Abstract
RATIONALE:
Quantitation and determination of antigen specificity of systemic and mucosal immune responses to influenza vaccination is beneficial for future vaccine development. Previous methods to acquire this information were costly, time consuming and sample exhaustive. The benefits of suspension microbead array (MBA) analysis are numerous. The multiplex capabilities of the system conserve time, money and sample, while generating statistically powerful data.
OBJECTIVE:
To demonstrate the use of the assay by comparing the humoral influenza-specific responses of two cohorts from two countries that differed in circulating influenza strains and rates of influenza vaccination.
METHODS:
Influenza hemagglutinin (HA) from different strains were coated on microbeads and incubated with serum samples to capture immunoglobulin (Ig) A1 and IgG1 host antibodies.
RESULTS:
Statistically significant differences in IgA1 and IgG1 exist between the serum samples from Winnipeg (Manitoba) donors and those from Kenyan (Africa) donors. Data were compared using Mann-Whitney nonparametric tests. The Winnipeg donors had higher mean fluorescence intensity values, with significant P values for anti-HA IgA1 to A/Wyoming/3/2003 (P=0.044), A/Vietnam/1203/2004 (P=0.0179), A/New Caledonia/20/99 (P<0.0001) and B/Tokyo/53/99 (P=0.0002). No differences were seen between the groups in their response to B/Jilin/20/2003. The Winnipeg donors had higher mean fluorescence intensity values, with significant P values for anti-HA IgG1 to A/Wyoming/3/2003 (P=0.0135), B/Tokyo/53/99 (P=0.006) and B/Jilin20/2003 (P=0.026).
CONCLUSION:
Influenza-specific IgA1 and IgG1 antibodies were successfully detected using MBA technology. A significant difference in antibody response was observed between Winnipeg and Kenyan donor serums. MBA analysis is a relatively quick and cost-effective method for serum antibody analysis. The potential to simultaneously assay small sample volumes for a multitude of antigens makes this method invaluable for future vaccine response monitoring.
Keywords: Hemagglutinin, Immunoglobulin, Influenza, Microbead array
Abstract
JUSTIFICATION :
La quantification et la détermination de la spécificité antigénique des réactions immunitaires systémiques et muqueuses au vaccin antigrippal servent à l’élaboration des futurs vaccins. Les méthodes précédentes d’acquisition de ce type de données étaient coûteuses, fastidieuses et exigeaient de nombreux d’échantillons. La technologie des microbilles comporte de nombreux avantages : tout en générant des données puissantes sur le plan statistique, les capacités multiplex du système permettent d’épargner temps et argent, en plus d’exiger moins d’échantillons.
OBJECTIF :
Démontrer l’utilisation de cette technologie en comparant les réponses humorales spécifiques au virus de la grippe dans deux cohortes provenant de deux pays où les souches grippales circulantes, de même que les taux de vaccination antigrippale, étaient différents.
MÉTHODES :
L’hémagglutinine (HA) de différentes souches du virus de la grippe a été appliquée à la surface de microbilles et mise en incubation avec des spécimens sérologiques afin de capter les immunoglobulines (Ig)A1 et IgG1 de l’hôte.
RÉSULTATS :
On note des différences statistiquement significatives entre les IgA1 et IgG1 des échantillons sérologiques provenant de donneurs de Winnipeg (Manitoba) et de donneurs kényans (Afrique). Les données ont été comparées à l’aide de tests non paramétriques de Mann-Whitney. Les donneurs de Winnipeg présentaient des valeurs plus élevées en termes d’intensité moyenne à la fluorescence, avec des valeurs p significatives pour l’anti-IgA1 dirigé contre A/Wyoming/3/2003 (p = 0,044), A/Vietnam/1203/2004 (p = 0,0179), A/New Caledonia/20/99 (p = 0,0001) et B/Tokyo/53/99 (p = 0,0002). Aucune différence n’a été observée entre les groupes quant à leur réponse à B/Jilin/20/2003. Les donneurs de Winnipeg ont présenté des valeurs plus élevées en termes d’intensité moyenne à la fluorescence, avec des valeurs p significatives pour l’anti-IgG1 dirigé contre A/Wyoming/3/2003 (p = 0,0135), B/Tokyo/53/99 (p = 0,006) et B/Jilin20/2003 (p = 0,026).
