Abstract
The majority of eukaryotic genes undergo alternative splicing, an evolutionarily conserved phenomenon, to generate functionally diverse protein isoforms from a single transcript. The fact that defective pre-mRNA splicing can generate non-functional and often toxic proteins with catastrophic effects, accurate removal of introns and joining of exons is vital for cell homeostasis. Thus, molecular tools that could either silence a disease-causing gene or regulate its expression in trans will find many therapeutic applications. Here we present two RNA-based approaches, namely RNAi and theophylline-responsive riboswitch that can regulate gene expression by loss-of-function and modulation of splicing, respectively. These strategies are likely to continue to play an integral role in studying gene function and drug discovery.
Keywords: Alternative splicing, spliceosome, RNAi, theophylline-responsive riboswitch
1. Introduction
The diversity of the human proteome is attributable in large part to the complexity and processing of pre-mRNAs via a process known as RNA splicing. This biochemical pathway leads to the assembly onto pre-mRNA of the mature spliceosome, which catalyzes the excision of non-coding sequences from precursor mRNA. A pre-mRNA is defined by four cis-acting elements: the 5′ and 3′ splice sites (ss), the branchpoint sequence, and the poly(Y)-tract. These cis-acting elements direct the assembly of the basal splicing machinery that leads to the removal of introns and joining of exons. In general, pre-mRNAs that undergo alternative splicing harbor suboptimal splicing signals and often require the help of auxiliary regulatory elements known as splicing enhancers and silencers for splice site pairing. These elements provide the binding surface for splicing regulatory proteins that communicate with the basic splicing machinery to define splice site choice (for review see Ref. 1-3).
Alternative splicing of pre-mRNA is now considered to be the most important source of protein diversity in vertebrates. It is estimated that more than 60% of human genes generate transcripts that are alternatively spliced (4, 5) and 70–90% of alternative splicing events affect coding capacity of genes (6). Importantly, deregulated splice variant expression has been identified as the cause of a number of genetic disorders, and certain forms of cancer have been linked to unbalanced isoform expression from genes involved in cell cycle regulation or apoptosis (7, 8). Given the critical role of alternative splicing in a variety of cellular processes (9), strategies that could influence pre-mRNA splicing decisions will have far-reaching effects in biotechnology and medicine. This chapter focuses on the design and construction of two RNA-based molecular approaches, namely RNA interference (RNAi) and theophylline riboswitch, with the ultimate aim of regulating gene expression. Whereas RNAi exerts its effect in trans to downregulating the expression of a target gene, theophylline-responsive riboswitch controls gene expression by modulating pre-mRNA splicing.
The use of RNAi as a gene-silencer strategy represents a powerful tool in the field of small-molecule nucleic acid-based therapeutics. Described first in Caenorhabditis elegans, this biological phenomenon has subsequently been studied in a wide range of organisms (for recent review, see (10)). RNAi involves double-stranded RNA molecules of approximately 20–25 nucleotides termed short interfering RNAs (siRNAs) that are processed by the endogenous RNAse III family member Dicer and are incorporated into an RNA-induced silencing complex (RISC) in a process that prevents the expression of a particular gene (11). The relative ease with which a siRNA can be designed and synthesized, its specificity and potency and, most importantly, the ability to preferentially suppress the expression of mutant alleles makes this approach highly appealing. Moreover, RNAi not only has the potential to be an effective therapeutic tool, but also enables the identification of genes that regulate alternative splicing (7, 12). However, RNAi is faced with limitations when compared with other approaches such as the theophylline-responsive riboswitch and antisense oligonucleotides, which unlike RNAi, can modulate mRNA isoform levels (13-16).
Riboswitches (17, 18) are natural RNA aptamers that regulate gene expression by binding to small-molecule ligands. As RNA structures (14, 19) are known to influence splice site choice, we hypothesized that sequestering of splicing regulatory elements within RNA secondary structure could influence splice site choice. By exploiting the ligand-induced conformational rearrangement property of theophylline riboswitch we have demonstrated the control of alternative splicing both in vitro and in vivo (13, 14, 20). This novel technology represents the possibility for controlling splicing of a trans gene in a gene therapy setting where the target gene expression could be controlled in a ligand-dependent manner.
2. Materials
2.1. RNA Interference
All buffers and solutions were made using filtered deionized, 18 MΩ water purified by a Barnstead MP-3A Megapure system.
Fetal bovine serum (Irvine Scientific, CA, USA).
MCF7 cells: This is a malignant mammary epithelial cell line (ATCC, Manassas, VA, http://www.atcc.org/).
Minimum essential medium (MEM): 2 mM L-glutamine and Earle's BSS adjusted to contain 1.5 g/l sodium bicarbonate, 0.1 mM non-essential amino acids, and 1 mM sodium pyruvate and supplemented with 0.01 mg/ml bovine insulin; and 10% fetal bovine serum (ATCC, Manassas, VA, http://www.atcc.org/).
Lipofectamine 2000 transfection reagent (Invitrogen, CA, USA).
Opti-MEM (Invitrogen, CA, USA).
siRNAs: We synthesized the following RNAi oligos against human CEA: sense 5′-CUGGCCAGUUCCGGGUAUA-3′ and antisense 5′-UAUACCCGGAACUGGCCAG-3′ (nucleotides 404–422, numbering from the initial start codon); sense 5′-CGGGACCUAUGCCUGUUUU-3′ and antisense 5′-AAAACAGGCAUAGGUCCCG-3′ (nucleotides 1950–1968, numbering from the initial start codon), (Qiagen, CA, USA). A scrambled control siRNA was synthesized at the City of Hope DNA/RNA core facility and is randomized with respect to its nucleotide distribution. The siRNAs were diluted in water to a stock concentration of 20 μM.
