Abstract
The transcription factor Broad-Complex (BR-C) is required for differentiation of adult structures as well as for the programmed death of obsolete larval organs during metamorphosis of the fruit fly Drosophila melanogaster. Whether BR-C has a similar role in other holometabolous insects could not be proven without a loss-of-function genetic test, performed in a non-drosophilid species. Here we use a recombinant Sindbis virus as a tool to silence BR-C expression in the silkmoth Bombyx mori. The virus expressing a BR-C antisense RNA fragment reduced endogenous BR-C mRNA levels in infected tissues (adult wing and leg primordia) via RNA interference (RNAi). The RNAi knock-down of BR-C resulted in the failure of animals to complete the larval–pupal transition or in later morphogenetic defects, including differentiation of adult compound eyes, legs, and wings from their larval progenitors. BR-C RNAi also perturbed the programmed cell death of larval silk glands. These developmental defects correspond to loss-of-function phenotypes of BR-C Drosophila mutants in both the morphogenetic and degenerative aspects, suggesting that the critical role of BR-C in metamorphosis is evolutionarily conserved. We also demonstrate that the Sindbis virus is a useful vehicle for silencing of developmental genes in new insect models.
Metamorphosis in the higher insects is a coordinate process that forms the structures of a flying adult while destroying organs needed only for the life of a crawling larva. This change takes place in a transitory stage of the pupa, where adult appendages differentiate from imaginal discs of epithelial cells (1), and obsolete tissues undergo programmed death (2). Metamorphosis is triggered by a steroid hormone ecdysone, which causes pupal commitment of tissues in the absence of the sesquiterpenoid juvenile hormone (3).
Genetic studies in the fruit fly Drosophila melanogaster have shown that ecdysone-induced transcription factors of the Broad-Complex (BR-C) are critical for metamorphosis. Multiple BR-C isoforms consist of a common core region, including a conserved protein–protein interaction domain BTB and one of four unique C-terminal C2H2 zinc-finger pairs that are added by alternative splicing (4, 5). Drosophila BR-C-null nonpupariating (npr1) mutants fail to initiate the larval–pupal transition (6). Mutants lacking only subsets of BR-C proteins belong to three complementation groups (br, rbp and 2Bc), in which morphogenesis of adult organs, such as legs, wings, nervous system, and compound eyes, is disrupted (6–9), whereas the larval salivary glands and midgut fail to undergo the programmed cell death (2, 10–12). Although distinct BR-C isoforms play specific roles during development, rescue experiments have suggested their partial functional redundancy (8).
Studies in the moth Manduca sexta have shown that BR-C expression in the epidermis coincide with pupal commitment (13, 14). Juvenile hormone (JH), given before the epidermis is pupally committed, prevents that commitment as well as the ecdysone-dependent expression of BR-C. This effect suggests that JH blocks metamorphosis by repressing BR-C, and thus that BR-C is required for metamorphosis also in non-drosophilid insects. It is of interest to know whether BR-C has a conserved role in insect development. However, causal evidence for such a role requires a loss-of-function genetic test, which has not been performed for any insect other than Drosophila.
Genetic studies in other insects are now possible because of new gene transfer techniques using virus-based transient expression systems. Recombinant double subgenomic Sindbis viruses (SINV) have been successfully used for ectopic gene expression in mosquitoes (15–17), a beetle Tribolium castaneum (18), a butterfly Precis coenia (18), and the commercial silkmoth Bombyx mori (19). In mosquitoes, foreign and endogenous genes have been silenced by SINV infection (20–22), presumably through the process of RNA interference (RNAi). Here we show that infection with a recombinant SINV transcribing RNA derived from Bombyx BR-C causes a knock-down of BR-C expression in vivo, and that this silencing is because of RNAi. With this tool, we demonstrate that loss of BR-C function disrupts two classes of metamorphic events in Bombyx, formation of the adult structures from their primordia and programmed death of larval silk glands, suggesting that the key role of BR-C in metamorphosis is common at least in moths (Lepidoptera) and flies (Diptera).
Materials and Methods
Experimental Animals and Virus Injection. The silkmoth larvae of a nondiapausing strain, Nistari, were reared on artificial diet (Nihon Nosankogyo, Yokohama) at 25°C under a 16-h light/8-h dark photoperiod. For viral infection, larvae on day 2 of the fourth instar or day 0 of the fifth instar were cold-anesthetized and injected with 15–20 μl of viral suspension between the first and second abdominal segments by using a glass needle.
Isolation of Bombyx BR-C cDNA. Total RNA was prepared from the epidermis of a day 6 fifth-instar larva by using the TRIzol reagent (Invitrogen). First-strand cDNA was synthesized with oligo(dT) primers and the SuperScript II reverse transcriptase (Invitrogen). The cDNA was used for nested PCR with degenerate primers, based on the BTB and Z4 zinc-finger domains conserved between M. sexta and D. melanogaster BR-C Z4 isoforms. The first pair of primers (5′-AARWSIACICCITGYAARCAYCC-3′ and antisense 5′-TGRTAIATISWYTTRTGRTTRTT-3′) were combined in a touch-down PCR with annealing temperatures declining from 60°C to 41°C over 20 cycles, followed by 39 cycles with annealing at 54°C. The entire reaction was then repeated with the nested primer set (5′-TAYCAYGGIGARGTIAAYGTNCA-3′ and antisense 5′-ARIGTICKRAAIACYTTRTGRCA-3′). Three independent PCR products were cloned into the pGEM-T vector (Promega) and sequenced by using the BigDye terminator kit (Perkin–Elmer).
