Abstract
Background
The non-pathogenic bacterium Mycobacterium smegmatis is widely used as a near-native expression host for the purification of Mycobacterium tuberculosis proteins. Unfortunately, the Hsp60 chaperone GroEL1, which is relatively highly expressed, is often co-purified with polyhistidine-tagged recombinant proteins as a major contaminant when using this expression system. This is likely due to a histidine-rich C-terminus in GroEL1.
Results
In order to improve purification efficiency and yield of polyhistidine-tagged mycobacterial target proteins, we created a mutant version of GroEL1 by removing the coding sequence for the histidine-rich C-terminus, termed GroEL1ΔC. GroEL1ΔC, which is a functional protein, is no longer able to bind nickel affinity beads. Using a selection of challenging test proteins, we show that GroEL1ΔC is no longer present in protein samples purified from the groEL1ΔC expression strain and demonstrate the feasibility and advantages of purifying and characterising proteins produced using this strain.
Conclusions
This novel Mycobacterium smegmatis expression strain allows efficient expression and purification of mycobacterial proteins while concomitantly removing the troublesome contaminant GroEL1 and consequently increasing the speed and efficiency of protein purification.
Background
Heterologous expression of recombinant proteins in Escherichia coli can result in the production of insoluble inclusion bodies. Recent statistics show that less than half of the M. tuberculosis (Mtb) proteins expressed in E. coli are soluble [1]. Therefore, the non-pathogenic bacterium Mycobacterium smegmatis is often used as an alternative, more closely related host for the expression of mycobacterial proteins. Furthermore, M. smegmatis may also provide mycobacterium-specific chaperones, which can help correct folding of Mtb proteins [1].
During nickel affinity purification, it has been observed that a protein of 56 kDa is co-purified with polyhistidine-tagged recombinant proteins while using M. smegmatis as an expression system. This contaminant was previously identified as the Hsp60 chaperone GroEL1 of M. smegmatis [1-3]. The protein sequence of GroEL1 shows a histidine-rich C-terminus (7 out of 11 amino acids are histidines), which is likely to be the reason for the observed nickel sepharose binding [1,2].
Unlike most other bacteria, mycobacteria possess two Hsp60 chaperone groEL genes, one of which is arranged in the bicistronic groESL operon [4]. M. smegmatis also encodes a third Hsp60 protein (Msmeg1978), which is more distantly related to GroEL1 (Msmeg1583) and GroEL2 (Msmeg0880) [3]. Although groEL1 of M. smegmatis can be found in the same operon as groES, an arrangement indispensable for the chaperone function in bacteria, its histidine-rich tail is distinct from the more typical glycine-methionine-rich C-terminal region found in GroEL2 [3]. Furthermore, groEL2 is an essential gene and exists in all actinobacteria, in contrast to groEL1 [3,5]. Recently, it has been shown that groEL2 and groES are expressed more strongly than groEL1, which might have arisen from a difference in stability of the predicted post-transcriptionally cleaved mRNAs for groES and groEL1 [5]. Consistent with the current chaperone model in mycobacteria, one chaperone, here GroEL2, would act as the main house keeping chaperone in M. smegmatis, with the other chaperones (GroEL1 and Msmeg1978) adopting more specialised functions. Indeed, GroEL1 of M. tuberculosis was recently identified as being associated with nucleotides, suggesting a role as a DNA chaperone, while GroEL1 of M. smegmatis was found to have a role in mycolic acid biosynthesis during biofilm formation [5,6,3].
The co-purification of GroEL1 with histidine-tagged recombinant proteins can be particularly problematic since native GroEL1 is expressed at relatively high levels, meaning that in the case of a low yield of recombinant protein, GroEL1 may well compete with the protein of interest for binding sites on nickel affinity beads. Minimal sample manipulation is recommended during protein purification to improve efficiency. Therefore, additional steps required to remove GroEL1 can result in a significant loss of the protein of interest.