2.2. Western Blot
ECL plus Western Blotting detection reagents (Amersham Biosciences, Buckinghamshire, England).
Blocking buffer: 150 mM NaCl, 5 mM EDTA, 50 mM Tris-HCl, pH 7.5, 0.05% Triton X-100, 0.25% gelatin. Blocking buffer can be made as a 10x stock and stored at room temperature without gelatin. Store the 1x dilution at 4°C.
Lysis buffer: 10 mM Tris-HCl, pH 8, 140 mM NaCl, 0.025% sodium azide, 1% Triton X-100, 1 mM EDTA, 1 mM PMSF, and 1 mM sodium vanadate.
2x Gel loading buffer: 0.125 M Tris-HCl, pH 6.8, 4% SDS, 20% glycerol, 10% 2-mercaptoethanol, 0.005% bromophenol blue.
Anti-β-actin Goat Polyclonal IgG (Santa Cruz Biotechnology, Inc, CA, USA). Dilution for immunoblots, 1:2000.
Donkey α-Goat IgG-HRP (Santa Cruz Biotechnology, Inc, CA, USA). Dilution for immunoblots, 1:5000.
Goat α-Mouse IgG-HRP (Santa Cruz Biotechnology, Inc, CA, USA). Dilution for immunoblots, 1:5000.
α-CEA T84.66: This is a chimeric monoclonal antibody of high specificity for tumor-associated CEA (22). Dilution for immunoblots, 1:2000.
Immun-blot PVDF/Filter Paper (BioRad, CA, USA).
2.3. Theophylline-Responsive Riboswitch
α-[32P] UTP: 3000 Ci/mmol, 10 μCi/μl (Perkin Elmer, MA, USA).
8% Denaturing polyacrylamide gel: From the Sequa-gel sequencing system kit (see Section 2.3, Step 17), mix 6.4 ml of Sequa-gel concentrate, 11.6 ml of Sequa-gel diluent, and 2 ml of Sequa-gel buffer. To this solution, add 200 μl of 10% ammonium persulfate and 20 μl of TEMED. Swirl gently to mix and cast the gel.
BC300: 20 mM HEPES, pH 8.0, 20% glycerol, 300 mM KCl, 0.2 mM EDTA.
Dithiothreitol (DTT): To prepare a 0.1 M stock, add 0.154 g of DTT in 10 ml RNAse-free water.
Dulbecco's Modified Eagle medium supplemented with L-glutamine and 10% fetal bovine serum (Omega Scientific, CA, USA).
Electrophoresis sample buffer: Add 2.4 ml 1 M Tris-HCl, pH 6.8, 3 ml 20% SDS, 3 ml 100% glycerol, 1.6 ml 2-mercaptoethanol, 0.006 g bromophenol blue. Store at 4°C.
Exon junction primers: The following primers were used to assess theophylline-mediated modulation of alternative splicing in vivo: #43573, 5′-GGGCCAGCTGTTGGGGTCGA-3′ and #43754, 5′-GGGCCAGCTGTTGGGCTCGC-3′ as forward primers and oligonucleotide #39368, 5′-TAGA GGATCCCCACTGGAAAGACCG-3′ as reverse primer.
5x First-strand buffer for cDNA synthesis: 250 mM Tris-HCl, pH 8.3, 375 mM KCl, 15 mM MgCl2.
Glycogen: Stock solution is 20 mg/ml (Roche Applied Science, Mannheim, Germany). Dilute to 5 mg/ml and store in small aliquots at −20°C.
HeLa nuclear extract for in vitro splicing: HeLa nuclear extract was prepared from 10 l HeLa-S3 cells (National Cell Culture Center, MN, USA) as described previously (21). HeLa nuclear extracts can also be purchased from 4C Biotech (Seneffe, Belgium).
Lipofectamine 2000 (Invitrogen, CA, USA).
Molecular Dynamics PhosphorImager and the ImageJ software (http://rsb.info.nih.gov/ij/) were used to quantitate the products of the in vitro RNA splicing reaction and RNAi experiments.
M-MLV reverse transcriptase (Invitrogen, CA, USA).
Opti-MEM Reduced Serum Medium (Invitrogen, CA, USA).
Proteinase K (Roche Applied Science, Mannheim, Germany).
RNasin: 10 units/10 μl reaction (Promega, WI, USA).
Sequa-gel sequencing system kit: Contains Sequa-gel concentrate, Sequa-gel diluent, and Sequa-gel buffer (National Diagnostics, Georgia USA).
Splicing loading buffer: Mix 2 mg of xylene cyanol and 2 mg of bromophenol blue in 10 ml of deionized formamide.
7mG(ppp)G RNA cap structure analog: Dilute the stock in 131 μl RNase-free water to give a working concentration of 10 mM. For transcription reactions, use a 5:1 ratio of 7mG(ppp)G RNA cap structure analog to GTP.
Splicing stop buffer: To stop in vitro splicing reaction, add 50 μl 2x PK buffer, 2.5 μl 10 mg/ml Proteinase K, 2 μlof 5 mg/ml glycogen, and RNase-free water to a total volume of 87.5 μl.
2x PK buffer: 200 mM Tris-HCl, pH 7.5, 25 mM EDTA, pH 8.0, 300 mM NaCl, 2% SDS.
10x RNA polymerase reaction buffer: 400 mM Tris-HCl, pH 7.9, 60 mM MgCl2, 20 mM spermadine, 100 mM DTT.
