Abstract
Previous studies of genome evolution usually have involved one or two genomes and have thus been limited in their ability to detect the direction and rate of evolutionary change. Here, we use complete genome data from 20 poxvirus genomes to build a robust phylogeny of the Poxviridae and to study patterns of genome evolution. We show that, although there has been little gene order evolution, there are substantial differences between poxviruses in terms of genome content. Furthermore, we show that the rate of gene acquisition is not constant over time and that it has increased in the orthopox lineage (which includes smallpox and vaccinia). We also tested for positive selection on 204 groups of genes and show that a disproportionately high proportion of genes in the orthopox clade are under positive selection. The association of an increased rate of gene gain and positive selection is indicative of adaptive genome evolution. Many of the genes involved in these processes are likely to be associated with host–parasite coevolution.
Our understanding of whole-genome evolution is in its infancy (1). Although researchers have studied genomewide patterns of base composition skew (2), codon usage bias (3), and chromosomal duplication (4–6), there is generally little information about the evolution of gene order and gene content. Although genome rearrangement has been well studied in mammals and some other genomes (7–9), relatively few studies have addressed the evolution of gene content (10, 11). With the exception of gross genomic effects, such as whole or segmental genome duplication, changes in gene content can be uncovered only by multiple genome comparisons. With only two genomes, it is not possible to distinguish ancestral from derived states, nor is it possible to examine rates of change in gene content. By combining genomic and phylogenetic data, one can infer whether gene content has changed through gain or loss and associate changes in gene content with branches of the tree, and thereby gain insights into potential adaptive genomic events.
Poxviruses are double-stranded DNA viruses with no RNA stage. They replicate in the host cytoplasm and so encode the genes necessary for their own replication, unlike other viruses that enter the nucleus and use the host cell machinery. Poxvirus genomes range from 145 to 290 kb in size, and each genome contains ≈200 genes. Many of these genes may have been acquired by horizontal transfer, as has been documented in another group of double-stranded DNA viruses with no RNA stage, the Baculoviridae (12–14). Poxviruses are broadly classified into insect-infecting viruses (entomopox) and vertebrate-infecting viruses (chordopox). The chordopox are further divided into eight genera. Despite extensive sequence data, there have been no comprehensive analyses of poxvirus genome evolution. To date, there have been few phylogenetic treatments of poxviruses (15–18). Previous analyses of gene content focused on few genomes, but identified several features of poxvirus genome evolution, such as broad conservation of gene order between chordopox genomes (15–19) and disruption of this conservation in the fowlpox (20) and the entomopox genomes (21).
The availability of 20 completely sequenced poxvirus genomes enables us to systematically characterize genome evolution in the context of phylogenetic history. We combined the data from all 20 genomes to build a robust phylogeny of the group and to address the following questions. First, to what extent has the conserved genome structure of the chordopox genomes been retained? Second, how many genes have been acquired during the history of the Poxviridae, where do these genes come from, and has the rate of gene acquisition been homogeneous throughout evolutionary history? Third, how many genes have been lost, and are some genes particularly liable to loss? Finally, is there a correlation between genome evolution and adaptive evolution of genes?
Methods
Data. We obtained genome sequences of 20 completely sequenced poxviruses from GenBank in October 2002 (Table 1). A total of 4,042 predicted protein sequences from these genomes were obtained from the Poxvirus Bioinformatic Resource Centre, www.poxvirus.org.
Table 1. Poxvirus genomes included in this study.