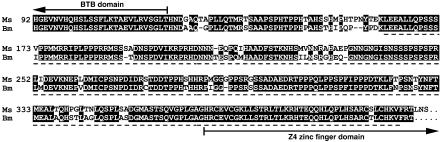
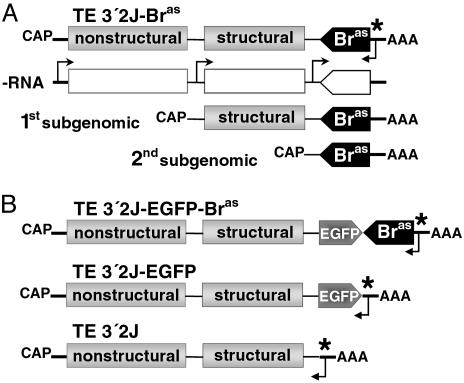
Construction of SINV Vectors. A 705-bp Bombyx BR-C cDNA fragment excluding the BTB domain (Fig. 1) was amplified with primers 5′-AGTCTAGACTCATCCAGCATCCCGCCCAT-3′ and 5′-AGTCTAGAAGACCTTGTGGCAGAGCGT-3′ containing XbaI sites (underlined). The fragment was subcloned into the TE 3′2J plasmid in the antisense orientation, producing TE 3′2J-Bras. The same primers with Sbf I sites were used to prepare TE 3′2J-EGFP-Bras (EGFP, enhanced GFP). Antisense orientation of the BR-C cDNA was confirmed by PCR and restriction analyses.
Fig. 1.
Amino acid identity between the BR-C Z4 isoform of M. sexta (top, AF0326761) and the protein sequence deduced from the B. mori BR-C cDNA fragment is 88%. The highly conserved C-terminal part of the BTB domain and the N-terminal portion of the Z4 zinc finger are indicated by arrows; between them is a variable part of the core, not conserved in Drosophila. The dashed line indicates which portion of the Bombyx cDNA was used for cloning into TE 3′2J-based SINV vectors (Fig. 2) and as a probe for hybridizations in Fig. 7.
Cell Lines and Virus Production. Hamster BHK-21, mosquito C6/36, and monkey Vero cells were all cultured as described (23). Recombinant SINV plasmids were linearized by using XhoI, and their genomic RNA was transcribed in vitro by using SP6 RNA polymerase and capped with 7-methylguanosine (Ambion, Austin, TX). The RNA was then electroporated into the BHK-21 cells. The BHK-21 medium was collected 48 h later, cell debris was removed by centrifugation, and the supernatant containing virus was stored in aliquots at –80°C. Deletion of the inserted BR-C sequence in the SINV genome often occurred with passage of the virus in cell culture. Therefore, SINV amplification in C6/36 cells was omitted in most cases. Viral titers were determined by plaque assays in Vero cells as described (23). Each aliquot used for larval injection was thawed only once.
Northern Blot Analysis. Total RNA was extracted with Trizol (Invitrogen) from wing discs and larval legs. Ten micrograms of total RNA were separated on a 1% agarose-formaldehyde gel and transferred onto a BrightStar-Plus nylon membrane (Ambion). The blots were hybridized and washed at 68°C with digoxigenin (DIG)-labeled antisense cRNA probes according to the DIG Northern kit protocol (Roche Diagnostics, Mannheim, Germany). Detection with an anti-DIG alkaline phosphatase antibody and the CDP-Star chemiluminescent substrate (Roche) was captured on x-ray films.
Small Interfering RNA (siRNA) Isolation and Detection. Total RNA, extracted from larval tissues as for Northern blots above, was heated to 65°C for 10 min and placed on ice for 30 min. High molecular-weight RNA was precipitated with polyethylene glycol (Mr 8,000) and NaCl, added to final concentrations of 5% and 0.5 M respectively (24). Low-molecular-weight RNAs were precipitated from the supernatant with 3 volumes of ethanol, separated on a 12% polyacrylamide-7 M urea gel, and transferred onto a BrightStar-Plus membrane (Ambion). Hybridization was performed at 42°C in DIG Easy-Hyb buffer (Roche) overnight. Membrane was washed at 45°C for 15 min twice in each, 2× SSC and 0.2× SSC containing 0.1% SDS. The size of the Bombyx BR-C-specific DIG-labeled antisense cRNA probe was reduced to 50–100 nt with an alkali treatment. Detection was the same as described above for Northern blots.
Results
The BR-C Sequence Is Conserved in the Silkmoth B. mori. The aim of our study was to show whether BR-C plays a role in metamorphosis of non-drosophilid insects by using RNAi knock-down, delivered by the SINV transformation system. Expression of BR-C isoforms corresponding to Drosophila Z2, Z3, and Z4, has been described in M. sexta (13, 14). However, we found recently that Manduca larvae were highly resistant to SINV infection (19) and therefore not suitable for such a study. In contrast, Bombyx larvae appeared susceptible to SINV, but a BR-C cDNA from Bombyx was not available.