In this article, we describe an M. smegmatis expression strain containing a mutant version of GroEL1, termed GroEL1ΔC, which consists of a groEL1 gene without a coding sequence for the histidine-rich C-terminal tail. We show that GroEL1ΔC is a functional protein, which no longer co-purifies when using nickel affinity purification and we provide evidence that proteins purified from this strain are correctly folded, active and that they behave identically to those purified from the original expression strain. Taken together, our data demonstrate that M. smegmatis groEL1ΔC is a competent protein expression strain, which allows the efficient removal of the troublesome contaminant GroEL1 without the requirement of additional purification steps.
Methods
Bacterial strains and media
The E. coli strains DH5α (Invitrogen) and HB101 (Promega) were used for cloning of expression constructs and the target substrate to generate the mutant version of groEL1 using standard procedures [7]. Transformants were selected in Luria Broth containing the appropriate antibiotics.
M. smegmatis mc2155 was used as the parent (wild type) strain for the groEL1ΔC strain. Both M. smegmatis strains were maintained in Middlebrook 7H9 or 7H10 medium supplemented with 0.2% (v/v) glycerol, 10% ADC, 0.05% (v/v) tween-80 and the appropriate antibiotics.
For biofilm formation, 10 ml of biofilm media was inoculated with 10 μl of saturated culture and incubated at 30°C without disturbance [3,8].
For the expression of the recombination proteins in M. smegmatis in order to create the mutant form of groEL1, 0.2% succinate (w/v) was added as a carbon source to 7H9 medium supplemented with 0.2% (v/v) glycerol, 0.05% (v/v) tween and the appropriate antibiotics. Expression of his-tagged recombinant proteins in M. smegmatis was performed in 7H9 medium supplemented with 0.2% (w/v) glucose as carbon source. Acetamide was added to a final concentration of 0.2% (w/v) at 0.5 OD600 and at 2.5 OD600 for the expression of the recombination proteins and his-tagged recombinant proteins, respectively.
Plasmids, constructs and oligonucleotides
All plasmids and constructs are summarised in Table 1 and oligonucleotides are listed in Table 2. pJV53 was used to express the recombination proteins [9]. pYUB854 was used for the preparation of the target substrate to create the groEL1ΔC strain [10]. pGH542, harbouring a δγ resolvase, was used to generate an unmarked deletion [11]. Using the primer pairs Msmeg1583-F1.2 & Msmeg1583-R1 and Msmeg1583-F2 & Msmeg1583-R2.1, two 500 bp fragments, homologous to the fragments +1067/+1587 and +1621/+2176 relative to the translational start of Msmeg1583, were amplified and subsequently ligated AflII-XbaI (F1.2-R1) and HindIII-SpeI (F2-R2.1) into pYUB854, creating pEN15.
Table 1.
Plasmids and constructs used in this study
| Plasmid/construct | Description | Reference |
|---|---|---|
| pJV53 | Che9c recombination proteins under control of the acetamidase promoter in pLAM12 | [9] |
| pYUB854 | HygR cassette flanked by γδ-res sites and 2 MCSs | [10] |
| pGH542 | Expressing an γδ resolvase and tetracycline resistant | [11] |
| pEN15 | pYUB854 with a 520 bp fragment harbouring groEL1 (+1067/+1587, relative to groEL1) inserted upstream of the HygR cassette and a 555 bp fragment downstream of groEL1 including the STOP codon of groEL1, inserted downstream of the HygR cassette | This paper |
| pMyNT | Mycobacterial overexpression vector | Geerlof et al., unpublished data |
| pMyNT/PrcA-B | Rv2109-2110 in pMYNT, Rv2110 is N-terminally his-tagged | [12] |
| pMyNT/AccD5E5 | Rv3280-3281 in pMYNT. Only his-tagged Rv3280 seems to express using this construct. | This paper |
| pMyNT/AccA3 | Rv3285 in pMyNT | This paper |
| pMyNT/CFP10-ESAT6 | Rv3874-3875 in pMYNT, Rv3874 is N-terminally his-tagged | [12] |
| pMyNT/ACPS | Rv2523 in pMYNT | This paper |
Table 2.