| Genome | Abbreviation | Genus | Host | Size, bp | GenBank accession no. | Ref. |
|---|---|---|---|---|---|---|
| Amsacta moorei entomopoxvirus | AMV-EPB | Entomopox | Red hairy caterpillar | 232,392 | AF250284 | 21 |
| Camelpox virus isolate M-96 from Kazakhstan | CMPV-M96 | Orthopox | Camel | 205,719 | AF438165 | 17, 18 |
| Cowpox virus strain Brighton Red | CPXV-BR | Orthopox | Cow | 224,501 | AF482758 | |
| Ectromelia virus Moscow strain | ECTV-MOS | Orthopox | Mouse | 209,771 | AF012825 | |
| Fowlpox virus | FPV-FCV | Avipox | Chicken and turkey | 288,539 | AF198100 | 20 |
| Lumpy skin disease virus strain Neethling isolate 2490 | LSD-NEE | Capripox | Cow | 150,773 | AF325528 | 38 |
| Molluscum contagiosum virus subtype 1 | MCU-SBI | Molluscipox | Human | 190,289 | U60315 | 19 |
| Monkeypox virus strain Zaire-96-I-16 | MPXV-ZRE | Orthopox | Monkey | 196,858 | AF380138 | 39 |
| Melanoplus sanguinipes entomopoxvirus | MSE-TUC | Entomopox | Grasshopper | 236,120 | AF063866 | 40 |
| Myxoma virus strain Lausanne | MYX-LAU | Leporipox | Rabbit | 161,774 | AF170726 | 41 |
| Rabbit fibroma virus | RFB-KAS | Leporipox | Rabbit | 159,857 | AF170722 | 42 |
| Sheeppox virus | SPPV | Capripox | Sheep | 149,955 | AY077832 | 43 |
| Swinepox virus isolate 17077-99 | SWPV-NEB | Suipox | Pig | 146,454 | AF410153 | 16 |
| Vaccinia virus strain Ankara | VAC-ANK | Orthopox | 177,923 | U94848 | 44 | |
| Vaccinia virus strain Copenhagen | VAC-COP | Orthopox | 191,737 | M35027 | 45 | |
| Vaccinia virus strain Tian Tan | VAC-TAN | Orthopox | 189,274 | AF095689 | ||
| Variola major virus strain Bangladesh-1975 | VAR-BSH | Orthopox | Human | 186,103 | L22579 | 32 |
| Variola major virus strain India-1967 | VAR-IND | Orthopox | Human | 185,578 | X69198 | 46 |
| Variola minor virus strain Garcia-1966 | VMN-GAR | Orthopox | Human | 186,986 | Y16780 | |
| Yaba-like disease virus | YAB-YLD | Yatapox | Monkey | 144,575 | AJ293568 | 15 |
Poxvirus Gene Family Definition. The complete set of 4,042 poxvirus proteins was compared by an all-against-all blastp similarity search (22) with the SEG filter (low complexity masking) (23), and using the blosum62 substitution matrix. Two proteins were considered similar if one hit the other in the blast search with an e-value of ≤10–5 and the maximal scoring pair alignment in the blastp search covered at least 40% of the longer protein. This method excludes proteins with similarity over only short regions (6).
A simple complete-linkage method of gene family classification (i.e., where all members of the group are similar to all others) is likely to be too conservative and to artefactually increase the number of families with a small taxonomic range. Conversely, a simple single-link clustering method (i.e., where each member of a group is similar to at least one other member) is prone to include distant homologues. We therefore introduced a method of gene family classification whereby genes were initially grouped by single link clustering, but groups were retained only if at least one member of the group was similar to all other members. The proteins in groups not meeting this criterion were excluded from further analysis. Our method balances the sensitivity of single-link clustering with the specificity of complete-linkage methods.
Poxvirus Phylogeny. Thirty-four proteins were identified that had one member in each of the 20 genomes. These 34 sets of orthologs were aligned independently by the t-coffee multiple alignment program (24). The 34 amino acid alignments were concatenated into a single alignment for phylogenetic inference. A neighbor-joining (NJ) tree was constructed by using Takezaki's njboot program (25) with the Poisson correction for multiple hits. A second data set consisting of 92 sets of orthologs present in all of the orthopox genomes (including the 34 present in all pox genomes) was used to build an orthopox-specific tree by using the same protocol. The trees were also inferred under maximum parsimony (MP) using paup 4.0b10 (26).
Gene Gain and Loss Events on the Phylogeny. The origin of a new family was assumed to be a unique event. The common ancestor of a given family was inferred as the most recent common ancestor of all genomes containing a member of that family. Gene loss events were assigned to branches in the topology under the assumption that there is a single origin of each family such that the number of loss events was parsimonious. If there are multiple origins of families, e.g., by horizontal transfer between pox genomes, then our assumption of a single origin will inflate both the number of gain events closer to the root of the tree and the number of loss events in that family.
We performed simulations of gene gain events by assigning events to each branch of the tree with a probability that was proportional to the branch length. This process was repeated 10,000 times; for each simulation the total number of gain events was distributed on the tree, and the number of gene gain events per branch was tallied. The observed number of gain events for each branch was considered significant at the 1% level if the number was >99.5% or <0.5% of the simulated values for that branch. Similarly, observed values were significant at the 5% level if they were in the 2.5 or 97.5 percentile. We investigated the type I error (false positives) of these simulations by evaluating 1,000 randomized data sets, with significance assessed by 10,000 simulations per data set. Over 1,000 data sets, type I error did not deviate significantly from the expected values on any branch of the phylogeny.
We applied this approach with branch lengths inferred from protein data and synonymous distances. The latter were calculated with mega2 (27), using the modified Nei–Gojobori method with Jukes–Cantor correction and a transition/transversion ratio of 2.0. The Molluscum contagiosum (MCU-SB1) genome, which has 90% GC at codon third positions for the set of 34 orthologs, was saturated for synonymous distances with every other genome. We therefore removed MCU-SB1 and all earlier branching taxa and estimated the branch lengths for the tree of the remaining 16 poxviruses by using fitch in the phylip software package.