10x transcription master mix: Combine 3.1 μl each of 4 mM GTP, 20 mM ATP, 20 mM CTP, 4 mM UTP, and 10 μl of 0.1 M DTT. The mixture is gently agitated and briefly micro-centrifuged to bring the mixture to homogeneity. Dispense 2.24 μl per transcription reaction. Store mix @ −20°C with limited freeze-thaw cycles.
25 mM Theophylline: Add 0.09 g theophylline (Sigma-Aldrich, MO, USA) to 20 ml of RNase-free water. For a final concentration of 2 mM, add 2 μltoa25 μl splicing reaction.
T7 RNA polymerase: 10 units/10 μl reaction (New England Biolabs, MA, USA).
13% denaturing polyacrylamide gel: From the Sequa-gel sequencing system kit (see Section 2.3, Step 17), mix 10.4 ml of Sequa-gel concentrate, 7.6 ml of Sequa-gel diluent, and 2 ml of Sequa-gel buffer. To this solution, add 200 μl of 10% ammonium persulfate and 20 μl of TEMED. Swirl gently to mix and cast the gel.
3. Methods
3.1. siRNA-Mediated Degradation of CEA mRNA
The siRNA inhibits gene expression by degrading the corresponding endogenous mRNA. RNAi has been successfully used in establishing the role of splicing regulatory proteins in human cancer (22-25). In addition, siRNA-mediated downregulation of BclxL isoform in TRAIL-resistant cells has been shown to inhibit cell proliferation and sensitize TRAIL-induced apoptosis in human cancer cells with both acquired and intrinsic TRAIL resistance (26). However, the design of siRNA that targets with high efficiency and specificity is critical to successful gene silencing. Here we show that RNAi can be used to downregulate carcinoembryonic antigen (CEA), a glycoprotein that is overexpressed in a variety of cancers (27, 28).
3.1.1. siRNA Design
Prior to finding an RNAi target on the gene of interest, the mRNA sequence or sequence accession number should be retrieved from Refseq at NCBI. The Entrez query tool can be located at http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Nucleotide. On the “Limits” tab, pull down the “Only from” menu and choose Refseq. This will restrict the query to the RefSeq database only. Other potential search routes are: (1) Search LocusLink by gene name or symbol at http://www.ncbi.nlm.nih.gov/LocusLink/ and (2) Search Entrez Gene at http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene (see (29), Table 1 for a list of other Web servers for siRNA design). To prevent off-target effects, check the gene of interest for significant homology to other genes or sequences in the genome using the NCBI Blast tool – Nucleotide–nucleotide BLAST (blastn), or Blat tool on UCSC Genome Website http://genome.ucsc.edu/cgi-bin/hgBlat, or Ensembl Blast at http://www.ensembl.org/Multi/blastview.
Many different algorithms and programs that depend on sequence characteristics or target mRNA secondary structures are available now for the design of siRNAs for post-transcriptional gene silencing (29-32). The whole length of an mRNA sequence can be targeted by RNAi though it is especially important to be cognizant that not all siRNAs target the mRNA with equal efficiency. It is crucial to perform a homology search for candidate siRNA targets against other mRNA sequences of the genome. In general, a siRNA should not have significant homology with unrelated sequences especially at its 3′ end. If a siRNA design program does not provide the function for homology sequence, a manual BLAST search is mandatory for avoiding off-target effects. The main challenge for developing siRNA in vivo is delivering duplex RNA intact to a target tissue. Many of these pharmacokinetic obstacles also confront antisense oligonucleotides. However, a crucial difference between the two is that antisense oligonucleotides comprise just one nucleic acid strand, whereas siRNA is made up of two strands. On a practical level, the mass of a synthetic duplex RNA is twice that of a traditional antisense oligonucleotide and can result in increased costs. The higher molecular weight of siRNA can also make uptake by cells more difficult. Furthermore, the presence of a second nucleic acid strand increases the potential for off-target effects. For effective delivery, high duplex stability is needed because duplex RNA must remain hybridized until it enters the RISC complex. The most effective siRNA is identified experimentally with rules that govern effective siRNA design dependent on the particular application. Here we present some sequence-characteristic-based guidelines to design high-efficiency siRNAs (33, 34):
Regions located 50–100 nt downstream of the start codon (ATG) make good targets for RNAi.
Sequence motifs that have AA(N19)TT, NA(N21), or NAR(N17)YNN where N is any nucleotide, R is purine (A, G), and Y is pyrimidine (C, U) make good targets for RNAi.
Avoid introns. As siRNAs are processed only in the cytoplasm and not within the nucleus, post-transcriptional processing would prevent targeting of the siRNA.
Sequences with high G + C content (more than 50%) should be avoided. The GC content of the sense strand should be between 30 and 50%.
Stretches of polynucleotide repeats should be avoided.