Primers used in this study
| Primer | Sequence (5'-3') | Location 5' | Relative to |
|---|---|---|---|
| Msmeg1583-F1.2 | GCGCCTTAAGCGACTGGGATCGCGAGAAGCTGC | +1067 | Msmeg1583 |
| Msmeg1583-R1 | GCGCTCTAGACTCGTCCTCGTCGGCCGGCTTG | +1587 | Msmeg1583 |
| Msmeg1583-F2 | GCGCAAGCTTGATCCATTTCACGCGACACCCCC | +1620 | Msmeg1583 |
| Msmeg1583-R2.1 | GCGCACTAGTGGTGTTCGATCGTCTGGCCGATG | +2176 | Msmeg1583 |
| accD5E5-F | GATCTCATGAGTATGACAAGCGTTACC G | +1 | Rv3280 |
| accD5E5-R | GTCAAAGCTTTTATCGGCGCATGTGCG | +2161 | Rv3280 |
| accA3-F | GATCCCATGGGTATGGCTAGTCACGCC | +2 | Rv3285 |
| accA3-R | GTCAAAGCTTTTACTTGATCTCGGCGAGC | +1803 | Rv3285 |
| Rv2523-F | CATGCCATGGGCATCGTCGGTGTGGGG | +1 | Rv2523 |
| Rv2523-R | CCCAAGCTTACGGGGCCTCCAGGATGGC | +391 | Rv2523 |
Restriction sites are presented in bold face. CTTAAG = EcoRI, TCTAGA = XbaI, CCATGG = NcoI, TCATGA = BspHI
AAGCTT = HindIII, ACTAGT = SpeI.
For the expression of M. tuberculosis proteins in M. smegmatis, the pMyNT expression vector was used [Geerlof et al., unpublished data]. pMyNT/ACPS, pMyNT/AccA3 and pMyNT/AccD5 were made as follows: PCR was performed with primer pair Rv2523-F & Rv2523-R for ACPS, accA3-F & accA3-R for AccA3 and accD5E5-F & accD5E5-R for AccD5 and the resulting fragments were digested with NcoI-HindIII and inserted into NcoI-HindIII digested pMyNT.
Creation of the groEL1ΔC mutant
The groEL1ΔC mutant was created using the mycobacterial recombineering method [9]. pEN15 was digested with AflII and SpeI to create the linear target substrate, which was introduced into mc2155 electrocompetent cells, expressing the recombinase genes on pJV53 and in this way creating hygromycin-resistant transformants. The hygromycin-resistance cassette was removed using δγ resolvase, expressed on pGH542, generating an unmarked deletion [11].
Southern blot analysis
Genomic DNA (5ug) was isolated as described [9], digested with the appropriate enzymes, separated on a 0.9% agarose gel and transferred to a positively charged nylon membrane (Roche). For DNA probe labelling, hybridisation and detection, the DIG high prime DNA labelling and detection starter kit 1 (Roche) was used.
Growth curves
Bacterial growth was followed by measuring the optical densities at a wavelength of 600 nm as a function of time. Cultures were prepared with 7H9 expression medium (0.2% (w/v) glucose as carbon source) in identical triplicates for each strain. Duplicate samples were taken every 4 hours for 40 hours. When the optical density at 600 nm exceeded 1.5, samples were diluted in order to remain within the linear range of the detector.
