Abstract
Several nuclear receptors (NRs) are still characterized as orphan receptors since ligands have not yet been identified for these proteins. The retinoic acid receptor-related receptors (RORs) have no well-defined physiological ligands. Here, we describe the identification of a selective RORα synthetic ligand, SR3335 (ML-176). SR3335 directly binds to RORα, but not other RORs, and functions as a selective partial inverse agonist of RORα in cell-based assays. Furthermore, SR3335 suppresses the expression of endogenous RORα target genes in HepG2 involved in hepatic gluconeogenesis including glucose-6-phosphatase and phosphoenolpyruvate carboxykinase. Pharmacokinetic studies indicate that SR3335 displays reasonable exposure following an i.p. injection into mice. We assess the ability of SR3335 to suppress gluconeogenesis in vivo using a diet induced obesity (DIO) mouse model where the mice where treated with 15 mg/kg b.i.d., i.p. for 6-days followed by a pyruvate tolerance test. SR3335 treated mice displayed lower plasma glucose levels following the pyruvate challenge consistent with suppression of gluconeogenesis. Thus, we have identified the first selective synthetic RORα inverse agonist and this compound can be utilized as a chemical tool to probe the function of this receptor both in vitro and in vivo. Additionally, our data suggests that RORα inverse agonists may hold utility for suppression of elevated hepatic glucose production in type 2 diabetics.
Several nuclear receptors (NRs) are still characterized as orphan receptors since ligands have not yet been identified for these proteins. NRs display a conserved domain structure with highly conserved DNA-binding and ligand-binding domains. The retinoic acid receptor-related orphan receptor α(RORα) is an orphan receptor that has been demonstrated to play an important role in regulation of metabolism (1, 2). Cholesterol and its sulfate derivative have been suggested to be natural ligands for RORα (3, 4). More recently, our work identified various oxygenated sterols that bind to both RORα and RORγ with high affinity and regulate their activity (5, 6). The RORs have been characterized as constitutively active receptors displaying the ability to activate transcription in the absence of a ligand; however, there is some controversy as to the nature of this constitutive activity. Our data indicate that RORs display the constitutive activity in biochemical assays under conditions where the receptor would be expected to have no ligand present (denatured and refolded receptor) (5), but others have suggested that endogenous ligands may copurifiy with the receptor leading to this activity (7). Although the physiological significance of these natural ligands for the RORs is unclear, the potential utility of synthetic ligands that modulate the activity of these receptors is apparent. For example, loss of RORα in the staggerer mice results in mice resistant to weight gain and hepatic steatosis when placed on a high fat diet suggesting that suppression of RORα activity may offer efficacy in treatment of obesity(8). RORα has also been shown to be a critical factor regulating the expression of key enzymes in the gluconeogenic pathway (9) and suppression of RORα activity may lead to suppression of elevated hepatic glucose output that is observed in type 2 diabetics.
We recently identified the first synthetic ligand that binds to and regulates the activity of RORα and RORγ, T0901317 (T1317) (Fig. 1A) (10). T1317 was originally identified as a liver X receptor agonist (LXR) (11), an NR that serves as a physiological receptor for oxysterols and plays key roles in regulation of lipogenesis and reverse cholesterol transport (12). Our group demonstrated that T1317 displays a degree of promiscuity and also activated another NR that serves as a receptor for bile acids, FXR (13). Interestingly, T1317 acts as a LXR agonist, but a ROR inverse agonist. We utilized the benzenesulfonamide scaffold as an initiation point for development of the first selective ROR ligand, SR1078 that behaves as a dual RORα/γ agonist (14).
Figure 1.
Identification of a selective RORα synthetic ligand, SR3335. A) Comparison of the chemical structure of T0901317 to SR3335 and SR1078. B) Scheme illustrating the synthesis of SR3335. C) Competition radioligand binding assay illustrating the ability of SR3335 to displace radiolabeled 25-hydroxycholesterol from RORα LBD. D) Competition radioligand binding assay illustrating the inability of SR3335 to displace radiolabeled 25-hydroxycholesterol from RORγ LBD.