Calculation of ω. We used the codeml program from the paml software package (28) to calculate the ω, the ratio of nonsynonymous to synonymous substitution, for all 204 gene families that contained at least three members. All families were aligned as proteins by using t-coffee (24). NJ trees were determined with clustalw (29) while correcting for multiple substitutions and ignoring gaps; the tree topology was used in paml analyses. We tested our data under models 7 and 8 of the paml package, which detects selection on individual codons. Significance was determined with the likelihood statistic, as described by Yang (30).
Identification of Nonviral Homologues. All pox proteins were compared with the complete GenBank protein database (October 2002) by using the blastp similarity search at the National Center for Biotechnology Information with the SEG filter switched on.
Results and Discussion
Poxvirus Gene Families. Pox genes were classified into families based on protein sequence similarity. Similarity was determined by the significance of the blast hit (blast e-value threshold), and the extent of the similar region (minimum aligned proportion in blast maximal scoring pair). Different values for these parameters have the potential to cause wide variation in the family definitions, but there are no apriori best values. We investigated the effect of these parameters on the distribution of families in the pox genomes. We chose the parameter combination of an e-value threshold of 1e-5 and a minimum blast alignment of 40% of the longer protein because they maximized the number of families with a single member in each pox genome for the parameters tested (Table 3, which is published as supporting information on the PNAS web site). Because these families can be used for phylogenetic inference, maximizing this count increases the information for phylogenetic inference.
Gene family classification was based on a variant of single-link clustering. Of the 4,042 proteins encoded by these genomes, 3,384 were unambiguously classified into 875 families of one or more members. In 813 cases our method had the same effect as the conservative complete linkage method. Families were assigned numbers for identification purposes. Family classification and other results can be browsed at http://titus.bio.uci.edu/pox.
We identified 521 and 150 families of one and two members, respectively, and 204 with three or more members. The largest family has 42 members and contains antithrombin III serine proteinase inhibitors (serpins). The serpin family (family50) is chordopox specific, shows strong homology to vertebrate serpins (see below), and is involved in inhibition of host cell apoptosis (31). These 875 families form the basis of our analyses; they mainly consist of groups of orthologs but may include coorthologs.
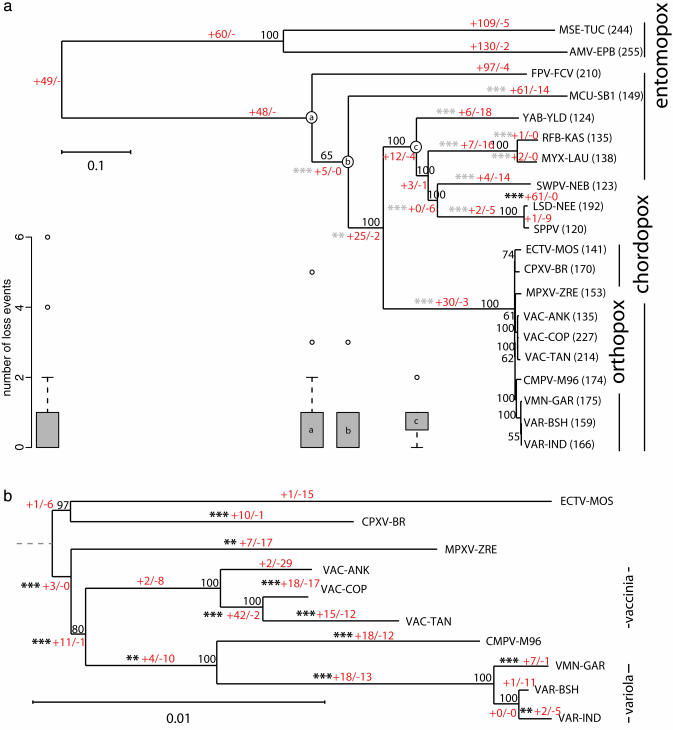
Poxvirus Phylogeny. Forty-nine families were present in all genomes, with 34 of these including only a single gene from each genome. Most of these genes are involved in basic replication processes and are thus a subset of the “core” pox genes. We constructed a NJ tree from their concatenated alignments (Fig. 1a). Entomopox was treated as the outgroup on the basis of a previous phylogenetic study of Poxviridae and Baculoviridae DNA polymerase genes (13).
Fig. 1.
NJ trees of the poxviruses. The classifications of the viruses are indicated to the right. Bootstrap values are shown for each branch. The numbers in red separated by/are the number of gain and loss events, respectively that occurred along each branch. At the base of the phylogeny it is not possible to distinguish a gaininone lineage from a loss in the other. In these cases the events were counted as gains and we use a lone hyphen (-) in place of a count of loss events to indicate this fact. *** indicates that the number of gains was significant at the 1% level, and ** indicates significance at the 5% level (see Methods). Gray and black asterisks indicate the number of events were below or above expectations, respectively. (a) NJ tree of all completely sequences poxviruses, based on 34 gene families. The number of genes from each genome that were assigned into families is shown after the genome name. The box plots show the distribution of the number of loss events in families dating back to the node of the tree above (indicated by a labeled circle). Only box plots with outliers are shown. The leftmost box plot refers to families ancestral to all poxvirus genomes analyzed. (b) NJ tree of the orthopoxviruses, based on 92 gene families. The dashed line shows the approximate position of the root of the orthopoxviruses inferred from tree a.