CONCLUSION :
Les anticorps IgA1 et IgG1 spécifiques au virus de la grippe ont été détectés avec succès à l’aide de la technologie des microbilles. Les auteurs ont constaté une réponse immunitaire significativement différente entre les sérums de donneurs de Winnipeg et du Kenya. La technologie des microbilles est une méthode relativement rapide et économique d’analyse des anticorps sériques. La possibilité de tester simultanément de petits volumes d’échantillons pour une multitude d’antigènes donne à cette méthode une valeur inestimable pour la surveillance de la réponse aux vaccinations futures.
Influenza viruses are the only members of the family Orthomyxoviridae; they are transmitted by respiratory droplets and are capable of causing seasonal infections and epidemics (1,2). These viruses have a segmented (typically eight units) single-stranded RNA genome, with RNA segments of 890 to 2341 nucleotides each, a helical nucleocapsid and an outer envelope comprised of lipoprotein (3,4). Three influenza virus groups (groups A, B and C) infect humans and have been characterized based on antigenic differences in internal proteins – nucleoprotein and matrix protein (3–5). These internal proteins, along with a viral replication protein (polymerase acidic protein), are highly conserved within groups. The group A influenza virus is further subtyped by the variation of its surface proteins – hemagglutinin (HA) and neuraminidase. These proteins cover the outer membrane and are important virulence factors involved in binding to host cells and viral budding (3,4).
Immunity to infection by influenza is conferred by anti-HA antibodies blocking initial viral attachment to host cells. Antibodies to neuraminidase do not prevent infection, but do lessen the severity of disease by limiting viral release from infected host cells (4,6). The antibodies generated are type specific and provide little cross-protection against diseases caused by heterotypic or heterosubtypic strains.
The main response to natural infection is local production of secretory immunoglobulin (Ig) A in the mucosal surfaces of the upper respiratory tract and, to a lesser extent, serum IgG, IgA and IgM (3). One of the problems with current vaccines consisting of killed virus or purified protein particles administered intramuscularly is that the main response to these vaccines is relatively low IgG and IgA production typically lasting only six months and requiring annual boosters. Another shortcoming of the inactivated vaccines is that the mucosal response at the respiratory tract, the site of viral entry, is limited following these vaccines.
Traditional methods to diagnose influenza infection relied on a fourfold increase in antibody titre, as measured by hemagglutination inhibition assay (HIA), between acute and convalescent serum samples. The HIA is a functional assay that measures the ability of the HA antibody to inhibit the agglutination of sheep red blood cells. Recent evidence (7) suggests that a single serum sample tested within the first week of symptoms can lead to a diagnosis if the antiflu IgA titre is high. Normally, the antiflu IgA is short lived, so its presence indicates recent infection (primary or secondary) (7).
Bead-based flow cytometry can be used to detect antibodies to multiple antigens from a single sample. This technique consists of a high throughput method that allows for the quantitation of the immune response to vaccinations that contain more than one antigen, without having to run individual assays for each antigen (8). The technology is based on 5.5 micron polysterene microspheres that contain a distinct ratio of two luminescent dyes – one red and the other infra-red – creating 100 bead sets with unique identifying spectral addresses. This allows for the simultaneous assay of up to 100 analytes within a single sample as small as 50 μL, although typical analyses are in the range of 10 to 20 beads. Antigens of interest are coupled to their own specific bead set; several sets are added together to form a master mix, which in turn, is incubated with diluted serum sample to allow complementary antibodies to bind. Biotinylated monoclonal antibodies against human Igs (eg, mouse antihuman IgA1) are added and bind to any human IgA1 attached to antigens on the beads. The beads are passed through a flow cytometer with two lasers. A classification laser detects the colour ratio of the bead and distinguishes the bead set, and thus the antigen bound to it. A second reporter laser detects the intensity of the fluorochrome bound to the monoclonal antibody distinguishing the type and amount of antibody attached to the antigen. This technology allows quantitative and qualitative determination of antibodies present, pre- and postinfluenza viral challenge.