Protein expression and purification
All methods related to protein expression in M. smegmatis were carried out as described [12,13]. Protein-protein complexes from operon-encoded proteins were expressed using the native operon structure [9]. In brief, pellets from 500 ml cultures were dissolved in 30 ml lysis buffer containing 50 mM Tris-HCl pH 8.0, 300 mM NaCl, 0.5 M urea with protease inhibitor cocktail (Sigma) and 1 mg/ml DNase I (Serva). Resuspended cells were sonicated four times, each for 5 min (with a 0.3 s pulse and 0.7 s rest) at 5 min intervals to prevent overheating, using a Bandelin VW3200 probe at 45% amplitude. The supernatant was collected after centrifugation (30,000 × g) for 1 h at 4°C, filtered through a 0.44 μm filter and loaded onto a nickel affinity sepharose (NiAC) column. After washing with 10 column volumes of 50 mM Tris-HCl pH 8.0, 300 mM NaCl and 20 mM imidazole, proteins were eluted in 50 mM Tris-HCl, 100-150 mM NaCl and 250-500 mM imidazole and subjected to size exclusion chromatography using either a Superdex 75 (16/60) column (GE Healthcare) or, for large protein complexes, a Superose 6 (10/300) (GE Healthcare) with 25 mM Tris-HCl pH 8.0, 150 mM NaCl and 1 mM DTT as buffer. The collected protein samples were analysed by SDS-PAGE and concentrated accordingly.
Circular Dichroism (CD) spectrum analysis
CD measurements were performed on a Jasco J-810 spectropolarimeter. Prior to measurement, samples were dialysed into 10 mM potassium phosphate, 150 mM NaCl, pH 7.4. Spectra were recorded between 182 and 260 nm in a 2 mm cuvette with machine settings as follows: 1 nm bandwidth, 1 sec response, 1 nm data pitch, 100 nm/min scan speed, cell length of 0.1 cm. Each curve presented is the average of three separate measurements.
Coupled enzyme assay
Enzymatic activity of the AccD5-AccA3 complex was estimated by a coupled enzyme assay that follows the rate of ATP hydrolysis spectrophotometrically [14]. The production of ADP during the reaction was coupled to pyruvate kinase and lactate dehydrogenase, and the oxidation of NADH was probed at 340 nm. The assay mixture contained 7 units of pyruvate kinase, 10 units of lactate dehydrogenase, 50 mM NaHCO3, 3 mM ATP, 0.5 mM phosphoenol pyruvate, 0.2 mM NADH, 0.3 mg/ml BSA, 100 mM K2HPO4 pH 7.6 and 5 mM MgCl2 and varying concentrations of propionyl-coenzyme A. Reactions were initiated by the addition of enzyme to the assay mixture and were maintained at 30°C. Data were acquired using a Tecan infinite M1000 microplate reader. The kinetic parameters Km and Vmax were determined by fitting the mean velocities versus the substrate concentration to the Michaelis-Menten equation of enzyme kinetics using nonlinear regression analysis, executed by the program Prism 5 (GraphPad Software™).
Results and Discussion
Creation of the groEL1ΔC strain
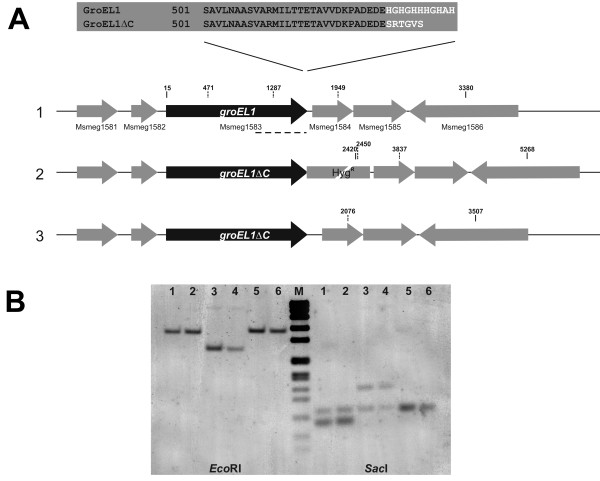
Currently, the role of GroEL1 in protein folding is uncertain. A closer look at the structure of E. coli GroEL [15] indicates that, although the C-terminal region of the protein is not easily accessible, pointing towards the central cavity of the wheel-like structure adopted by oligomeric GroEL, the extreme C-terminal 20 amino acids are absent from the model. Similarly, the GroEL structure of Paracoccus denitrificans also lacks these residues [16]. These observations suggest that the C-terminal region of GroEL is highly flexible and could reach out of the central cavity, allowing in this way M. smegmatis GroEL1 to bind nickel affinity beads. Additionally, as native GroEL1 from M. tuberculosis is oligomeric [17], nickel binding would require only one accessible histidine-rich region. Therefore, we decided to change only the last eleven amino acids of the protein, rather than to make a full knock out strain, in order to minimise changes to the expression strain. A precise chromosomal deletion of fragment 1588-1620, relative to the translational start of groEL1 (Msmeg1583), was created using the mycobacterial recombineering technique [9] (Figure 1A). Southern hybridisation (Figure 1B) was used to verify that correct homologous recombination had taken place in the hygromycin-resistant and the unmarked deletion strain (Figure 1B). The latter strain, in which the hygromycin resistance cassette has been removed, has the C-terminal eleven residues of GroEL1, containing seven histidines, replaced by six non-histidine residues, which are part of the "scar" sequence left behind after removal of the resistance cassette (Figure 1A). The stop codon of this recombinant version of GroEL1 is TAA, which although rare, is recognised in high G+C mycobacteria [18]. This unmarked deletion strain, referred to as M. smegmatis groEL1ΔC, is used in all further experiments.