To resolve low bootstrap values in the orthopox clade, another NJ tree (Fig. 1b) was inferred from the concatenated protein alignments of 92 orthologous genes identified in all orthopox genomes. The orthopox phylogeny inferred from the 92 orthologs differed from that in the tree of all pox viruses inferred from the 34 orthologs only in the placement of monkeypox (MPXV-ZRE). As all of the branches in the orthopox-only tree were supported by bootstrap values of at least 80%, the topology was used for the orthopox clade.
Phylogenetic trees were also inferred by MP. The NJ and MP topologies were identical with the exception of the placement of monkeypox (MPXV-ZRE) and the relative position of cowpox (CPXV-BR) and ectromelia (ECTV-MOS). In both cases the parsimony tree had a bootstrap value <80% (63% and 54%, respectively), but the NJ tree had bootstrap values >80%. We thus chose the NJ topology for further analyses.
The phylogeny reveals three important features of Poxviridae evolution. First, both the orthopox and the chordopox are monophyletic clades, with the former a subgroup of the latter. Second, the analysis confirms that camelpox (CMPV-M96) is the closest relative of variola (VMN-GAR, VAR-BSH, VAR-IND), the causative agent of smallpox (18). Finally, neither the phylogeny of the chordopox nor the subphylogeny of the orthopox follows the host phylogeny. For example, the variola virus, which causes smallpox in humans, is more closely related to camelpox than monkeypox. Thus, phylogenetic evidence clearly indicates that these viruses have changed hosts several times in their history.
Pox Genome Structure. The physical arrangement of the 92 orthologous families used for phylogenetic inference is shown in Fig. 2. Gene order and gene spacing are highly conserved within the chordopox genomes (Fig. 2). The lone exception to this pattern is the fowlpox genome (FPV-FCV) (20), where there have been several rearrangements. By contrast, there is no apparent conservation of gene order between the two entomopox genomes nor between the entomopox genomes and the chordopox genomes (Fig. 2) (21). Genome rearrangement data are consistent with the phylogenetic tree both in terms of the long branch length to the entomopox viruses and in the placement of fowlpox externally to other chordopox viruses.
Fig. 2.
The genomes of all 20 poxviruses showing the taxonomic breadth and the genome arrangement of 92 gene families used in the phylogeny inference. Genomes are shown in the same order as in Fig. 1a, and names are shown on the right. All genes are indicated by black outline boxes. Horizontal distances are proportional to base pair distances. The 34 orthologs present in all poxviruses are shown in red. The 29 orthologs present in all of the chordopox genomes, but not in the entomopox genomes, are shown in blue. The 29 orthologs present in all orthopox viruses are shown in yellow. Lines along the vertical dimension link orthologs. The orthologs represented are only those that were present in all of the genomes in each classification [i.e., in all pox genomes (red); in all chordopox genomes (blue); or in all orthopox genomes (yellow)].
Notably, even in the fowlpox and the entomopox genomes the orthologs retain their central location, which could be functionally important. These conserved genes are involved in basic life cycle processes and include, for example, family 109 (RNA polymerase subunit rpo147), family 131 (mRNA capping enzyme large subunit), family 144 (NTPase; DNA replication), family 156 (virion core protein P4b), as well as other genes involved in virus replication and assembly.
Most (17 of 29) of the families conserved uniquely in orthopox (colored yellow in Fig. 2) are located closer to the chromosome ends than the 34 orthologs present in all genomes. These 17 include some families involved in virulence such as family 259 (CD47-like membrane glycoprotein) and family 26 (putative virulence factor). These gene families are relatively recent acquisitions, and their distribution is consistent with the observation that species-specific genes, such as virulence factors and genes controlling host-range specificity, tend to be located toward the ends of the chromosome (32).
Genome Content Evolution. Given the high level of genome arrangement conservation between poxvirus genomes, we investigated other possible mechanisms of genome divergence between these viruses. It is clear from the number of genes that are not shared by all members of the major virus subgroups (uncolored genes in Fig. 2) that there has been substantial evolution of gene content.
We counted gene gain and loss events along each branch of the phylogeny (Fig. 1). In contrast to genome arrangement, which has been strongly conserved, there has been substantial gene loss and gain throughout the history of the poxviruses. For example, we estimate that each of the two entomopox genomes has gained >100 genes since their divergence, and fowlpox has gained 94 genes since its divergence from the common ancestor of the chordopox.