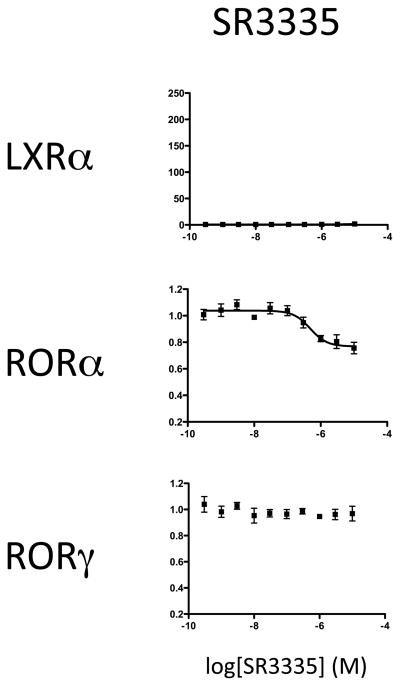
Continued evaluation of this scaffold led to the identification of a RORα selective inverse agonist that is characterized in this study, SR3335 (ML-176) (Fig. 1A). The synthetic scheme for SR3335 is shown in Fig. 1B. This compound was initially identified based on its ability to inhibit the constitutive activity of RORα in a GAL4-RORα ligand binding domain (LBD) cotransfection assay. In a biochemical radioligand binding assay using [3H]25-hydroxycholesterol as a label (5, 6, 10) it is clear that unlabeled SR3335 dose-dependently competes for binding to the RORα LBD (Fig. 1C). The Ki was calculated as 220 nM using the Cheng-Prusoff equation. As shown in Fig. 1D, SR3335 did not compete well for binding when the LBD of RORγ was utilized. In a cell-based chimeric receptor Gal4 DNA-binding domain – NR ligand binding domain cotransfection assay, SR3335 significantly inhibited the constitutive transactivation activity of RORα (IC50=480 nM)(partial inverse agonist activity), but had no effect on the activity of LXRα and RORγ (Fig. 2). Although T1317 shows considerably more efficacy than SR3335 in terms of suppression of RORα activity, the RORα selectivity of SR3335 is clear. SR3335 also displays no activity on RORβ (radioligand binding or cotransfection assays), FXR (cotransfection assays) or any other receptors in a selectivity panel for human nuclear receptors (10) (data not shown). We also observed no effect on the enzymatic activity of kinases (JNK or MAPK). These data clearly demonstrate that we developed a compound that selectively targets RORα.
Figure 2.
SR3335 is a selective RORα partial inverse agonist. Cotransfection of HEK293 cells with RORα, RORγ or LXRα LBD fused to a GAL4 DNA binding domain and a reporter containing 5 copies of the GAL4 UAS upstream of a luciferase reporter. The effect of T1317 is compared to SR3335 in each assay.
In order to examine the activity of SR3335 in more detail, we performed additional cotransfection assays where we transfected cells with full-length RORα and a luciferase reporter gene driven by a promoter derived from a known ROR target gene, glucose-6-phosphatase (G6Pase). G6Pase is a well-characterized RORα target gene that plays a critical role in the gluconeogenesis pathway (5, 9, 15). As shown in Fig. 3A, in a RORα cotransfection assay, treatment of cells with SR3335 resulted in a significant suppression of transcription driven by the G6Pase promoter. Consistent with these cotransfection data, treatment of HepG2 cells with SR3335 lead to suppression of expression of both G6Pase and phosphoenolpyruvate carboxykinase (PEPCK) mRNA expression (Fig. 3B). Given the critical roles that these two enzymes play in regulation of gluconeogenesis, we hypothesized that SR3335 may offer utility in suppression of hepatic glucose output, which is elevated in type 2 diabetics and contributes to the hyperglycemic state. In order to investigate this, we pursued additional studies in mice.