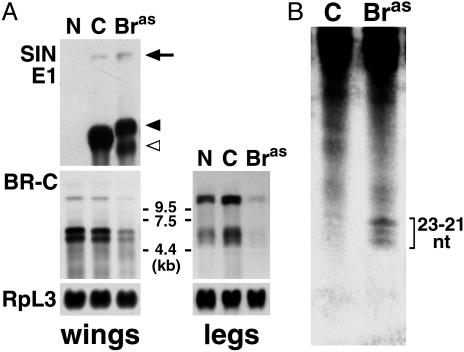
We have cloned Bombyx BR-C cDNA by using RT-PCR with degenerate primers based on conservation between Drosophila and Manduca BR-C Z4 proteins. Conceptual translation of the obtained cDNA fragment revealed 88% amino acid identity with Manduca BR-C Z4 from the C-terminal part of the BTB domain to the zinc-finger region (Fig. 1). The high homology of the variable core clearly identified the Bombyx cDNA as BR-C. A probe derived from the core and Z4 region (Fig. 1) detected three BR-C transcripts of ≈10.5, 6.5, and 5.5 kb on Northern blots from the wing imaginal discs and leg epidermis of final-instar Bombyx larvae (see Fig. 7A). As in Manduca, these mRNAs likely encode BR-C isoforms with common core and alternatively spliced zinc-finger regions (13, 14).
Fig. 7.
SINV mediates specific BR-C silencing via RNAi. (A) Northern blot hybridization shows that infection with TE 3′2J-Bras reduced the levels of BR-C mRNAs. Total RNA was isolated from wing discs (Left) and legs (Right) of fifth-instar larvae, either uninfected or 6 days after virus injection. The Bombyx BR-C-specific probe detected three transcripts. To show equal RNA loading, the membranes were rehybridized with a probe for a constitutive ribosomal protein RpL3. Hybridization with a probe for the Sindbis E1 structural protein (for wing discs) showed the full-length viral genomic RNA (arrow) and the first subgenomic viral RNA before (black arrowhead) and after (open arrowhead) deletion, which probably removed RNA at the 3′ end. N, uninfected larvae; C, control TE 3′2J virus; Bras, TE3′2J-Bras virus. (B) Hybridization of small RNAs, isolated from mixed tissues of fifth-instar larvae 6 days after infection with TE 3′2J-Bras (right lane) but not the control TE 3′2J virus (left lane) shows BR-C-specific siRNAs of the indicated size range.
Design of SINV Vectors for RNAi and Viral Dissemination in Bombyx. Sindbis is an RNA virus whose nonstructural genes are translated on host cell ribosomes immediately after infection. Then, as diagrammed in Fig. 2A, a viral RNA-dependent RNA polymerase produces a negative sense RNA copy of the genome, which serves as a template for first and second subgenomic RNAs and also to make new positive sense RNA. Double-stranded RNAs, assembled from the positive and negative strands during viral replication, may be recognized by the DICER enzymatic complex, thus triggering the RNAi process in infected cells (20, 25).
Fig. 2.
Design and replication of recombinant SINV. Recombinant TE 3′2J-based viruses contain a full Sindbis viral genome (13.8 kb in length), represented by single-stranded positive sense RNA with a second subgenomic promoter added at the 3′ end (17). Parts of the construct are not to scale in this scheme. (A) The 705-bp BR-C cDNA fragment was cloned in antisense orientation downstream of the second subgenomic promoter, thus generating TE 3′2J-Bras (top). On infection, a negative sense RNA copy of the genome (–RNA) is made by viral RNA-dependent RNA polymerase from a signaling sequence (*) in the 3′ noncoding region of the viral genome. The–RNA serves as a template for first and second subgenomic RNAs produced from two internal promoters and for new full-length positive sense RNA. (B) The chimeric TE 3′2J-EGFP-Bras virus contains a fusion of the EGFP coding region and the antisense Bombyx BR-C cDNA. The TE 3′2J-EGFP and TE 3′2J (bottom) viruses without the BR-C insert were used for controls.
To prepare a vector for RNAi silencing of all BR-C isoforms, we cloned the BR-C cDNA behind the second subgenomic promoter of TE 3′2J SINV, generating TE 3′2J-Bras (Fig. 2 A). The sequence encoding the conserved BTB domain was eliminated to avoid a possibility that genes other than BR-C might be affected. Although RNAi can potentially result from the viral RNA replication regardless of the insert orientation (21), the cDNA was placed in the antisense orientation to preclude any fortuitous expression of BR-C protein fragments. We also prepared a chimeric construct TE 3′2J-EGFP-Bras, where the antisense BR-C insert followed the EGFP coding region (Fig. 2B). Translation of EGFP from the second subgenomic RNA should therefore mark tissues where RNAi silencing of BR-C occurs. The same viruses lacking the BR-C insert served as controls. To obtain high viral titers, SINV is routinely propagated first in the BHK-21 cell line, then by passage through mosquito C6/36 cells. Consistent with previous reports (16, 23), we observed SINV genome instability due to deletions at the 3′ end, causing the loss of the foreign gene (see Fig. 7A below). To decrease the risk of injecting larvae with virus lacking the BR-C fragment, we preferred to omit amplification in C6/36 cells. Although this resulted in lower viral titers, parenteral injections into Bombyx larvae allowed us to deliver volumes high enough for efficient virus dissemination.