Heterogeneous Rates of Gene Acquisition in Pox Genomes. In total, 24 of 37 branches exhibited significant deviation from expectation based on homogenous rates (P < 0.05), indicating the rate of gene gain has been statistically heterogeneous relative to the rate of protein evolution. Eleven of these branches showed a relative dearth of gene gain events and 13 demonstrated a relative excess of gene gain. Surprisingly, 12 of the 13 branches with excess gene gain were located in the orthopox clade; these branches represent 66% (12 of 18) of orthopox branches. The clustering of 12 branches with excess gain events in a single clade of 18 branches is highly unlikely (P = 0.005; G test). The pattern of branches on the tree with a deficit or excess of gene acquisition events indicates that the pattern in the orthopox clade is derived.
The apparent excess of gene gain within the orthopox could be explained by one of several phenomena: a slowdown in the rate of protein evolution in the orthopox lineage, a sampling effect caused by the overrepresentation of orthopox genomes, increased selection for the retention of gained genes, or an increased rate of gene acquisition. A rate slowdown in the orthopox lineage would cause the branch lengths in this clade to be shorter in relation to time than branches in the rest of the tree. We investigated this possibility by applying Takezaki et al.'s two-cluster test (25), which examines relative rates of amino acid substitution, to the Poxviridae phylogeny. The null hypothesis of rate homogeneity was rejected overall, but it was not rejected on any branch implying a change of rate in the whole orthopox clade. More importantly, we reanalyzed gene gain events by using branch lengths based on synonymous distances, reasoning that synonymous rates may be less susceptible to major rate fluctuations. The qualitative results were identical with synonymous distances, because the same branches were significant at the 5% level.
A second possibility for excess gene gain events in the orthopox clade is a sampling effect driven by the inclusion of several closely related orthopox genomes. We therefore investigated the effect of removing genomes from within the same species. When VAR-IND, VAR-BSH, VAC-TAN, and VAC-COP were excluded from analysis, the significance of the remaining branches remained un-changed. Altogether, our results indicate an increase in the rate of gene acquisition or retention in the orthopox clade; this increase does not appear to be driven by either a sampling effect or heterogeneous rates of protein substitution.
Mechanisms of Gene Gain. There are at least three possible mechanisms of gene gain: extensive sequence divergence, which could push homologous genes below our similarity threshold; recombination between genes, which produces novel protein products; and horizontal transfer. All three types of event may be at work in pox genomes. Of the 875 families described here, 238 (27%) include members that have similarities to other pox proteins that were not included in the family because the length of the similar region was too short. These below-threshold similarities may be the result of either recombination with portions of other genes or extensive sequence divergence. However, examination of patterns of similarity in these 238 families did not reveal any clear examples of recombination.
The third possibility, horizontal transfer, is expected to be detectable through similarity to genes from phylogenetically distinct species. We compared all 4,042 pox proteins to the entire GenBank database and examined the resulting lists of hits for nonviral sequences. This approach is limited by the presence of sequences in GenBank, and the absence of a hit cannot be considered as a negative result. Nonetheless, 482 proteins have similarity to some nonviral GenBank entry with an e-value ≤1e-20, including members of 57 (7%) of the 874 families in our analysis. Of these 482 hits, 62% were to an organism of the same taxonomic class and 16% to an organism of the same taxonomic family as the viral host. These numbers increase to 70% and 22%, respectively for orthopox proteins. Many of these are plausible cases of horizontal transfer. For example, all members of the viral DNA ligase family (family 184) have similarity to a mouse or human DNA ligase III with a blast e-value of at least 1e-146. This gene was acquired in the common ancestor of the chordopox; it is tempting to surmise that poxviruses have acquired much of the machinery to replicate autonomously in the host cytoplasm via horizontal transfer. Horizontally transferred genes may also be important in evading host defenses. For example, the Amsacta moorei entomopoxvirus gene AMV-EPB_034 codes for an inhibitor of apoptosis. Its top non-self hit in GenBank is an inhibitor of apoptosis from Bombyx mori (with a blast e-value of 9e–81), which is an insect of the order Lepidoptera, the same order as the normal host of the virus (the red hairy caterpillar Amsacta moorei). This gene has probably been acquired independently several times in the history of Baculoviridae (13).
Analysis of Gene Loss Events. The genome complement is also affected by gene loss. A simulation test similar to the one we performed for gene gain events is not possible for loss events, because loss events depend on gain and loss events that precede them in time. However, gene families with a common ancestral node are expected to have broadly similar histories; at the very least they have been retained in equally distant clades of the tree. We grouped loss events in each family according to the ancestral node of the family and plotted them in a box plot. Several of these plots contained outlying families (Fig. 1a), i.e., gene families that have an extraordinary amount of gene loss with respect to other families with similar taxonomic depth. These families include at least two that are thought to be involved in virulence, and several others whose description indicates that they may be antigenic (Table 4, which is published as supporting information on the PNAS web site). Notably, one of the families in this group (family 190) was also found to have evolved under positive selection, as determined by ω (see below).