Figure 3.
SR3335 suppresses the expression of RORα target genes. A) Treatment of HepG2 cells with 5 μM SR3335 results in suppression of transcription in a full-length RORα, G6Pase promoter-luciferase reporter cotransfection assay. G6Pase expression was normalized to cyclophilin. B) Treatment of HepG2 cells with 5 μM SR3335 results in suppression of G6Pase and PEPCK mRNA expression. *, indicates p<0.05.
We examined the pharmacokinetic properties of SR3335 in mice and noted significant in vivo exposure. Plasma concentrations reached nearly 9 μM 0.5h after a 10 mg/kg i.p. injection of SR3335 and levels were sustained above 360 nM even 4h after the single injection (Fig. 4A). These levels were sufficient to perform a proof-of-principle experiment to determine if SR3335 treatment could suppress gluconeogenesis in vivo. Diet induced obese mice were treated with SR3335 (15 mg/kg b.i.d., i.p.) for 6-days and a pyruvate tolerance test was performed on day 6 to estimate gluconeogenesis. As shown in Fig. 4B plasma glucose levels were slightly lower in SR3335 treated animals at time 0, but after injection of the pyruvate the SR3335 treated animals displayed significantly lower plasma glucose levels at each time point vs. vehicle treated animals (15, 30, and 60 min) indicating suppression of hepatic gluconeogenesis and an improvement in glucose homeostasis by the RORα inverse agonist. Expression of hepatic pepck and g6pase expression in the mice revealed ~50% decrease in pepck expression (the enzyme that catalyzes the rate limiting step in gluconeogenesis); however, g6pase expression was not significantly affected (Fig. 4C). It is unclear why only pepck expression was suppressed in vivo and not g6pase since both were repressed in cell culture experiments, but the suppression of only pepck in vivo may be responsible for the moderate effects on pyruvate stimulated gluconeogenesis. Clearly SR3335 is effectively targeting RORα since a well characterized RORα target gene (nr1d1) (16,17) is also repressed in the livers (Fig. 4C). Importantly, mice treated with SR3335 displayed no difference in body weight or food intake after 7-days of treatment with the compound (data not shown). The effects on glucose homeostasis are thus not secondary to weight loss and represent a metabolic response to the compound.
Figure 4.
SR3335 suppresses gluconeogenesis in vivo. A) Pharmcokinetic profile SR3335 following a single injection of 10 mg/kg i.p. B) Pyruvate tolerance test in diet induced obese (DIO) mice (C57Bl/6) following 1 week of b.i.d. dosing (i.p.) 15 mg/kg. C) Gene expression in mice following administration of SR3335 as inidicated in 4B. Gene expression was normalized to cyclophilin. *, indicates p<0.05.
Several crystal structures of the LBD of both RORα and RORγ bound to sterol ligands have been solved (3, 4, 7). In all of these cases the LBD appears to be in an active conformation with helix 12 positioned in such a manner to allow for coactivator protein recruitment. It is unclear whether binding of a sterol is required for the transcriptional activity of these receptors since we have clearly observed that RORα and RORγ expressed in a sterol free environment retain constitutive ability to recruit coactivators (5,10). We do observe that SR3335 is able to displace 25-hydroxycholesterol in a radioligand binding assay, thus whether it be by displacing an endogenous agonist or by binding to a receptor that has a basal conformation that is already active, SR3335 appears to limit the receptor’s ability to activate transcription. This is most likely due to SR3335 inducing a conformation that reduces the affinity of the LBD for coactivators.