3.1.2. RNAi Against CEA
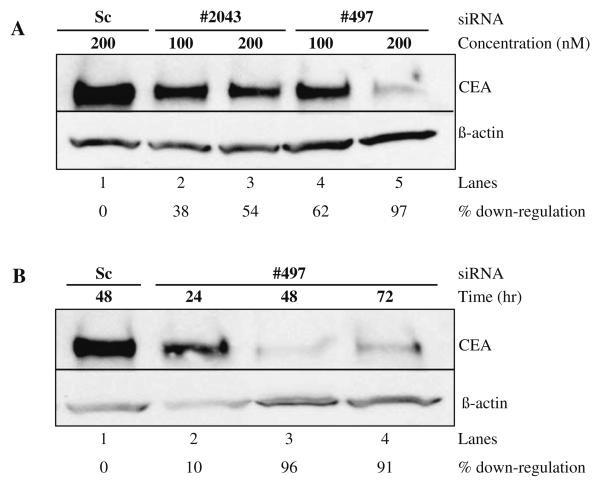
To investigate the relationship between overexpression of CEA and breast cancer progression, we employed two chemically synthesized siRNAs (siRNAs #497 and #2043) to downregulate CEA mRNA (see Note 1). To monitor the cellular uptake, a scrambled control siRNA (Sc) bearing fluorescein at the 5′ end of the antisense strand was also synthesized. MCF7 cells that are known to overexpress CEA were transiently transfected with different siRNAs (see Section 3.1.3 for details on animal cell transfection, see Note 2). After indicated time (we recommend performing a time course), cells were harvested and protein lysates were resolved on a 4–20% SDS-polyacrylamide gradient gel followed by transfer to a polyvinylidene fluoride (PVDF) membrane. Transferred proteins were probed with either anti-CEA or anti-β-actin antibody. Our results demonstrate that compared to Sc control, siRNA #497 significantly downregulated CEA protein (Fig. 10.1A)(see Note 3). By contrast, siRNA #2043 showed moderate downregulation of CEA protein (54%). This observed difference could be due to the accessibility of the target region and not unequal loading of sample as evidenced by the levels of probes to β-actin protein (Fig. 10.1A). We note that high dosages of siRNA #497 are more effective at gene silencing; suggesting that nuclease digestion likely limits the efficacy at the lower concentration (Fig. 10.1A, compare lane 4 with lane 5). A time course presented in Fig. 10.1B demonstrates that knockdown of CEA protein occurs as early as 24 h and is effective at least up to 72 h post-treatment. The slight increase in protein levels at 72 h is apparently the effect of siRNA depletion (Fig. 10.1B, compare lane 3 with lane 4).
Fig. 10.1. Gene silencing of CEA by RNAi.
(A) Western blot of MCF7 cells treated for 48 h with siRNAs #2043, #497, and a scrambled (Sc) control at a concentration of 100 nM (lanes 2 and 4) or 200 nM (lanes 1, 3, and 5). (B) MCF7 cells treated after 24, 48, and 72 h with 200 nM siRNAs #497 or 200 nM Sc as indicated. Upper panels in both A and B show a Western blot probed with α-CEA T84.66 while the lower panels were treated with anti-β-actin antibody. The percentage of downregulation is indicated below each panel and calculated by comparing CEA downregulation to β-Actin and normalizing to the scrambled control.
3.1.3. Transient Transfection of MCF7 Cells
In 2 ml MEM supplemented with 10% fetal bovine serum, seed the following number of cells in each well of a six-well plate 24 h prior to transfection: 6 × 105 cells (70% confluency for harvest time at 24 h), 5 × 105 cells (60% confluency for harvest time at 48 h), and 4 × 105 cells (50% confluency for harvest time at 72 h).
Cells should be maintained under standard incubation conditions (humidified atmosphere, 5% CO2,37°C) (see Note 4).
Next day, prepare two mixtures in separate eppendorf tubes (A and B). Tube A contains either 100 or 200 nM siRNA diluted with Opti-MEM (see Note 5) to a final volume of 250 μl. Tube B contains 2 μl Lipofectamine 2000 (see Note 6) diluted with Opti-MEM to a final volume of 250 μl. A master mix is recommended if all the wells to be transfected contain the same siRNA.
After an incubation period of 5 min, both tubes A and B are combined and incubated for 20 min to allow complex formation between siRNA and the Lipofectamine 2000.
Prior to the completion of the 20-min incubation period, aspirate the media from cells and replace with 2 ml fresh media to each well.
Add RNAi–Lipofectamine 2000 complex (500 μl per well) dropwise. Distribute around the well and gently swirl the plate.
Cells should be maintained under standard incubation conditions (humidified atmosphere, 5% CO2,37°C) for an additional 24–72 h depending on the harvest time.
Assay the cells using the appropriate post-treatment protocol (see Section 3.1.4 for Western Blot Protocol and Section 3.2.7 for RT-PCR analysis; see Note 7).
3.1.4. Western Blot Analysis
After the indicated time, cells are harvested and the lysate is prepared by adding 150–200 μl of lysis buffer.
The cells are homogenized by the passage (approximately 10 times) through a 1-ml syringe with a 22-gauge needle.
Next, spin the homogenized cell lysate @ 10,000 × g for 20 min at 4°C. Remove the supernatant and to the cell pellet add 2x Gel loading buffer.
4. The protein lysates were resolved on a 4–20% SDS-polyacrylamide gradient gel essentially as described earlier (35). Next, the proteins were transferred to an Immun-Blot PVDF membrane and incubated in blocking buffer for 60 min at room temperature on a shaking platform (with a change of buffer after 30 min).
5. The primary antibodies, either α-CEA T84.66 or anti-β-actin Goat Polyclonal IgG, are next added to the blocking buffer and incubated in the presence of the membrane overnight at room temperature on a shaking platform (see Note 8).
6. Next day, the membrane is washed three times (20 min each rinse) in blocking buffer followed by an additional incubation with the secondary antibody (either Goat α-Mouse IgG-HRP or Donkey α-Goat IgG-HRP) for 1 h in blocking buffer.
7. After washing, develop the membrane using Amersham ECL Plus Western Blotting detection reagents, following the manufacturer's recommendation (see Note 9).
3.2. Theophylline-Responsive Riboswitch
It is widely accepted that deregulated alternative splicing is the cause of a number of human diseases. It has been estimated that approximately 15% of all mutations that cause genetic diseases result in defective splicing of pre-mRNA: defects in splicing has been shown to be associated with genetic disorders such as β-thalassemia, cystic fibrosis, Duchenne muscular dystrophy, etc. In addition, certain forms of cancer have been linked to unbalanced isoform expression from genes involved in cell cycle regulation or angiogenesis (reviewed in Refs. (12, 36)). Given the significance of alternative splicing in generating protein diversity and its link to human diseases, mRNA splicing has become an important target of therapeutic intervention (37).