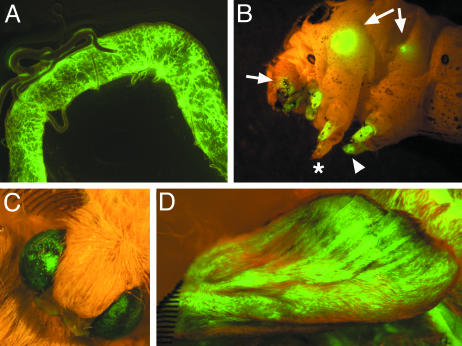
Examples of tissues infected by the control TE 3′2J-EGFP virus in animals injected as fourth-instar larvae are shown in Fig. 3. The first signs of viral replication were visible via EGFP fluorescence 3 d postinfection in the fat body, larval muscles, and circulating hemocytes (not shown). Among tissues infected later were the nervous system (ventral ganglia and the brain), larval eyes (stemmata), silk glands, and primordia of adult organs (wings, compound eyes, and legs). In moths, the epithelium for the adult legs is inside the existing legs of the larva, whereas wings originate from typical imaginal discs as in Drosophila (26). The virus did not seem to enter the gonads, Malpighian tubules, and larval epidermis. Infected animals underwent metamorphosis and emerged as adults with EGFP-positive tissues (Fig. 3).
Fig. 3.
Various tissues of the silkmoth are SINV targets. The TE 3′2J-EGFP virus was injected into fourth-instar larvae. Expression of EGFP serves as a marker of successful viral replication and shows the temporal spreading of the infection. (A) The middle part of the silk gland in a fifth-instar larva. (B) Strong EGFP signals in wing discs (arrows), imaginal leg primordia (arrowhead), and eye primordia in the head (arrow) are visible through the cuticle of a fifth-instar larva. Asterisk shows a leg apparently not infected with the virus. (C and D) Infected animals survive metamorphosis and show the viral EGFP expression in adult organs such as the compound eye (C) and forewing (D), both derived from infected imaginal progenitors.
Loss of BR-C Function Disrupts Metamorphic Events in Bombyx. That Drosophila npr1 mutants lacking all BR-C isoforms survive until pupariation shows that the BR-C function is not essential for earlier postembryonic development (6). The rapid dissemination of TE 3′2J in Bombyx larvae thus offers an opportunity to trigger the RNAi process in time to interfere with BR-C function as it becomes required. Initially, we injected viruses into silkmoth larvae shortly after ecdysis into the final (fifth) instar and looked for specific phenotypes during metamorphosis in pupae and after eclosion in adults (Table 1). Although there was ≈10% higher pupal lethality with the BR-C RNAi constructs compared to control vectors, most injected animals formed adults.
Table 1. Effects of SINV-mediated BR-C RNAi on Bombyx development after virus injection on day 0 of the final (fifth) larval instar.
| Adults
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Viral construct | Number of trials | Average viral titer, pfu/ml | Injected larvae | Complete pupae | Pupa—adult arrest, % | Total | Leg defects, % | Eye defects, % | Normal, % |
| TE 3′2J or TE 3′2J-EGFP | 4 | 1.1 × 107 | 55 | 53 | 2 | 52 | 17 | 0 | 83 |
| TE 3′2J-EGFP-Bras | 4 | 1.9 × 106 | 69 | 60 | 13 | 52 | 33 | 23 | 52 |
| TE 3′2J-Bras | 5 | 1.2 × 106 | 67 | 67 | 12 | 59 | 56 | 66 | 15 |
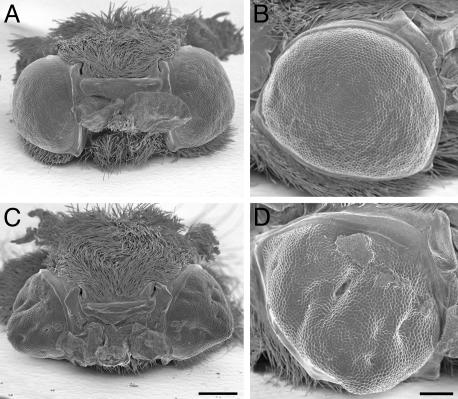
Differentiation of the compound eye was strongly affected by BR-C RNAi but not by control SINV viruses (Table 1). Compared to controls, which all developed round eyes with regularly organized ommatidia, 66% of the adults carrying TE 3′2J-Bras displayed collapsed conical shaped eyes with invaginations, holes, and ommatidial disarray (Fig. 4). Similar effects were seen in 23% of the adults carrying TE 3′2J-EGFP-Bras.
Fig. 4.
Differentiation of compound eyes is disrupted by BR-C RNAi. Shown are scanning electron micrographs of eyes from adults infected as day 0 fifth-instar larvae with the control TE 3′2J virus (A and B) and with TE 3′2J-Bras (C and D). Compared to controls, animals carrying TE 3′2J-Bras show cone-shaped eyes with invaginations and folds. The bars correspond to 500 μm in A and C and to 200 μm in B and D.
