobacterium nucleatum tandem aptamer glycine riboswitch was crystallized by hanging drop vapor diffusing using 25% PEG MME550, 0.05M HEPES pH 7.0, 0.01M MgCl2, 0.025M sodium acetate pH 4.5. The crystals were cryoprotected in a stabilization solution containing 32% PEG MME550, 0.05M HEPES pH 7.0, 0.02M MgCl2, 0.025M sodium acetate pH 4.5 before freezing into liquidnitrogen. Native data were collected at beamline X25 at BNL. Derivative data were collected at beamline 24-ID-C at APS from an iridium hexamine soaked crystal. Data were processed with HKL2000 (Otwinowski, 1997). Eleven Ir atom sites were located in SHELXD using a two wavelength MAD dataset as well as a SAD dataset (peak) collected at a different position of the same crystal (Sheldrick, 2008; Terwilliger and Berendzen, 1999). Initial maps were improved by multi-domain multicrystal averaging in DMMulti using the native dataset, the nonisomorphous iridium soak dataset (Riso=57%), a native data set, and the U1A region from the hairpin ribozyme dataset (3HHN) (Cowtan and Main, 1996; Potterton et al., 2002). The resulting maps were of sufficient quality to build an initial model, which was refined in Refmac v.5.5.0110 (Murshudov et al., 1997). Model building was done in Coot with the aid of the B-factor sharpening function (Emsley et al., 2010). Electron density attributed to be the ligand was relatively weak using calculated model phases. To improve this, the binding pocket density was averaged between the two aptamers in both the native and iridium datasets, resulting in significant improvement in density for Apt1 (Cowtan and Main, 1996; Potterton et al., 2002).
Supplementary Material
Acknowledgments
We thank A. Heroux and the beamline staff at X25, X12C, and X29 at the National Synchrotron Light Source at Brookhaven National Laboratory; F. Murphy and J. Schuermann and the beamline staff at 24-ID-C and 24-ID-E at the Advanced Photon Source at Argonne National Laboratory; M. Strickler, D. Keller, and the Yale Center for Structural Biology core staff; M. Kwon, K. Smith, and T. Erion for help and advice; N. Carrasco, D. Hiller, T. Erion, K. Smith and other members of the Strobel lab for helpful discussions; V. Singh (Strobel lab) for synthesis of iridium hexammine. This work was supported by NIH grant GM02278 to S.A.S.
Footnotes
ACCESSION NUMBERS
Coordinates and structure factors for the glycine riboswitch are available in the Protein Data Bank (http://www.rcsb.org/) under ID code 3P49.
AUTHOR CONTRIBUTIONS
E.B.B. determined the crystal structure and wrote the manuscript. X.Y. gave critical help in phasing and refinement. J.W. gave critical help in model building and refinement. S.A.S. advised on the project and wrote the manuscript.
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests.
Contributor Information
Ethan B. Butler, Email: ethan.butler@yale.edu, Department of Molecular Biophysics and Biochemistry Yale University, New Haven CT 06520-8114, USA
Yong Xiong, Email: yong.xiong@yale.edu, Department of Molecular Biophysics and Biochemistry Yale University, New Haven CT 06520-8114, USA.
Jimin Wang, Email: jimin.wang@yale.edu, Department of Molecular Biophysics and Biochemistry Yale University, New Haven CT 06520-8114, USA.
Scott A. Strobel, Email: scott.strobel@yale.edu, Department of Molecular Biophysics and Biochemistry Yale University, New Haven CT 06520-8114, USA
References
- Ackers GK, Doyle ML, Myers D, Daugherty MA. Molecular code for cooperativity in hemoglobin. Science. 1992;255:54–63. doi: 10.1126/science.1553532. [DOI] [PubMed] [Google Scholar]
- Barrick JE, Corbino KA, Winkler WC, Nahvi A, Mandal M, Collins J, Lee M, Roth A, Sudarsan N, Jona I, et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc Natl Acad Sci U S A. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowtan KD, Main P. Phase combination and cross validation in iterated density-modification calculations. Acta Crystallogr D Biol Crystallogr. 1996;52:43–48. doi: 10.1107/S090744499500761X. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erion TV, Strobel SA. Identification of a tertiary interaction important for cooperative ligand binding by the glycine riboswitch. RNA; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Serganov A, Patel DJ. Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch. Mol Cell. 2010;40:774–786. doi: 10.1016/j.molcel.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M, Strobel SA. Chemical basis of glycine riboswitch cooperativity. RNA. 2008;14:25–34. doi: 10.1261/rna.771608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ER, Baker JL, Weinberg Z, Sudarsan N, Breaker RR. An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science. 2010;329:845–848. doi: 10.1126/science.1190713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipfert J, Das R, Chu VB, Kudaravalli M, Boyd N, Herschlag D, Doniach S. Structural transitions and thermodynamics of a glycine-dependent riboswitch from Vibrio cholerae. J Mol Biol. 2007;365:1393–1406. doi: 10.1016/j.jmb.2006.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, Breaker RR. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004;306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc Natl Acad Sci U S A. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudler E, Mironov AS. The riboswitch control of bacterial metabolism. Trends Biochem Sci. 2004;29:11–17. doi: 10.1016/j.tibs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Otwinowski ZMW. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Perutz MF. Stereochemistry of cooperative effects in haemoglobin. Nature. 1970;228:726–739. doi: 10.1038/228726a0. [DOI] [PubMed] [Google Scholar]
- Potterton E, McNicholas S, Krissinel E, Cowtan K, Noble M. The CCP4 molecular-graphics project. Acta Crystallogr D Biol Crystallogr. 2002;58:1955–1957. doi: 10.1107/s0907444902015391. [DOI] [PubMed] [Google Scholar]
- Royer WE, Jr, Knapp JE, Strand K, Heaslet HA. Cooperative hemoglobins: conserved fold, diverse quaternary assemblies and allosteric mechanisms. Trends Biochem Sci. 2001;26:297–304. doi: 10.1016/s0968-0004(01)01811-4. [DOI] [PubMed] [Google Scholar]
- Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- Sudarsan N, Hammond MC, Block KF, Welz R, Barrick JE, Roth A, Breaker RR. Tandem riboswitch architectures exhibit complex gene control functions. Science. 2006;314:300–304. doi: 10.1126/science.1130716. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner GJ, Galacteros F, Doyle ML, Hedlund B, Pettigrew DW, Turner BW, Smith FR, Moo-Penn W, Rucknagel DL, Ackers GK. Mutagenic dissection of hemoglobin cooperativity: effects of amino acid alteration on subunit assembly of oxy and deoxy tetramers. Proteins. 1992;14:333–350. doi: 10.1002/prot.340140303. [DOI] [PubMed] [Google Scholar]
- Welz R, Breaker RR. Ligand binding and gene control characteristics of tandem riboswitches in Bacillus anthracis. RNA. 2007;13:573–582. doi: 10.1261/rna.407707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.