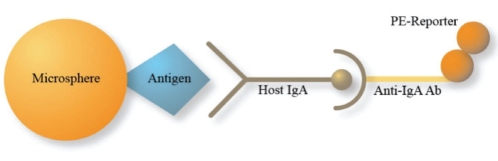
A schematic representation of microbead array (MBA) is shown in Figure 1.
Figure 1).
A schematic representation of microbead array. Latex micro-spheres are coupled to specific antigens (hemagglutinin or p24). Host antigen-specific antibodies (Ab) bind the antigens on the beads. Mouse antihuman immunoglobulin A (IgA) with phycoerythrin (PE) reporter binds any host antigen-specific IgA. PE intensity quantifies host IgA in the sample
The benefits of MBA analysis are numerous. The multiplex capabilities of the system conserve time, money and sample (8–10). The machine reports the average of 100 individual beads per region, equivalent to 100 ELISA replicates, lending statistical power to the data. To generate the same amount of data using traditional ELISA technology would require heavy investment in time, money and sample (10). The present swine-origin influenza A (H1N1) outbreak has caused 94,512 cases and 429 fatalities as of July 6, 2009 (11), and 10,156 laboratory-confirmed cases leading to 1115 hospitalizations and 45 deaths in Canada as of July 15, 2009 (12). The number of individuals affected by the virus and the distribution within provinces is unknown; seroprevalence is labour intensive and hampered by the lack of a high-throughput method. Assessing the prevalence of antibodies against various influenza strains is of particular importance during an interepidemic phase because the results may identify unexposed populations and guide allocation of vaccine or antiviral therapy. MBA can be used for rapid detection of infection or previous exposure. In the present study, we compared samples from two countries differing in influenza epidemiology and rate of influenza immunization. The distinctly different origin of the samples serves to illustrate the use of multiplex suspension-bead array for detection of antibody response to multiple influenza strains in a high-throughput system requiring low sample quantity. IgA1 was surveyed because it is the primary mucosal antibody responsible for neutralization of the virus on challenge and, due to its relatively low yield in serum, to showcase the sensitivity of the assay.
METHODS
Samples
Frozen plasma samples from 60 adult donors enrolled in a cohort study in Nairobi (Kenya, Africa) were analyzed for IgA1 and IgG1 antibodies specific to influenza and control antigens. The study population included North American controls (30 individuals from Winnipeg [Manitoba]) and 30 women from Kenya. The study was approved by local ethics boards in Winnipeg and Nairobi.
Materials
Latex microspheres used in the assay, and assay reagents (wash buffer, assay buffer, reporter antibody and filter well plate) were purchased from Bio-Rad Laboratories (USA). Detection antibody (Mouse Anti-Human IgA1-Biot clone B3506B4, and IgG1-Biot clone 4e3) was purchased from SouthernBiotech (USA). Recombinant influenza HA proteins (A/Wyoming/3/2003, A/New Caledonia/20/99, A/ Vietnam/1203/2004 and B/Jilin/20/2003) were purchased from Protein Sciences Corporation (USA). Recombinant HA from B/ Tokyo was purchased from Advanced ImmunoChemical Inc (USA). As a positive control, recombinant HIV p24 was purchased from Fitzgerald Industries International (USA).
Microsphere coupling
Unconjugated, carboxylated microspheres were dispersed with sonication and vortexed for 60 s, and 5×106 microspheres (400 μL) were dispensed into a 1.5 mL centrifuge tube and spun down. The pellet was resuspended in 80 μL of activation buffer, and sonicated until homogeneous. Ten microlitres of Sulfo-NHS solution (10 mg Sulfo-NHS per 2 mL activation buffer) was added to the microsphere suspension and vortexed, and 10 μL of EDC solution (10 mg EDC per 2 mL activation buffer) was added to the solution and vortexed. The beads were incubated for 20 min in the dark at room temperature. The activated microspheres were centrifuged and the supernatant aspirated. The pellet was resuspended in 250 μL of coupling buffer, centrifuged and supernatant aspirated. The step was repeated. Each specific bead set was resuspended with 250 μL (100 μg/mL) of its assigned antigen preparation. The solutions were incubated at room temperature for 1 h to mix and then washed twice. After the second wash cycle, 250 μL of phosphate-buffered saline of pH 7.4, 1% bovine serum albumin and 0.05% sodium azide (PBSBN) was added. The tube was incubated for 30 min in the dark at room temperature. After incubation, centrifugation and removal of the supernatant, the pellet was resuspended in 100 μL of PBSBN. The concentration of conjugated beads in each set was enumerated by a hemocytometer. The bead sets were kept at 4°C in the dark until needed.