In summary, we report the identification of the first selective synthetic RORα ligand that functions as an inverse agonist. In cotransfection assays, SR3335 suppresses transcription in both GAL4-RORα LBD and full-length RORα contexts. Furthermore, treatment of HepG2 cells with SR3335 results in suppression of RORα target gene expression. Suppression of the expression of G6Pase and PEPCK mRNA suggested that SR3335 might be able to suppress gluconeogenesis. After determining that SR3335 displayed reasonable pharmacokinetics in mice, we tested this hypothesis and in vivo using a DIO mouse model and showed that SR3335 did indeed suppress gluconeogenesis. These data clearly define SR3335 as a valid chemical tool to evaluate the in vitro and in vivo actions of RORα and suggest that compounds like SR3335 may hold utility in treatment of type 2 diabetes.
Methods
Synthesis of SR3335
To a solution of 2-(4-aminophenyl)-1,1,1,3,3,3-hexafluoropropan-2-ol (18) (1.5M in THF, 2.90 mL, 4.35 mmol) in acetone (4.3 mL) were successively added at room temperature 2,6-lutidine (658 μL, 5.65 mmol) and 2-thiophenesulfonyl chloride (910 mg 4.78 mmol). The mixture was heated overnight at 60°C, then diluted by ethyl acetate (EtOAc) and quenched at room temperature by the addition of saturated NaHCO3 solution. The aqueous phase was extracted two times with EtOAc. The combined organic phases were dried over Na2SO4, filtrated and concentrated on a rotary evaporator. The residue was purified by silica gel column and eluted with hexane-EtOAc (70/30) to obtain 1.1 g of SR3335 (62%, purity >98%) as a white powder: 1H NMR (400 MHz, (CD3)2SO) d 6.64 (dd, J = 5.0, 3.8 Hz, 1H), 6.77 (d, J = 8.8 Hz, 2H), 7.08 (d, J = 8.8 Hz, 2H), 7.14 (dd, J = 3.7, 1.4 Hz, 1H), 7.43 (d, J = 5.0, 1.4 Hz, 1H), 8.11 (s, 1H), 10.30 (s, 1H); 13C NMR (100 MHz, (CD3)2SO) d 119.1 (2C), 125.9, 127.7, 127.9 (2C), 132.7, 133.7, 139.2, 139.9; the three carbon resonances of the hexafluoropropanol unit are not observed in the 13C spectrum of SR3335. The fluorine coupling with these carbons gives multiplets which were difficult to detect even with increased number of scans; FTIR 3362, 3228, 1614, 1519, 1472, 1404, 1341, 1308, 1280, 1257, 1230, 1186, 1146, 1098, 1067, 1027, 963, 946, 928, 832, 821, 730, 708 cm−1; MS (ES-) m/z = 404 (found for C13H9F6NO3S2H+).
Cell Culture and Cotransfections
HEK293 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum at 37 °C under 5% CO2. HepG2 cells were maintained and routinely propagated in minimum essential medium supplemented with 10% fetal bovine serum at 37 °C under 5% CO2. 24 h prior to transfection, cells were plated in 96-well plates at a density of 15 × 103 cells/well. Transfections were performed using LipofectamineTM 2000 (Invitrogen). 16 h post-transfection, the cells were treated with vehicle or compound. 24 h post-treatment, the luciferase activity was measured using the Dual-GloTM luciferase assay system (Promega). The values indicated represent the means ± S.E. from four independently transfected wells. The experiments were repeated at least three times. The ROR and reporter constructs have been previously described (5,10).
cDNA Synthesis and Quantitative PCR
Total RNA extraction and cDNA synthesis as well as the QPCR were performed as previously described (19, 20).
Radioligand Binding Assay
The radioligand binding assay for RORα and RORγ using [3H]25-hydroxycholesterol has been previously described (5, 6,10).