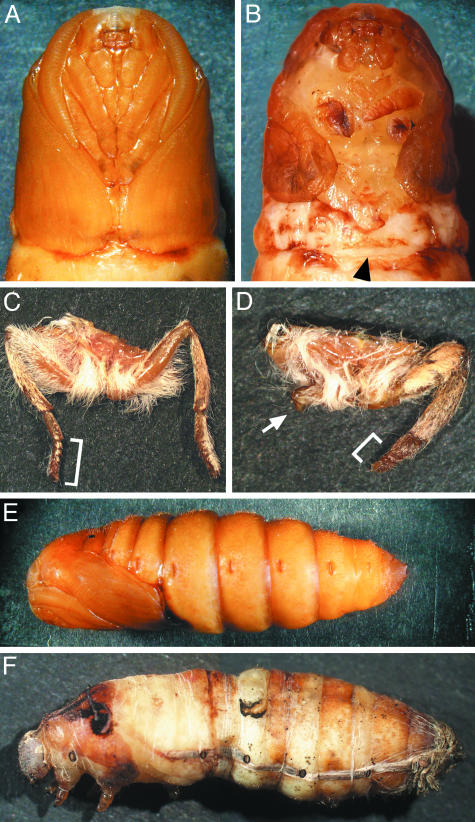
In addition to the eye phenotypes, a large proportion of animals injected with TE 3′2J-Bras (56%) and TE 3′2J-EGFP-Bras (33%) developed short legs with reduced number of tarsal segments; in some cases, other parts of legs were also malformed or missing (Fig. 5 B and D). Slightly shorter tarsi (but no more severe defects) were seen in 17% of animals injected with ≈10-fold higher titers of the control viruses (Table 1), indicating that SINV infection alone may perturb leg formation to some extent. The failure of legs to elongate was already visible in BR-C RNAi but not in control pupae (Fig. 5B). The affected pupae also displayed short wings. This phenotype, however, could not be scored in adults, because they usually failed to emerge from the pupal case and had pieces of the pupal cuticle attached to their wings. In both Manduca (13, 14) and Bombyx (not shown), BR-C expression ensues in the wing discs soon after ecdysis to the fifth instar, before the injected virus could spread widely. To achieve a more efficient BR-C knock-down, we injected larvae on day 2 of the fourth instar. Over 50% of animals carrying TE 3′2J-EGFP-Bras or TE 3′2J-Bras arrested during the pupal molt (Fig. 5F) compared to 97% of controls that pupated successfully (Fig. 5E and Table 2). In the case of TE 3′2J-Bras, all pupae showed severely shortened wings and short or undeveloped legs, confirming the results with larvae infected in the fifth instar (Table 1). Although 86% of animals injected with 10-fold higher TE 3′2J titer emerged as adults, only one adult infected with TE 3′2J-Bras eclosed (Table 2). The same phenotypes with lower penetrance (67%) were observed with TE 3′2J-EGFP-Bras. The leg and wing defects corresponded with the sites of SINV activity in the imaginal primordia (Fig. 3B) and indicated that BR-C is necessary for the normal development of adult appendages in Bombyx.
Fig. 5.
BR-C function is required for pupation, elongation, and differentiation of adult legs and wings. Most larvae infected on day 0 of the fifth instar undergo the larval–pupal transition. Compared to a control pupa carrying the TE 3′2J virus (A), a pupa infected with TE 3′2J-Bras displays short forewings and short malformed legs (B). The arrowhead in B indicates where the wings should extend and meet normally. A mesothoracic leg of a control TE 3′2J infected adult (C) shows the normal number and size of segments, whereas infection with TE 3′2J-Bras leads to deletions of segments and overall leg malformation (D). Brackets show the normal (C) and shortened (D) tarsi; arrow in D points to an undeveloped mesothoracic leg. (E and F) Although a majority of animals injected as day 2 fourth-instar larvae with control viruses form normal pupae (E), most of those infected with TE 3′2J-Bras die when trying to ecdyse (F).
Table 2. Effects of SINV-mediated BR-C RNAi on Bombyx development after virus injection on day 2 of the fourth larval instar.
| Pupae
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Viral construct | Number of trials | Average viral titer, pfu/ml | Injected larvae | Larva-pupa arrest, % | Total | Short wings, % | Leg defects, % | Normal, % | Total adults |
| TE 3′2J | 2 | 3.6 × 107 | 30 | 3 | 29 | 0 | 14 | 86 | 27 |
| TE 3′2J-EGFP-Bras | 2 | 1.4 × 106 | 23 | 61 | 9 | 67 | 67 | 33 | 4 |
| TE 3′2J-Bras | 3 | 0.7 × 106 | 44 | 55 | 20 | 100 | 100 | 0 | 1 |
Besides formation of adult structures, metamorphosis also involves programmed death of larval organs, no longer needed for adult life. For example, in Bombyx, the silk glands degenerate on cocoon completion (27). However, animals that had been infected with BR-C RNAi viruses as day 2 fourth-instar larvae spun cocoons but still retained largely intact glands 12 h after pupation, when control silk glands showed a high degree of degeneration (Fig. 6). Infection with TE 3′2J-EGFP-Bras showed that of a pair of silk glands within one pupa, only the gland infected with the virus and expressing EGFP failed to degenerate (Fig. 6C). The gland vestiges were still visible 3 d later, by which time the silk glands completely disappeared in pupae infected with control viruses. Interestingly, the silk glands in affected pupae eventually histolysed, indicating that the cell death program could be delayed but not averted.