Master bead mix
The master bead mix containing the bead sets coupled to the proteins of interest was mixed to ensure the total concentration of beads was 68 beads/μL. Based on the volume of the master bead mix required, each individual bead set was added to achieve the desired final concentration in PBSBN.
Plate preparation
The wells of a filter plate were prewetted with 100 μL of Bio-Plex Assay Buffer (Bio-Rad Laboratories) and vacuum dried. The master bead mix was vortexed and sonicated for 2 min. One hundred microlitres of master bead mix were pipetted into each well followed by vacuum removal. The wells were washed twice with 100 μL of Bio-Plex Wash Buffer (Bio-Rad Laboratories) and vacuumed. The serum samples were diluted to 10−1, 10−2, 10−3 and 10−4 concentration with PBSBN. One hundred microlitres of sample for each dilution and a duplicate were added to the wells. The plate was covered and placed on a shaker to incubate for 30 min at room temperature. The wells were then vacuum dried and washed three times with Bio-Plex Wash Buffer.
Secondary biotinylated detection antibody
One hundred microlitres at a concentration of 0.5 mg/mL of detection antibody (Mouse Anti-Human IgA1-Biot) (1:400 PBSBN) was added to each well. The plate was covered, incubated and shaken at room temperature for 30 min. Following incubation, the wells were vacuum dried and washed three times with Bio-Plex Wash Buffer.
Reporter antibody
One hundred microlitres of Bio-Plex Streptavidin-PE (Bio-Rad Laboratories), 1 mg/mL (1:500 PBSBN), was added to each well. The plate was covered and incubated on a shaker at room temperature for 10 min. Following incubation, the wells were vacuum dried and washed three times with Bio-Plex Wash Buffer. One hundred twenty-five microlitres of Bio-Plex Assay Buffer was used to resuspend the samples. The plate was covered on a shaker for 20 min. The samples were then transferred from the filter plate to a flat-bottom 96-well plate just before analysis.
Plate analysis
Before each plate calibration and validation, beads (Bio-Rad Laboratories) were used to ensure quality control. The flat-bottom 96-well plate was run on the Bio-Plex Protein Array System (Bio-Rad Laboratories) using Luminex xMap technology. The system was managed with Bio-Plex Manager 4.0 software (Bio-Rad Laboratories).
Titration curve
Serum samples were run at 10−1, 10−2, 10−3, 10−4 and 10−5 to determine the optimal dilution for the detection of IgA1 and IgG1 responses.
Controls
Quality control using Bio-Rad calibration and validation kits was performed before every plate run. As a positive control, a bead coupled to p24 with a known HIV-positive serum sample was used. The negative controls included beads incubated with phosphate-buffered saline only; uncoupled beads were included in the master mix and, thus, tested with each sample.
RESULTS
Titration curve
Titration data indicated that 10−3 was optimal for IgA1 detection and 10−4 for IgG1. This concentration allows for quantitation of IgA1 and IgG1 by avoiding the saturation of response seen at higher concentrations. Figure 2 depicts the titration curves for IgA1 and IgG1.
Figure 2).