There is no obvious relationship between the amount of gene loss for a given family and the dispensability of that gene. For example, the thymidine kinase (TK) gene is nonessential in tissue culture (33), but our results indicate that the TK gene family (family 104) was present in the common ancestor of all poxviruses and has been lost in only two genomes.
Adaptive Evolution. It is not clear from the pattern and frequency of gene gain and loss events whether the events are neutral or are an adaptive response to the host immune system or other environmental factors. We tested for selection acting on the evolution of genes (as opposed to genomes) by comparing the number of synonymous and nonsynonymous substitutions. A significantly higher rate of nonsynonymous substitution indicates that the gene is evolving by adaptive evolution.
Twenty-six of 204 families had sites with a ratio of nonsynonymous to synonymous substitutions (ω) significantly >1.0 (Table 2). Many pox genes evolving under positive selection are either known to be involved in virulence [e.g., family 190, the schlafen-like genes (34); and family 187, coding for hemaglutinin, which is a known antigenic agent under positive selection in the influenza virus (35)] or their protein products' location in the infecting agent makes them plausible candidates for host–parasite interaction (e.g., families 173, 187, 358, and 391 are all predicted to be membrane associated).
Table 2. Gene families with ω > 1.
| Gene family ID | ω | Aligned protein length | No. of members | Description | Taxonomic range* |
|---|---|---|---|---|---|
| 409 | 75.57 | 83 | 3 | Hypothetical | G |
| 402 | 69.12 | 98 | 4 | Hypothetical | I |
| 398 | 26.22 | 63 | 3 | Hypothetical | H |
| 345 | 16.92 | 141 | 3 | Hypothetical | F |
| 389 | 14.14 | 90 | 4 | Hypothetical | F |
| 399 | 13.7 | 99 | 3 | Hypothetical | H |
| 366 | 12.64 | 234 | 4 | Hypothetical | G |
| 85 | 9.03 | 92 | 10 | Hypothetical | B |
| 391 | 6.34 | 142 | 5 | Semaphorin-like | G |
| 699 | 5.78 | 63 | 4 | Hypothetical | B |
| 60 | 5.68 | 157 | 18 | DUTPase | C |
| 18 | 4.6 | 120 | 4 | Hypothetical | D |
| 334 | 4.45 | 166 | 7 | 17k myristylprotein | B |
| 93 | 3.51 | 99 | 15 | Hypothetical | E |
| 8 | 3.07 | 209 | 8 | Kelch protein | B |
| 115 | 2.55 | 165 | 17 | Hypothetical | A |
| 190 | 2.36 | 505 | 7 | Schlafen-like | C |
| 187 | 2.33 | 319 | 10 | Hemagluttinin | B |
| 173 | 1.95 | 215 | 10 | Membrane glycoprotein | B |
| 358 | 1.77 | 235 | 5 | Similar to cell surface glycoprotein | F |
| 33 | 1.51 | 771 | 11 | Ribonucleotide reductase, large subunit | E |
| 105 | 1.43 | 143 | 4 | Hypothetical | F |
| 13 | 1.32 | 228 | 19 | Hypothetical | C |
| 233 | 1.19 | 301 | 20 | Holiday junction resolvase | C |
| 129 | 1.1 | 237 | 16 | Late transcription factor | E |
| 7 | 1.03 | 122 | 18 | DNA-binding virion core protein | A |
Taxonomic specificity of a given gene family based on the clade defined by the branch along which the family arose. A, chordopox specific; B, orthopox specific; C, ancestral to all pox; D, arose within orthopox; E, specific to chordopox, excluding FPV-FCV and MCU-SBI; F, specific to clade containing YAB-YLD, RFB-KAS, MYX-LAU, SWPV-NEB, LSD-NEE, AND SPPV; G, specific to clade containing vaccinia, camelpox, and variola; H, variola specific; I, variola and camelpox specific.
ω is measured on genes, not genomes; yet these results can be related to the flux in genome content. For example, the schlafen family (family 190) has been under positive selection and is also one of the families exhibiting extraordinary gene loss. More importantly, gene acquisition rates have increased within the orthopox clade and, surprisingly, 50% (13 of 26) of the gene families found to be under positive selection are found only within this clade. Overall, positive selection is detected in a far higher proportion of orthopox-specific gene families (21%; 13 of 62) than in gene families that are not limited to the orthopox clade (9%; 13 of 142), and these proportions differ significantly (Fisher's exact test, P = 0.04). One must interpret this result cautiously, because the statistical power to detect positive selection varies from gene family to gene family and is a complex function of sample size and sequence divergence (36).
Conclusions
We uncovered four features of pox genome evolution. First, genome structure is highly conserved throughout the chordopox, despite substantial fluctuation in gene content. Second, gene loss and gain have been consistent characteristics of pox genome evolution, but the rate of gene loss varies among gene families and the rate of gene acquisition (or retention) has increased in the orthopox clade. Third, much of the gene acquisition is through identifiable horizontal transfer events.