We have recently demonstrated that a theophylline-responsive riboswitch, an RNA aptamer that binds to theophylline with high affinity and specificity, can modulate alternative splicing both in vitro and in cultured cells (13, 14, 20). This approach can be modified to regulate the expression of virtually any trans-genebycontrolling its splicing. Here we describe the experimental details for controlling pre-mRNA splicing with the theophylline-responsive riboswitch.
3.2.1. Design of Theophylline-Responsive Riboswitch
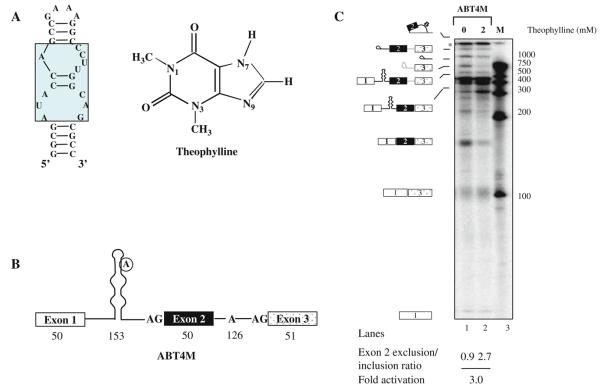
Theophylline riboswitch is an RNA aptamer consisting of a 15-nt core sequence, which provides affinity and specificity for RNA–theophylline interaction (Fig. 10.2A). Remarkably, caffeine that differs from theophylline by only a methyl group at the N-7 position in the imidazole ring binds to theophylline aptamer with 10,000-fold less affinity (38). Theophylline aptamer is unstructured; however, ligand-induced conformational rearrangement of RNA allows theophylline binding by formation of thermodynamically stable RNA–theophylline complex. Although theophylline riboswitch can be adapted to control pre-mRNA splicing by targeting a variety of cis-acting regulatory elements, here in this section we focus our attention at the branchpoint sequence. We constructed a model pre-mRNA comprising of three exons interrupted by two introns (ABT4M, Fig. 10.2B). Ourdataindicate that sequestering of intron 1 branchpoint by theophylline allows intron 2 branchpoint to choose between the 5′ ss of exon 1 and 2 for the first step of splicing. Thus, depending upon which of the two 5′ ss is utilized determines the amount of exon 2 included/excluded mRNA (see Sections 3.2.2 and 3.2.5). Since theophylline aptamer's lower stem affects thermodynamic stability of RNA-theophylline complex, splicing can be fine-tuned by simply altering the size of the stem (14, 19)(see Notes 10, 11).
Fig. 10.2. Ligand-dependent modulation of alternative splicing using theophylline-binding aptamer in vitro.
(A) Diagram of the theophylline RNA aptamer sequence (left panel) and the chemical structure of theophylline (right panel ). The residues that are conserved for theophylline binding are enclosed in the rectangular box. (B) Schematic representation of a theophylline-binding aptamer sequestering the branchpoint of a model pre-mRNA. The encircled adenosine residue represents branch nucleotide. Open boxes represent exon sequences and horizontal lines between exons indicate introns. The numbers indicate size of the exon or intron. (C) In vitro splicing of ABT4M pre-mRNA. 32P-labeled ABT4M pre-mRNA was subjected to in vitro splicing in the absence (lane 1) or presence of 2 mM theophylline (lane 2). The extracted RNAs were fractionated on a 13% polyacrylamide denaturing gel. Schematic representations of the various RNA species are indicated on the left. Molecular Dynamics PhosphorImager and the ImageJ software were used to quantitate the products of the in vitro and in vivo RNA splicing reactions (see Fig. 10.3).
3.2.2. Theophylline-Responsive Riboswitch for the Modulation of Pre-mRNA Splicing In Vitro
32P-labeled ABT4M pre-mRNA was transcribed (see Section 3.2.3) and incubated in HeLa nuclear extract in the absence or presence of theophylline (Fig. 10.2C). Splicing of ABT4M substrate gave rise to two spliced products, a slower migrating band that represents full-length mRNA and a faster moving band which represents exon 2-skipped mRNA generated as a result of alternative splicing. Notably, theophylline shifted the splicing of ABT4M pre-mRNA in favor of exon 2-excluded isoform by repressing the activation of intron 1 branchpoint.
3.2.3. In Vitro Transcription
T7 RNA polymerase transcription reactions were prepared in 12.5 μl volumes containing 1× RNA polymerase reaction buffer: 0.5 mM (ATP, CTP, GTP), 0.1 mM UTP, 10 mM DTT, 500 ng of linearized DNA template, 20 μCi α-[32P] UTP (3000 Ci/mmol), 0.5 unit RNasin, and 0.5 units of T7 RNA polymerase. Assemble this reaction by preparing a 10× Transcription master mix (see Materials Section 2.3). To each pre-chilled sample eppendorf tube, the following is added in order:
2.5 μl of RNase-free water
2.2 μl of 10x Transcription master mix
2.5 μl 7mG(ppp)G RNA cap structure analog
1.3 μl RNA polymerase reaction buffer
1 μl of DNA template
0.5 μl of T7 RNA polymerase
0.5 μl of RNasin
2 μl α-[32P] UTP
Incubate at 37°C for 120 min.
Reactions are terminated by adding an equal volume of splicing loading buffer and heated at 65°C for 5 min. Briefly incubate the tube on ice and resolve by 8% denaturing PAGE gel electrophoresis.