A Immunoglobulin (Ig) A1 titration curve of Winnipeg donor (D97) responses to influenza hemagglutinin (HA) antigens Jilin (B) and Wyoming (H3N2). B IgG1 titration curve of Winnipeg donor (D19) responses to influenza HA antigens Wyoming, Vietnam and New Caledonia
Serum assay
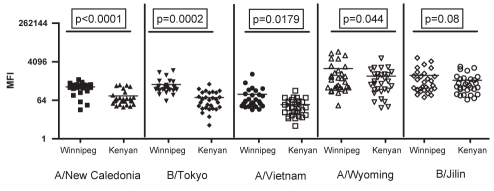
Statistically significant differences in IgA1 exist between the serum samples from Winnipeg donors and those from Kenyan donors. Data were compared using an unpaired t test with Welch’s correction for samples with unequal variances. The Winnipeg donors had higher mean fluorescence intensities (MFI) with significant P values for anti-HAIgA1 to A/Wyoming/3/2003 (P=0.044), A/Vietnam/1203/2004 (P=0.0179), A/New Caledonia/20/99 (P<0.0001) and B/Tokyo/53/99 (P=0.0002) (Figure 3). No significant differences were seen between the groups in their response to B/Jilin/20/2003.
Figure 3).
Significant differences in immunoglobulin A1 response to influenza antigens as measured by mean fluorescence intensity (MFI)
IgG1 from Winnipeg donors and those from Kenyan donors were also compared. Data were compared using an unpaired t test with Welch’s correction for samples with unequal variances. The Winnipeg donors had higher MFI with significant P values for A/Wyoming, B/ Tokyo and B/Jilin (P=0.0135, P=0.006, P=0.026, respectively) (Figure 4).
Figure 4).
Significant differences (denoted by asterisks) in immunoglobulin G1 response to flu antigens as measured by mean fluorescence intensity (MFI)
The results of IgG1 MBA to HIA were compared in a subset of individuals. When an MFI threshold of 500 was compared with an HIA titre of greater than 40, the agreement between the assays was 70%. The HIA titre was greater than 40 in 10% of MBA-negative individuals and, conversely, an MBA of greater than 500 MFI was detected in 10% of HIA-negative individuals.
DISCUSSION
MBA analysis has several benefits over traditional HIA methodology for the detection of anti-influenza IgA1 and IgG1. By simultaneously assaying for different type-specific anti-influenza IgA1 and IgG1, a full influenza workup can be performed on a small sample volume. This high-throughput capability is necessary for rapid identification of influenza strains during an epidemic. Second, the ability of MBA to simultaneously assay for multiple antiflu antibodies is invaluable for measuring vaccine response toward the vaccine strains and assessing cross-response to strains not included in the vaccine formulation. Another advantage of this flexible platform is the ability to couple different fragments of influenza proteins, thus enabling the detection of more specific targets within these proteins. The usefulness of the assay for seroepidemiology was demonstrated by the data presented. This semiquantitative measure allows for better characterization of the antibody response to multiple influenza strains simultaneously, and determination of different Ig classes by the use of diverse secondary antibodies.
Observed differences in IgG1 and IgA1 antibody responses to influenza antigens may be due to several factors. Circulating strains of influenza can vary geographically, creating different antibody profiles among distinct populations. Another possibility is that because Kenya does not have an influenza immunization program equivalent to the one in Canada, the Kenyan population does not have the exposure that vaccinations afford. As expected, the response to A/ New Caledonia/20/99 was significantly higher in the Winnipeg population. The finding is probably the result of this strain circulating in the community and being a part of the trivalent vaccine for the past six years in North America. Winnipeg donors also showed higher levels to B/Tokyo/53/99, but no difference in B/Jilin/20/2003. Again, this can be explained by the belief that although type B viruses have circulated in Kenya, the expected higher responses by Kenyan donors were offset and even exceeded by the vaccination programs in Canada.
Unexpectedly, a response by Winnipeg donors to A/Vietnam/1203/2004 (avian flu) without any illness suggests that current vaccination programs may confer some heterotypic immunity. It has been noted that pre-existing IgA to one type of influenza can cross-react with other types of virus much more strongly than IgG. This may be due to the dimeric nature of secretory IgA found in the respiratory epithelium (13,14). These factors may explain the cross-reactions seen with the H5N1 virus.
The limitation of the present study is the lack of correlation with clinical protection from influenza-like illness.
CONCLUSIONS
Serum IgA1 and IgG1 antibodies specific for influenza antigens were successfully detected using this method. Significant differences in the amount of flu-specific IgA1 and IgG1 antibodies were demonstrated between Winnipeg and Kenyan donors, reflecting geographical differences and the lack of influenza immunization in Kenya.