The final feature of pox evolution is the pattern of selection on individual gene families coupled with their phylogenetic distribution. Of the 49 gene families present in the common ancestor of all pox genomes, 42 were found to be under conservative selection (ω significantly <1.0). These 42 families represent 50% of the families with ω significantly <1.0. Their conservative evolution, coupled with their distribution in all 20 genomes, is consistent with their functional annotations as proteins involved in basic life cycle processes.
By contrast, genes acquired and lost during poxvirus evolution are likely to have host-specific effects. An acquired gene may facilitate evasion of host defenses, and a lost gene may coincide either with the loss of an antigenic signal to the host cell or continued evolutionary persistence by the attenuation of disease (31, 37). Thus, fluxes in genome content undoubtedly present opportunities for adaptation. Together, high rates of gene acquisition (or retention) coupled with a high incidence of positive selection suggest that the orthopox experienced a substantial shift in evolutionary dynamics that included several adaptive events. At present, it is not clear what has fueled these adaptive episodes, but it is possible they are associated with unique features of orthopox infection, replication, and virulence. As such, the genes listed in Table 2 comprise a list of genes that could be considered as targets of drug design.
Supplementary Material
Acknowledgments
We thank M. Mondragón-Palomimo, K. H. Wolfe, L. Villarreal, L. Zhang, R. Bush, S. Hampson, and two anonymous reviewers for discussion and comment.
Abbreviations: NJ, neighbor-joining; MP, maximum parsimony.
References
- 1.Wolfe, K. H. & Li, W. H. (2003) Nat. Genet. 33, 255–265. [DOI] [PubMed] [Google Scholar]
- 2.McLean, M. J., Wolfe, K. H. & Devine, K. M. (1998) J. Mol. Evol. 47, 691–696. [DOI] [PubMed] [Google Scholar]
- 3.Sharp, P. M. & Li, W. H. (1986) J. Mol. Evol. 24, 28–38. [DOI] [PubMed] [Google Scholar]
- 4.Wolfe, K. H. & Shields, D. C. (1997) Nature 387, 708–713. [DOI] [PubMed] [Google Scholar]
- 5.Vision, T. J., Brown, D. G. & Tanksley, S. D. (2000) Science 290, 2114–2117. [DOI] [PubMed] [Google Scholar]
- 6.McLysaght, A., Hokamp, K. & Wolfe, K. H. (2002) Nat. Genet. 31, 200–204. [DOI] [PubMed] [Google Scholar]
- 7.Nadeau, J. H. & Taylor, B. A. (1984) Proc. Natl. Acad. Sci. USA 81, 814–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hannenhalli, S., Chappey, C., Koonin, E. V. & Pevzner, P. A. (1995) Genomics 30, 299–311. [DOI] [PubMed] [Google Scholar]
- 9.Pevzner, P. & Tesler, G. (2003) Genome Res. 13, 37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin, W., Rujan, T., Richly, E., Hansen, A., Cornelsen, S., Lins, T., Leister, D., Stoebe, B., Hasegawa, M. & Penny, D. (2002) Proc. Natl. Acad. Sci. USA 99, 12246–12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daubin, V., Moran, N. A. & Ochman, H. (2003) Science 301, 829–832. [DOI] [PubMed] [Google Scholar]
- 12.Hughes, A. L. (2002) J. Mol. Evol. 54, 90–101. [DOI] [PubMed] [Google Scholar]
- 13.Hughes, A. L. (2002) Infect. Genet. Evol. 2, 3–10. [DOI] [PubMed] [Google Scholar]
- 14.Hughes, A. L. & Friedman, R. (2003) Mol. Biol. Evol. 20, 979–987. [DOI] [PubMed] [Google Scholar]
- 15.Lee, H. J., Essani, K. & Smith, G. L. (2001) Virology 281, 170–192. [DOI] [PubMed] [Google Scholar]
- 16.Afonso, C. L., Tulman, E. R., Lu, Z., Zsak, L., Osorio, F. A., Balinsky, C., Kutish, G. F. & Rock, D. L. (2002) J. Virol. 76, 783–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Afonso, C. L., Tulman, E. R., Lu, Z., Zsak, L., Sandybaev, N. T., Kerembekova, U. Z., Zaitsev, V. L., Kutish, G. F. & Rock, D. L. (2002) Virology 295, 1–9. [DOI] [PubMed] [Google Scholar]
- 18.Gubser, C. & Smith, G. L. (2002) J. Gen. Virol. 83, 855–872. [DOI] [PubMed] [Google Scholar]
- 19.Senkevich, T. G., Bugert, J. J., Sisler, J. R., Koonin, E. V., Darai, G. & Moss, B. (1996) Science 273, 813–816. [DOI] [PubMed] [Google Scholar]
- 20.Afonso, C. L., Tulman, E. R., Lu, Z., Zsak, L., Kutish, G. F. & Rock, D. L. (2000) J. Virol. 74, 3815–3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bawden, A. L., Glassberg, K. J., Diggans, J., Shaw, R., Farmerie, W. & Moyer, R. W. (2000) Virology 274, 120–139. [DOI] [PubMed] [Google Scholar]
- 22.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wootton, J. C. & Federhen, S. (1996) Methods Enzymol. 266, 554–571. [DOI] [PubMed] [Google Scholar]
- 24.Notredame, C., Higgins, D. G. & Heringa, J. (2000) J. Mol. Biol. 302, 205–217. [DOI] [PubMed] [Google Scholar]
- 25.Takezaki, N., Rzhetsky, A. & Nei, M. (1995) Mol. Biol. Evol. 12, 823–833. [DOI] [PubMed] [Google Scholar]
- 26.Swofford, D. L. (2000) paup*: Phylogenetic Analysis Using Parsimony (* and Other Methods) (Sinauer, Sunderland, MA).