3.2.4. In Vitro Splicing Assay
The extract preparation and in vitro splicing assay has been described previously (21, 39). Assemble the splicing reaction in the following order:
1 μl BC300
0.5 μl 160 mM MgCl2
1 μl of 10,000 c.p.m labeled RNA substrate
1 μl 0.5 M Creatine phosphate
0.5 μl25mMATP
0.25 μl0.1 MDTT
0.25 μl RNasin
2 μl 25 mM Theophylline or RNase-free water in control splicing reaction
RNase-free water to a volume of 12.5 μl
12.5 μl HeLa nuclear extract
Incubate at 30°C for 2 h and terminate by the addition of 87.5 μl splicing stop buffer.
Extract the RNA from the reaction mixtures by phenol-chloroform treatment followed by ethanol precipitation.
The RNA pellet is washed with 70% aqueous ethanol, dried, and dissolved in 10 μl loading buffer.
Splicing intermediates and products are analyzed by electrophoresis on a 13% denaturing polyacrylamide gel run at 50 W for 6 h.
3.2.5. Theophylline-Dependent Control of Alternative Splicing In Vivo
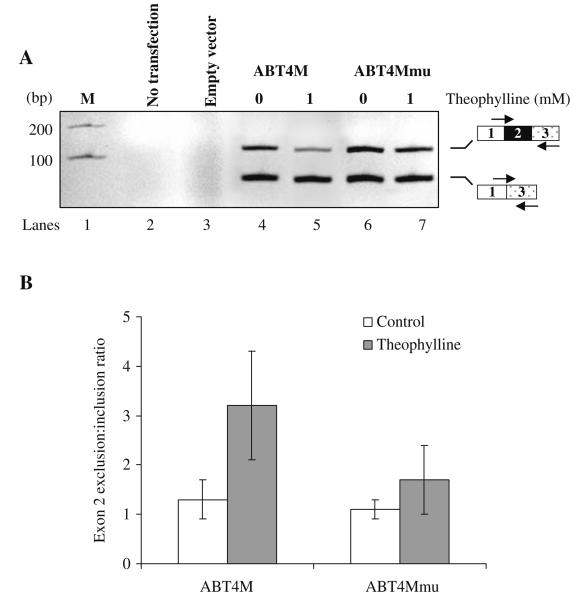
To determine whether theophylline-induced sequestering of branchpoint can control splicing in living cells, we inserted the DNA that encodes ABT4M pre-mRNA into the mammalian expression vector pcDNA Myc/His yielding pcABT4M. A mutant (pcABT4Mmu) that does not bind to theophylline was also constructed. Next, HeLa cells were transfected transiently (see Section 3.2.6) with pcABT4M, pcABT4Mmu, or empty vector and treated with theophylline or buffer. After 24 h, cells were harvested, and total RNA was isolated. Reverse transcription followed by PCR shown in Fig. 10.3A demonstrates that theophylline can regulate the alternative splicing of ABT4M premRNA (compare lane 4 with 5). In contrast, theophylline had a less significant effect on the splicing of ABT4Mmu pre-mRNA (Fig. 10.3A, compare lane 6 with 7; also see Fig. 10.3B), suggesting that binding of theophylline to its cognate RNA is necessary for controlling alternative splicing.
Fig. 10.3. Theophylline-mediated modulation of alternative splicing in vivo.
(A) HeLa cells were transiently transfected with empty vector, pcABT4M or pcABT4Mmu (containing mutations within core theophylline-binding aptamer). Cells were treated with buffer (lanes 4 and 6) or with 1 mM theophylline (lanes 5 and 7). The product of RT-PCR assays of total RNA isolated from each well was analyzed on a 2.5% agarose gel to estimate the effect of theophylline on the efficiency of exon 2 alternative splicing. The PCR-amplified bands corresponding to exon 2-included or -excluded mRNA is indicated. (B) The internal exon 2 exclusion: inclusion ratio for ABT4M or ABT4Mmu in the absence (open box) or presence of theophylline (shaded box) is shown. The data represent mean ± SEM. P < 0.04 versus mutant is significant.
3.2.6. Transient Transfection of HeLa Cells
HeLa cells were grown in Dulbecco's modified Eagle medium.
Cells should be maintained under standard incubation conditions (humidified atmosphere, 5% CO2, 37°C).
Seed 4.5 × 105 cells in a six-well plate a day prior to transfection.
Cells were grown to approximately 60–70% confluency and transfected with 1 μg of DNA by using Lipofectamine 2000 according to the manufacturer's protocol except that prior to the addition of DNA–Lipofectamine 2000 complex, the media was replaced by fresh media containing the indicated concentration of theophylline.
A freshly prepared solution of theophylline was used. In control samples, theophylline was replaced by buffer medium (see Note 12).
3.2.7. First-Strand cDNA Synthesis (RT-PCR)
In a sterile eppendorf tube we add 5 μg (total RNA), 500 ng oligo (dT)12-18, 10 mM dNTP, and nuclease-free H2O to final volume of 12 μl.
The mixture is heated for 5 min at 65°C. Spin briefly and place promptly on ice.
- Add in the following order:
- 4 μl 5x First-Strand Buffer
- 2 μl0.1 MDTT
- 1 μl RNasin
We then gently mix the contents of the tube and incubate for 2 min at 37°C.
Add 1 μl M-MLV Reverse Transcriptase (see Note 13).
Incubate at 37°C for 50 min.
Inactivate enzyme at 70°C for 15 min.
Store products at −20°C or proceed to amplification step using PCR.
Acknowledgments
We thank members of the Gaur laboratory for helpful discussions; Marieta Gencheva for valuable suggestions; and Faith Osep for administrative assistance. This work was supported in part by a Department of Defense (DOD; CDMRP) grant to RKG (BC023235), Beckman Research Institute excellence award to RKG, and NIH grant (CA 84202) to JES.