Fig. 6.
BR-C plays a role in the programmed cell death of larval silk glands. Animals infected as day 2 fourth-instar larvae were dissected 12 h after pupation. Silk glands found in control TE 3′2J-infected pupae displayed late phase of histolysis in the anterior and middle parts (A). In contrast, a middle gland dissected from a TE 3′2J-Bras-infected animal showed no signs of degeneration (B). In a day 1 pupa infected with TE 3′2J-EGFP-Bras, only one gland from the pair failed to degenerate (C); shown is the posterior part where the EGFP fluorescence indicates viral infection. The middle parts of silk glands were still visible in a day 3 pupa infected with TE 3′2J-Bras (D) but not in control pupae (not shown).
SINV Reduced BR-C mRNA Levels via RNAi. To determine whether the observed aberrant phenotypes correlated with a decrease in BR-C expression, we examined BR-C mRNA levels by using Northern blot hybridization. Bombyx larvae were injected with viruses on day 0 of the fifth instar, and RNA from their wing discs and legs was analyzed 6 days postinfection. Fig. 7A clearly shows reduction of all three BR-C transcripts in both tissues isolated from TE 3′2J-Bras infected animals compared to TE 3′2J controls. By using the viral SIN E1-specific probe, we confirmed that silencing of BR-C in the wing discs coincided with SINV RNA replication. In the case of TE 3′2J-Bras, two different lengths of the first subgenomic viral RNA were detected, indicating a deletion at the 3′ end. However, there was also a major population of the full-length virus containing the BR-C cDNA insert in the infected tissue.
To provide direct evidence that the observed reduction of BR-C transcripts involved RNAi, we tested infected larvae for the presence of siRNAs, a hallmark of the mechanism (28). Fig. 7B shows that BR-C-specific RNAs 21–23 nt in length were present in animals infected 6 days earlier with TE 3′2J-Bras but not with the control TE 3′2J virus, strongly suggesting that the observed developmental defects represent specific loss-of-function phenotypes, caused by SINV-mediated BR-C RNAi.
Discussion
The data presented here have two important implications. First, we show that the Sindbis viral system can be used for effective RNAi silencing in vivo and thus allows genetic studies on species in which gene-specific mutations are difficult to realize. Second, by using RNAi mediated by SINV, we have shown that the transcription factor BR-C is necessary for normal metamorphosis of the silkmoth B. mori, similarly to its role in Drosophila.
Metamorphosis in moths differs from that in Drosophila in that most of the epidermis of the moths is reprogrammed to make pupal and adult structures. Only the wings originate from true imaginal discs and the eyes and legs, from precursor cells that begin to proliferate in the final larval instar (26, 29). In contrast, the entire head and thorax of adult Drosophila derive from imaginal discs that proliferate during most of larval life then differentiate shortly before and after pupariation in response to ecdysone. At this time, BR-C is induced in the imaginal discs, and its absence causes defects seen in the adult (5, 9). Drosophila mutants lacking the distinct BR-C isoforms die during the larval–pupal transition (6). The frequent lethality we have seen during pupation after infection of fourth-instar larvae thus indicates that BR-C is similarly required for this transition in Bombyx and Drosophila despite their different developmental strategy. Importantly, more of the Bombyx pupal body was formed (Fig. 5F), because it derives from the uninfected epidermal cells rather than from imaginal precursors.
Drosophila eye discs lacking all BR-C functions display defects in the progressive differentiation of ommatidial cells (the morphogenetic furrow) that occurs at the end of the final larval instar (9). The progression of the furrow in both Drosophila (30) and Manduca (29) requires ecdysone, and BR-C is present in the eye disk in both species at this time (ref. 9; D. T. Champlin, C. A. Nelson, J. W. Truman, and L.M.R., unpublished work), suggesting that the mechanism is conserved. The lesions observed in Bombyx compound eyes therefore are likely a result of local arrest of the eye-forming furrow, caused by the loss of BR-C in foci of SINV activity. The mosaic character of this effect is probably due to the uneven virus distribution in the eye (Fig. 3C).
The wing and leg anomalies seen in the infected Bombyx are also similar to those of the hypomorphic Drosophila BR-C mutants (6, 8, 31). For instance, the lack of normal pupal leg elongation resulting in stunted or deformed adult legs is very similar to leg elongation defects visualized by using transgenic GFP in the br5 mutant (31). Thus, BR-C seems to be universally required for differentiation of imaginal disk structures during insect metamorphosis.