Pharmacokinetic Studies
Plasma levels of SR3335 were evaluated in C57BL6 mice (n = 3 per time point) administered by i.p. injection. After 0.25, 0.5,1, 2, 4, and 8h blood was taken. Plasma was generated using standard centrifugation techniques, and the plasma and tissues were frozen at −80°C. Plasma and tissues were mixed with acetonitrile (1:5 v/v or 1:5 w/v, respectively), sonicated with a probe tip sonicator, and analyzed for drug levels by liquid chromatography/tandem mass spectrometry. All the procedures were conducted in the Scripps vivarium, which is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care, and were approved by the Scripps Florida Institutional Animal Care and Use Committee.
Diet Induced Obesity Model
30 week old Diet induced obese (DIO) C57BL/6 male mice were purchased from Jackson Laboratories that were maintained on a 65% Kcal high-fat diet from weaning.. DIO mice were treated twice per day (07:00h and 18:00h) with 15 mg/kg SR3335 or vehicle for 6 days i.p. Pyruvate tolerance test was conducted on day 6 of the treatment. Food was removed from mice in the morning after SR3335 injection, fasted for 6 hours and the pyruvate tolerance test was conducted at 13:00h. Time 0 blood glucose was measured taken from the tail nip and the pyruvate challenge was initiated by injection of 2g/kg of pyruvate i.p. followed by measuring blood glucose at 15, 30 and 60 min following the injection. Blood glucose was measured by one touch ultra glucose-meter.
Acknowledgments
The efforts of P.R.G. and W.R.R. were supported by the National Institutes of Health (NIH) Molecular Library Screening Center Network (MLSCN) grant U54MH074404 (Hugh Rosen, Principal Investigator). This work was also supported by NIH grants DK080201 (T.P.B.) and GM084041 (P.R.G).
References
- 1.Jetten AM. Retinoid-related orphan receptors (RORs): critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nucl Recept Signal. 2009;7:e003. doi: 10.1621/nrs.07003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Solt LA, Griffin PR, Burris TP. Ligand regulation of retinoic acid receptor-related orphan receptors: implications for development of novel therapeutics. Current Opinion in Lipidology. 21:204–211. doi: 10.1097/MOL.0b013e328338ca18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kallen J, Schlaeppi JM, Bitsch F, Delhon I, Fournier B. Crystal structure of the human ROR alpha ligand binding domain in complex with cholesterol sulfate at 2.2 angstrom. Journal of Biological Chemistry. 2004;279:14033–14038. doi: 10.1074/jbc.M400302200. [DOI] [PubMed] [Google Scholar]
- 4.Kallen JA, Schlaeppi JM, Bitsch F, Geisse S, Geiser M, Delhon I, Fournier B. X-ray structure of the hROR alpha LBD at 1.63 angstrom: Structural and functional data that cholesterol or a cholesterol derivative is the natural ligand of ROR alpha. Structure. 2002;10:1697–1707. doi: 10.1016/s0969-2126(02)00912-7. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Kumar N, Solt LA, Richardson TI, Helvering LM, Crumbley C, Garcia-Ordonez RA, Stayrook KR, Zhang X, Novick S, Chalmers MJ, Griffin PR, Burris TP. Modulation of RORalpha and RORgamma activity by 7-oxygenated sterol ligands. Journal of Biological Chemistry. 2010;285:5013–5025. doi: 10.1074/jbc.M109.080614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Kumar N, Crumbley C, Griffin PR, Burris TP. A second class of nuclear receptors for oxysterols: Regulation of RORalpha and RORgamma activity by 24S-hydroxycholesterol (cerebrosterol) Biochim BiophysActa. 2010;1801:917–923. doi: 10.1016/j.bbalip.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin LH, Martynowski D, Zheng SY, Wada T, Xie W, Li Y. Structural Basis for Hydroxycholesterols as Natural Ligands of Orphan Nuclear Receptor ROR gamma. Molecular Endocrinology. 2010;24:923–929. doi: 10.1210/me.2009-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lau P, Fitzsimmons RL, Raichur S, Wang SCM, Lechtken A, Muscat GEO. The orphan nuclear receptor, ROR alpha, regulates gene expression that controls lipid metabolism - Staggerer (sg/sg) mice are resistant to diet-induced obesity. Journal of Biological Chemistry. 2008;283:18411–18421. doi: 10.1074/jbc.M710526200. [DOI] [PubMed] [Google Scholar]