Footnotes
Custom siRNA synthesis should be designed with high purity for animal studies and preclinical applications. Our siRNAs are obtained from Qiagen (Valencia, CA). We note that some companies (Santa Cruz Biotechnology) are now offering siRNA gene silencers targeted to all human genes. The RNAi molecules are provided as pools of three targets specific to downregulate the expression of the gene of interest.
Always keep RNA solutions on ice to prevent degradation. Store unused siRNA duplex solution frozen at −20°C. The siRNA duplex solution can be frozen and thawed many times.
Although 200 nM of siRNA #497 was required to knockdown CEA, we recommend titration of siRNA to identify the optimum dose.
Healthy and sub-confluent cells are required for successful transfection experiments. Adjust cell and reagent amounts proportionately for wells or amount of DNA transfected. The objective is to have 90–100% confluency of cells at the time of harvest. The number of cells seeded varies depending on cell line.
Opti-MEM is a versatile, chemically defined medium, formulated to significantly reduce the amount of serum required for cultivating mammalian cells in vitro.
Lipofectamine 2000 Transfection Reagent forms stable complexes with nucleic acid molecules, permitting efficient transfection into eukaryotic cells. Lipofectamine 2000 can be used for nuclear and cytoplasmic targets and transfects a wide variety of cell lines including CHO, HEK-293, NIH 3T3, and HeLa. Store at 4°C. See manufacturer's protocol when necessary.
Low efficiency of siRNA delivery with standard techniques could be related to material toxicity. The transfection efficiency of siRNAs can be improved by using transfection reagents developed for antisense oligonucleotides. These reagents can be less toxic than plasmid delivery reagents and also result in higher transfection efficiencies than conventional reagents.
Working rabbit sera stocks should be kept at 4°C supplemented with 0.02% sodium azide. For monoclonal antibodies such as purified IgG, diluted antibody solutions should not be stored unless detergent or carrier proteins such as goat serum or BSA are added. Diluted IgG solutions (less than 0.1 mg/ml protein) quickly adsorb, denature, and lose activity. Avoid repetitive freeze–thawing of dilute purified IgG.
Multiple bands on a Western blot could be indicative of a poor-quality antibody (e.g., not specific for protein of interest). It also indicates proteolytic breakdown of the antigen, excessive protein loaded per lane, a highly sensitive detection system, inefficient blocking, or the concentration of antigen is too low.
We have observed lower aptamer stem size between 4 and 10 bp to be optimum. The stem size >10 bp inhibits splicing even without addition of theophylline. The upper aptamer stem size can vary between 2 and 6 bp. The premRNA in which the branchpoint sequence is embedded in the upper stem displayed a strong response to theophylline-dependent control of alternative splicing.
We constructed pre-mRNA substrates containing the theophylline-responsive element using overlap extension PCR (40). This strategy removes the limitation placed by lack of restriction sites or the potential secondary effect of adding nucleotides to the pre-mRNA. In theory, two half-amplicons are generated in the first PCR reaction containing an overlap region of 20–30 nucleotides. In the second PCR, we generate the full-length product. Gel-purify each PCR product as parental DNA template can interfere with generation of the chimeric sequence. We note that success in creating the overlap extension increases when the two half-amplicons are equal in length and the same numbers of moles are used in the second PCR reaction.
Theophylline has clinical applications that are well documented (e.g., in the treatment of respiratory diseases with bronchospasm) but it can be toxic in high dosages (41). HeLa cells can tolerate dosages of theophylline from a range of 0.05 −1 mM without a significant growth defect. Our in vitro pre-mRNA splicing assays show good results with theophylline from a range of 1–2 mM.
Thoroughly mix reaction by repeated but gentle pipetting. We have observed inconsistent results occur from improper mixing of reverse transcriptase at this step.
References
- 1.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 2.House AE, Lynch KW. Regulation of alternative splicing: more than just the ABCs. J. Biol. Chem. 2008;283:1217–1221. doi: 10.1074/jbc.R700031200. [DOI] [PubMed] [Google Scholar]
- 3.Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mironov AA, Fickett JW, Gelfand MS. Frequent alternative splicing of human genes. Genome Res. 1999;9:1288–1293. doi: 10.1101/gr.9.12.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson JM, et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 6.Kan Z, et al. Gene structure prediction and alternative splicing analysis using genomically aligned ESTs. Genome Res. 2001;11:889–900. doi: 10.1101/gr.155001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Blanco MA. Alternative splicing: therapeutic target and tool. Prog. Mol. Subcell. Biol. 2006;44:47–64. doi: 10.1007/978-3-540-34449-0_3. [DOI] [PubMed] [Google Scholar]
- 8.Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 9.Benz EJ, Jr., Huang SC. Role of tissue specific alternative pre-mRNA splicing in the differentiation of the erythrocyte membrane. Trans. Am. Clin. Climatol. Assoc. 1997;108:78–95. [PMC free article] [PubMed] [Google Scholar]