MBA analysis is a relatively quick and cost-effective method for serum antibody analysis. The potential to simultaneously assay small sample volumes for a multitude of antigens makes this method invaluable for the study of influenza epidemiology and, potentially, other infectious agents, as well as for monitoring vaccine response. The rapid detection of anti-influenza IgA1 can be diagnostic of recent infection. Although correlation with HIA titres and protection from clinical illness caused by influenza would be important to establish, the assay may be useful in assessing the extent to which a population has been exposed to a specific strain and may assist in targeting preventive interventions to nonimmune populations.
Acknowledgments
The authors thank Catherine Card, Paul McLaren, Sandy Koesters, Judy Lawrence, Conar O’Neil, The University of Manitoba and The Gates Grand Challenges in Global Health.
REFERENCES
- 1.Doherty PC, Turner SJ, Webby RG, Thomas PG. Influenza and the challenge for immunology. Nat Immunol. 2006;7:449–55. doi: 10.1038/ni1343. [DOI] [PubMed] [Google Scholar]
- 2.Nicholson KG, Wood JM, Zambon M. Influenza. Lancet. 2003;362:1733–45. doi: 10.1016/S0140-6736(03)14854-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cox RJ, Brokstad KA, Ogra P. Influenza virus: Immunity and vaccination strategies. Comparison of the immune response to inactivated and live, attenuated influenza vaccines. Scand J Immunol. 2004;59:1–15. doi: 10.1111/j.0300-9475.2004.01382.x. [DOI] [PubMed] [Google Scholar]
- 4.Levinson W. Medical Microbiology & Immunology. 8th edn. Toronto: McGraw Hill; 2004. [Google Scholar]
- 5.Tamura S, Kurata T. Defense mechanisms against influenza virus infection in the respiratory tract mucosa. Jpn J Infect Dis. 2004;57:236–47. [PubMed] [Google Scholar]
- 6.Jeon SH, Ben-Yedidia T, Arnon R. Intranasal immunization with synthetic recombinant vaccine containing multiple epitopes of influenza virus. Vaccine. 2002;20:2772–80. doi: 10.1016/s0264-410x(02)00187-1. [DOI] [PubMed] [Google Scholar]
- 7.Rothbarth PH, Groen J, Bohnen AM, de Groot R, Osterhaus AD. Influenza virus serology – a comparative study. J Virol Methods. 1999;78:163–9. doi: 10.1016/s0166-0934(98)00174-8. [DOI] [PubMed] [Google Scholar]
- 8.Pickering JW, Martins TB, Schroder MC, Hill HR. Comparison of a multiplex flow cytometric assay with enzyme-linked immunosorbent assay for quantitation of antibodies to tetanus, diphtheria, and Haemophilus influenzae Type b. Clin Diagn Lab Immunol. 2002;9:872–6. doi: 10.1128/CDLI.9.4.872-876.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morgan E, Varro R, Sepulveda H, et al. Cytometric bead array: A multiplexed assay platform with applications in various areas of biology. Clin Immunol. 2004;110:252–66. doi: 10.1016/j.clim.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 10.Elshal MF, McCoy JP. Multiplex bead array assays: Performance evaluation and comparison of sensitivity to ELISA. Methods. 2006;38:317–23. doi: 10.1016/j.ymeth.2005.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.WHO influenza update. < www.who.int/csr/don/2009_07_06/en/index.html> (Accessed on June 2, 2010).
- 12.Public Health Agency of Canada. Cases of Pandemic (H1N1) 2009 virus in Canada. < www.phac-aspc.gc.ca/alert-alerte/swine-porcine/surveillance> (Accessed on June 2, 2010).
- 13.Asahi-Ozaki Y, Yoshikawa T, Iwakura Y, et al. Secretory IgA antibodies provide cross-protection against infection with different strains of influenza B virus. J Med Virol. 2004;74:328–35. doi: 10.1002/jmv.20173. [DOI] [PubMed] [Google Scholar]
- 14.Tamura S, Tanimoto T, Kurata T. Mechanisms of broad cross-protection provided by influenza virus infection and their application to vaccines. Jpn J Infect Dis. 2005;58:195–207. [PubMed] [Google Scholar]