Although the infected Bombyx spun normal cocoons, the ecdysone-induced programmed histolysis of the silk glands (27) was severely delayed. In Drosophila, two consecutive pulses of ecdysone at the onset of metamorphosis trigger a dramatic change in gene expression (32–34), followed by programmed cell death of the salivary gland. Several transcription factors, including BR-C, Ftz-F1, E74, E75, E93, and the ecdysone receptor EcR, all appear to function in salivary gland destruction (8, 10, 11, 35, 36). In Drosophila rbp mutants, which lack the BR-C Z1 isoform, the salivary glands are still present 22 h after puparium formation (10), and proapoptotic factors such as Rpr, Hid, and the DRONC caspase are not properly expressed (11, 37). The silk glands of the Lepidoptera are salivary gland derivatives, in which the ecdysone-induced transcription factors are expressed in a similar manner (38–40). Our results suggest that the critical role of BR-C in the process of programmed cell death is conserved in Bombyx. The eventual gland degeneration after a delay is likely due to an incomplete knock-down of BR-C by RNAi.
The observed effects of BR-C RNAi were confined to the tissues targeted by the virus, particularly the imaginal primordia. A similar pattern of SINV dissemination was previously reported for another lepidopteran, Precis coenia (18). The molecular basis of the tissue tropism is currently unknown. Expressivity of the abnormal phenotypes in infected Bombyx pupae and adults varied among individuals and within body parts. These differences were likely caused by the mosaic SINV distribution. Some of the variability was probably contributed by the loss of the BR-C insert due to viral genome deletion at the 3′ end, occurring in the target organs (Fig. 7A). As judged from the loss of EGFP during passage in C6/C36 cells, this deletion was more frequent in viruses with longer 3′ inserts. This explains why the shorter TE 3′2J-Bras vector was more potent than TE 3′2J-EGFP-Bras in eliciting developmental defects.
Recombinant SINV were successfully used to suppress genes in mosquitoes (20–22), and we hypothesized that this suppression involved the RNAi mechanism. By detecting BR-C-specific siRNAs, we have demonstrated here that infection with SINV carrying a heterologous sequence confers RNAi silencing of the corresponding endogenous gene and thus provides causal evidence for its function.
Acknowledgments
We thank Irma Sanchez-Vargas for advice on siRNA detection and the SINV E1 probe and David Chandler for scanning electron microscopy. This work was supported by National Institutes of Health Fogarty International Research Collaboration Grant R03TWO1209-01 (to L.M.R. and M.J.) and Grant IAA5007305 from the Czech Academy of Sciences (to M.J.). M.U. was supported by a North Atlantic Treaty Organization Science Fellowship Program. The work of B.D.F and K.E.O. was supported by National Institutes of Health Grants R01 AI46753 and R01 AI46435.
Abbreviations: BR-C, Broad-Complex; SINV, Sindbis virus; EGFP, enhanced GFP; RNAi, RNA interference; DIG, digoxigenin; siRNA, small interfering RNA.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY380796).
References
- 1.Fristrom, D. & Fristrom, J. W. (1993) in The Development of Drosophila melanogaster, eds. Bate, M. & Martinez Adrias, A. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 843–897.
- 2.Jiang, C., Baehrecke, E. H. & Thummel, C. S. (1997) Dev. Biol. 186, S13. [Google Scholar]
- 3.Riddiford, L. M. (1996) Arch. Insect Biochem. Physiol. 32, 271–286. [DOI] [PubMed] [Google Scholar]
- 4.DiBello, P. R., Withers, D. A., Bayer, C. A., Fristrom, J. W. & Guild, G. M. (1991) Genetics 129, 385–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bayer, C. A., Holley, B. & Fristrom, J. W. (1996) Dev. Biol. 177, 1–14. [DOI] [PubMed] [Google Scholar]
- 6.Kiss, I., Beaton, A. H., Tardiff, J., Fristrom, D. & Fristrom, J. W. (1988) Genetics 118, 247–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Restifo, L. L. & White, K. (1991) Dev. Biol. 148, 174–194. [DOI] [PubMed] [Google Scholar]
- 8.Bayer, C. A., vonKalm, L. & Fristrom, J. W. (1997) Dev. Biol. 187, 267–282. [DOI] [PubMed] [Google Scholar]
- 9.Brennan, C. A., Li, T. R., Bender, M., Hsiung, F. & Moses, K. (2001) Development (Cambridge, U.K.) 128, 1–11. [DOI] [PubMed] [Google Scholar]