- 10.Kurreck J. siRNA Efficiency: Structure or sequence – that is the question. J. Biomed. Biotechnol. 2006;2006:83757. doi: 10.1155/JBB/2006/83757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leuschner PJ, et al. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006;7:314–320. doi: 10.1038/sj.embor.7400637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaur RK. RNA interference: a potential therapeutic tool for silencing splice isoforms linked to human diseases. Biotechniques. 2006;(Suppl):15–22. doi: 10.2144/000112165. [DOI] [PubMed] [Google Scholar]
- 13.Kim DS, et al. Ligand-induced sequestering of branchpoint sequence allows conditional control of splicing. BMC Mol. Biol. 2008;9:23. doi: 10.1186/1471-2199-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim DS, et al. An artificial riboswitch for controlling pre-mRNA splicing. RNA. 2005;11:1667–1677. doi: 10.1261/rna.2162205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kole R, Vacek M, Williams T. Modification of alternative splicing by antisense therapeutics. Oligonucleotides. 2004;14:65–74. doi: 10.1089/154545704322988067. [DOI] [PubMed] [Google Scholar]
- 16.Dominski Z, Kole R. Restoration of correct splicing in thalassemic premRNA by antisense oligonucleotides. Proc. Natl. Acad. Sci. USA. 1993;90:8673–8677. doi: 10.1073/pnas.90.18.8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tucker BJ, Breaker RR. Riboswitches as versatile gene control elements. Curr. Opin. Struct. Bio. 2005;15:342–348. doi: 10.1016/j.sbi.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 18.Nudler E, Mironov AS. The riboswitch control of bacterial metabolism. Trends Biochem. Sci. 2004;29:11–17. doi: 10.1016/j.tibs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 19.Goguel V, Wang Y, Rosbash M. Short artificial hairpins sequester splicing signals and inhibit yeast pre-mRNA splicing. Mol. Cell. Biol. 1993;13:6841–6848. doi: 10.1128/mcb.13.11.6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gusti V, Kim DS, Gaur RK. Sequestering of the 3′ splice site in a theophylline-responsive riboswitch allows ligand-dependent control of alternative splicing. Oligonucleotides. 2008;18:93–99. doi: 10.1089/oli.2007.0107. [DOI] [PubMed] [Google Scholar]
- 21.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fischer DC, et al. Expression of splicing factors in human ovarian cancer. Oncol. Rep. 2004;11:1085–1090. [PubMed] [Google Scholar]
- 23.Ghigna C, et al. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol. Cell. 2005;20:881–890. doi: 10.1016/j.molcel.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 24.He X, et al. Alternative splicing of the multidrug resistance protein 1/ATP binding cassette transporter subfamily gene in ovarian cancer creates functional splice variants and is associated with increased expression of the splicing factors PTB and SRp20. Clin. Cancer Res. 2004;10:4652–4660. doi: 10.1158/1078-0432.CCR-03-0439. [DOI] [PubMed] [Google Scholar]
- 25.Karni R, et al. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat. Struct. Mol. Biol. 2007;14:185–193. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu H, et al. Enhancing TRAIL-induced apoptosis by Bcl-X(L) siRNA. Cancer Biol. Ther. 2005;4:393–397. doi: 10.4161/cbt.4.4.1616. [DOI] [PubMed] [Google Scholar]
- 27.Chevinsky AH. CEA in tumors of other than colorectal origin. Semin. Surg. Oncol. 1991;7:162–166. doi: 10.1002/ssu.2980070309. [DOI] [PubMed] [Google Scholar]
- 28.Hammarstrom S. The carcinoembryonic antigen (CEA) family: structures, suggested functions and expression in normal and malignant tissues. Semin. Cancer Biol. 1999;9:67–81. doi: 10.1006/scbi.1998.0119. [DOI] [PubMed] [Google Scholar]
- 29.Li W, Cha L. Predicting siRNA efficiency. Cell. Mol. Life Sci. 2007;64:1785–1792. doi: 10.1007/s00018-007-7057-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yiu SM, et al. Filtering of ineffective siRNAs and improved siRNA design tool. Bioinformatics. 2005;21:144–151. doi: 10.1093/bioinformatics/bth498. [DOI] [PubMed] [Google Scholar]
- 31.Patzel V, et al. Design of siRNAs producing unstructured guide-RNAs results in improved RNA interference efficiency. Nat. Biotechnol. 2005;23:1440–1444. doi: 10.1038/nbt1151. [DOI] [PubMed] [Google Scholar]
- 32.Huesken D, et al. Design of a genome-wide siRNA library using an artificial neural network. Nat. Biotechnol. 2005;23:995–1001. doi: 10.1038/nbt1118. [DOI] [PubMed] [Google Scholar]
- 33.Tuschl T. Targeting genes expressed in mammalian cells using siRNAs. Nat. Methods. 2004;X:13–17. [Google Scholar]
- 34.Reynolds A, et al. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 35.Daoud R, et al. Activity-dependent regulation of alternative splicing patterns in the rat brain. Eur. J. Neurosci. 1999;11:788–802. doi: 10.1046/j.1460-9568.1999.00486.x. [DOI] [PubMed] [Google Scholar]
- 36.Venables JP. Aberrant and alternative splicing in cancer. Cancer Res. 2004;64:7647–7654. doi: 10.1158/0008-5472.CAN-04-1910. [DOI] [PubMed] [Google Scholar]
- 37.Tazi J, Durand S, Jeanteur P. The spliceosome: a novel multi-faceted target for therapy. Trends Biochem. Sci. 2005;30:469–478. doi: 10.1016/j.tibs.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 38.Jenison RD, et al. High-resolution molecular discrimination by RNA. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- 39.Mayeda A, Krainer AR. Mammalian in vitro splicing assays. Methods Mol. Biol. 1999;118:315–321. doi: 10.1385/1-59259-676-2:315. [DOI] [PubMed] [Google Scholar]
- 40.Ge L, Rudolph P. Simultaneous introduction of multiple mutations using overlap extension PCR. Biotechniques. 1997;22:28–30. doi: 10.2144/97221bm03. [DOI] [PubMed] [Google Scholar]
- 41.Visitsunthorn N, Udomittipong K, Punnakan L. Theophylline toxicity in Thai children. Asian Pac. J. Allergy Immunol. 2001;19:177–182. [PubMed] [Google Scholar]