- 27.Kumar, S., Tamura, K., Jakobsen, I. B. & Nei, M. (2001) Bioinformatics 17, 1244–1245. [DOI] [PubMed] [Google Scholar]
- 28.Yang, Z. (1997) Comput. Appl. Biosci. 13, 555–556. [DOI] [PubMed] [Google Scholar]
- 29.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang, Z. (2001) in Handbook of Statistical Genetics, eds. Balding, D., Bishop, M. & Cannings, C. (Wiley, London), pp. 327–350.
- 31.Everett, H. & McFadden, G. (2002) Curr. Opin. Microbiol. 5, 395–402. [DOI] [PubMed] [Google Scholar]
- 32.Massung, R. F., Esposito, J. J., Liu, L. I., Qi, J., Utterback, T. R., Knight, J. C., Aubin, L., Yuran, T. E., Parsons, J. M., Loparev, V. N., et al. (1993) Nature 366, 748–751. [DOI] [PubMed] [Google Scholar]
- 33.Moyer, R. W. & Turner, P. C. (1990) Poxviridae (Springer, Berlin).
- 34.Schwarz, D. A., Katayama, C. D. & Hedrick, S. M. (1998) Immunity 9, 657–668. [DOI] [PubMed] [Google Scholar]
- 35.Bush, R. M., Fitch, W. M., Bender, C. A. & Cox, N. J. (1999) Mol. Biol. Evol. 16, 1457–1465. [DOI] [PubMed] [Google Scholar]
- 36.Anisimova, M., Bielawski, J. P. & Yang, Z. (2001) Mol. Biol. Evol. 18, 1585–1592. [DOI] [PubMed] [Google Scholar]
- 37.Zuniga, M. C. (2002) Virus Res. 88, 17–33. [DOI] [PubMed] [Google Scholar]
- 38.Tulman, E. R., Afonso, C. L., Lu, Z., Zsak, L., Kutish, G. F. & Rock, D. L. (2001) J. Virol. 75, 7122–7130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shchelkunov, S. N., Totmenin, A. V., Babkin, I. V., Safronov, P. F., Ryazankina, O. I., Petrov, N. A., Gutorov, V. V., Uvarova, E. A., Mikheev, M. V., Sisler, J. R., et al. (2001) FEBS Lett. 509, 66–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Afonso, C. L., Tulman, E. R., Lu, Z., Oma, E., Kutish, G. F. & Rock, D. L. (1999) J. Virol. 73, 533–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cameron, C., Hota-Mitchell, S., Chen, L., Barrett, J., Cao, J. X., Macaulay, C., Willer, D., Evans, D. & McFadden, G. (1999) Virology 264, 298–318. [DOI] [PubMed] [Google Scholar]
- 42.Willer, D. O., McFadden, G. & Evans, D. H. (1999) Virology 264, 319–343. [DOI] [PubMed] [Google Scholar]
- 43.Tulman, E. R., Afonso, C. L., Lu, Z., Zsak, L., Sur, J. H., Sandybaev, N. T., Kerembekova, U. Z., Zaitsev, V. L., Kutish, G. F. & Rock, D. L. (2002) J. Virol. 76, 6054–6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Antoine, G., Scheiflinger, F., Dorner, F. & Falkner, F. G. (1998) Virology 244, 365–396. [DOI] [PubMed] [Google Scholar]
- 45.Goebel, S. J., Johnson, G. P., Perkus, M. E., Davis, S. W., Winslow, J. P. & Paoletti, E. (1990) Virology 179, 247–266. [DOI] [PubMed] [Google Scholar]
- 46.Shchelkunov, S. N., Totmenin, A. V. & Sandakhchiev, L. S. (1996) Virus Res. 40, 169–183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.