- 10.Restifo, L. L. & White, K. (1992) Roux's Arch. Dev. Biol. 201, 221–234. [DOI] [PubMed] [Google Scholar]
- 11.Jiang, C. A., Lamblin, A. F. J., Steller, H. & Thummel, C. S. (2000) Mol. Cell 5, 445–455. [DOI] [PubMed] [Google Scholar]
- 12.Lee, C. Y., Cooksey, B. A. K. & Baehrecke, E. H. (2002) Dev. Biol. 250, 101–111. [DOI] [PubMed] [Google Scholar]
- 13.Zhou, B. H., Hiruma, K., Shinoda, T. & Riddiford, L. M. (1998) Dev. Biol. 203, 233–244. [DOI] [PubMed] [Google Scholar]
- 14.Zhou, B. H. & Riddiford, L. M. (2001) Dev. Biol. 231, 125–137. [DOI] [PubMed] [Google Scholar]
- 15.Olson, K. E., Higgs, S., Hahn, C. S., Rice, C. M., Carlson, J. O. & Beaty, B. J. (1994) Insect Biochem. Mol. Biol. 24, 39–48. [DOI] [PubMed] [Google Scholar]
- 16.Higgs, S., Olson, K. E., Klimowski, L., Powers, A. M., Carlson, J. O., Possee, R. D. & Beaty, B. J. (1995) Insect Mol. Biol. 4, 97–103. [DOI] [PubMed] [Google Scholar]
- 17.Higgs, S., Traul, D., Davis, B. S., Kamrud, K. I., Wilcox, C. L. & Beaty, B. J. (1996) BioTechniques 21, 660–664. [DOI] [PubMed] [Google Scholar]
- 18.Lewis, D. L., DeCamillis, M. A., Brunetti, C. R., Halder, G., Kassner, V. A., Selegue, J. E., Higgs, S. & Carroll, S. B. (1999) Curr. Biol. 9, 1279–1287. [DOI] [PubMed] [Google Scholar]
- 19.Foy, B. D., Myles, K. M., Pierro, D. J., Sanchez-Vargas, I., Uhlirova, M., Jindra, M. & Olson, K. E. (2003) Insect. Mol. Biol., in press. [DOI] [PubMed]
- 20.Johnson, B. W., Olson, K. E., Allen-Miura, T., Rayms-Keller, A., Carlson, J. O., Coates, C. J., Jasinskiene, N., James, A. A., Beaty, B. J. & Higgs, S. (1999) Proc. Natl. Acad. Sci. USA 96, 13399–13403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adelman, Z. N., Blair, C. D., Carlson, J. O., Beaty, B. J. & Olson, K. E. (2001) Insect Mol. Biol. 10, 265–273. [DOI] [PubMed] [Google Scholar]
- 22.Shiao, S. H., Higgs, S., Adelman, Z., Christensen, B. M., Liu, S. H. & Chen, C. C. (2001) Insect Mol. Biol. 10, 315–321. [DOI] [PubMed] [Google Scholar]
- 23.Pierro, D. J., Myles, K. M., Foy, B. D., Beaty, B. J. & Olson, K. E. (2003) Insect Mol. Biol. 12, 107–116. [DOI] [PubMed] [Google Scholar]
- 24.Hamilton, A. J. & Baulcombe, D. C. (1999) Science 286, 950–952. [DOI] [PubMed] [Google Scholar]
- 25.Li, H. W., Li, W. X. & Ding, S. W. (2002) Science 296, 1319–1321. [DOI] [PubMed] [Google Scholar]
- 26.Svacha, P. (1992) Dev. Biol. 154, 101–117. [DOI] [PubMed] [Google Scholar]
- 27.Terashima, J., Yasuhara, N., Iwami, M. & Sakurai, S. (2000) Dev. Genes Evol. 210, 545–558. [DOI] [PubMed] [Google Scholar]
- 28.Zamore, P. D., Tuschl, T., Sharp, P. A. & Bartel, D. P. (2000) Cell 101, 25–33. [DOI] [PubMed] [Google Scholar]
- 29.Champlin, D. T. & Truman, J. W. (1998) Development (Cambridge, U.K.) 125, 269–277. [DOI] [PubMed] [Google Scholar]
- 30.Brennan, C. A., Ashburner, M. & Moses, K. (1998) Development (Cambridge, U.K.) 125, 2653–2664. [DOI] [PubMed] [Google Scholar]
- 31.Ward, R. E., Reid, P., Bashirullah, A., D'Avino, P. P. & Thummel, C. S. (2003) Dev. Biol. 256, 389–402. [DOI] [PubMed] [Google Scholar]
- 32.Karim, F. D., Guild, G. M. & Thummel, C. S. (1993) Development (Cambridge, U.K.) 118, 977–988. [DOI] [PubMed] [Google Scholar]
- 33.vonKalm, L., Crossgrove, K., Vonseggern, D., Guild, G. M. & Beckendorf, S. K. (1994) EMBO J. 13, 3505–3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Renault, N., King-Jones, K. & Lehmann, M. (2001) Development (Cambridge, U.K.) 128, 3729–3737. [DOI] [PubMed] [Google Scholar]
- 35.Broadus, J., McCabe, J. R., Endrizzi, B., Thummel, C. S. & Woodard, C. T. (1999) Mol. Cell 3, 143–149. [DOI] [PubMed] [Google Scholar]
- 36.Kucharova-Mahmood S., Raska, I., Mechler, B. M. & Farkas, R. (2002) J. Struct. Biol. 140, 67–78. [DOI] [PubMed] [Google Scholar]
- 37.Cakouros, D., Daish, T., Martin, D., Baehrecke, E. H. & Kumar, S. (2002) J. Cell Biol. 157, 985–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun, G. C., Hirose, S. & Ueda, H. (1994) Dev. Biol. 162, 426–437. [DOI] [PubMed] [Google Scholar]
- 39.Jindra, M. & Riddiford, L. M. (1996) Dev. Genes Evol. 206, 305–314. [DOI] [PubMed] [Google Scholar]
- 40.Kamimura, M., Tomita, S., Kiuchi, M. & Fujiwara, H. (1997) Eur. J. Biochem. 248, 786–793. [DOI] [PubMed] [Google Scholar]