- 9.Chopra AR, Louet JF, Saha P, An J, DeMayo F, Xu JM, York B, Karpen S, Finegold M, Moore D, Chan L, Newgard CB, O’Malley BW. Absence of the SRC-2 Coactivator Results in a Glycogenopathy Resembling Von Gierke’s Disease. Science. 2008;322:1395–1399. doi: 10.1126/science.1164847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar N, Solt LA, Conkright JJ, Wang Y, Istrate MA, Busby SA, Garcia-Ordonez R, Burris TP, Griffin PR. The benzenesulfonamide T0901317 is a novel ROR{alpha}/{gamma} Inverse Agonist. Molecular Pharmacology. 2010;77:228–236. doi: 10.1124/mol.109.060905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schultz JR, Tu H, Luk A, Repa JJ, Medina JC, Li LP, Schwendner S, Wang S, Thoolen M, Mangelsdorf DJ, Lustig KD, Shan B. Role of LXRs in control of lipogenesis. Genes & Development. 2000;14:2831–2838. doi: 10.1101/gad.850400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michael LF, Schkeryantz JM, Burris TP. The pharmacology of LXR. Mini Rev Med Chem. 2005;5:729–740. doi: 10.2174/1389557054553767. [DOI] [PubMed] [Google Scholar]
- 13.Houck KA, Borchert KM, Hepler CD, Thomas JS, Bramlett KS, Michael LF, Burris TP. T0901317 is a dual LXR/FXR agonist. Molecular Genetics and Metabolism. 2004;83:184–187. doi: 10.1016/j.ymgme.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Kumar N, Nuhant P, Cameron MD, Istrate MA, Roush WR, Griffin PR, Burris TP. Identification of SR1078, a Synthetic Agonist for the Orphan Nuclear Receptors RORα and RORG. ACS Chemical Biology. 2010:null–null. doi: 10.1021/cb100223d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang YJ, Solt LA, Burris TP. Regulation of FGF21 Expression and Secretion by Retinoic Acid Receptor-related Orphan Receptor alpha. Journal of Biological Chemistry. 2010;285:15668–15673. doi: 10.1074/jbc.M110.102160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delerive P, Chin WW, Suen CS. Identification of Reverb alpha as a novel ROR alpha target gene. Journal of Biological Chemistry. 2002;277:35013–35018. doi: 10.1074/jbc.M202979200. [DOI] [PubMed] [Google Scholar]
- 17.Raspe E, Mautino G, Duval C, Fontaine C, Duez H, Barbier O, Monte D, Fruchart J, Fruchart JC, Staels B. Transcriptional regulation of human Rev-erb alpha gene expression by the orphan nuclear receptor retinoic acid-related orphan receptor alpha. Journal of Biological Chemistry. 2002;277:49275–49281. doi: 10.1074/jbc.M206215200. [DOI] [PubMed] [Google Scholar]
- 18.Farah BS, Gilbert EE, Sibilia JP. Perhalo ketones. V. Reaction of perhaloacetones with aromatic hydrocarbons. Journal of Organic Chemistry. 1965;30:998–1001. [Google Scholar]
- 19.Kumar N, Solt LA, Wang Y, Rogers PM, Bhattacharyya G, Kamenecka TM, Stayrook KR, Crumbley C, Floyd ZE, Gimble JM, Griffin PR, Burris TP. Regulation of Adipogenesis by Natural and Synthetic REV-ERB Ligands. Endocrinology. 2010;151:3015–3025. doi: 10.1210/en.2009-0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, McClure DB, Burris LL, Khorasanizadeh S, Burris TP, Rastinejad F. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat Struct Mol Biol. 2007;14:1207–1213. doi: 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]