Figure 1.
Construction of the groEL1ΔC strain. (A) A schematic representation of the genomic organisation of groEL1 (Msmeg1583) in M. smegmatis mc2155 (1), the hygromycin-resistant groEL1ΔC mutant (2) and the unmarked groEL1ΔC deletion strain (3). Msmeg1582 is groES and Msmeg1581, 1584, 1585 and 1586 encode for proteins of unknown function. Zoom sequence of the C-terminal 40 amino acids of the groEL1 gene product, showing the histidine-rich C-terminal region. In the groEL1ΔC strain, the eleven C-terminal amino acids (white) were replaced by six different residues. HygR = hygromycin-resistance cassette. (B) Southern blot analysis performed with genomic DNA of M. smegmatis mc2155 (1), mc2155 carrying pJV53 (2), two correct hygromycin-resistant groEL1ΔC mutants (3-4) and two correct unmarked groEL1ΔC deletions (5-6). The genomic DNA was digested with EcoRI or SacI. The positions of the restriction sites EcoRI and SacI, relative to the start of groEL1, are presented as a full and dotted vertical line in A, respectively. The PCR product of primer pair Msmeg1583-F1.2 & Msmeg1583-R1 was used as a probe, shown as a dashed line under the genes in A1. (M) DNA marker VII, DIG labelled (Roche).
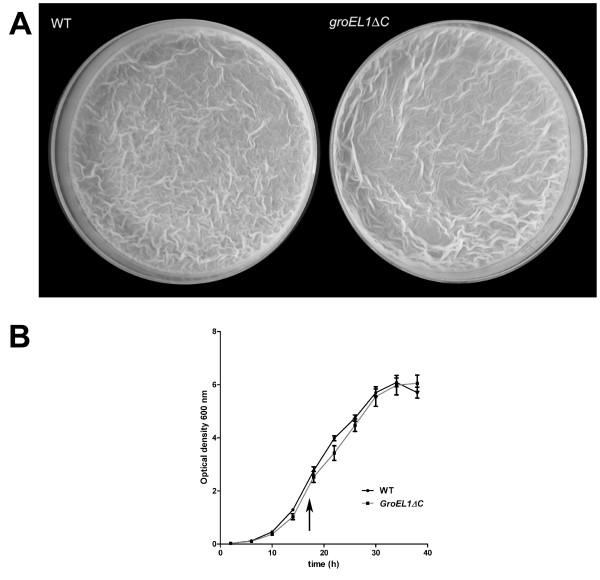
Ojha et al. reported that the last 18 amino acids of GroEL1 are essential for the formation of mature biofilms [3]. Therefore, to test the functionality of the GroEL1ΔC protein, we compared biofilm formation in both the wild type and groEL1ΔC strains. Both strains were able to form mature biofilms after an incubation time of 7 days at 30°C, indicating that GroEL1ΔC is indeed fully functional (Figure 2A). Taking into account the data from Ojha et al., our results could suggest that either the amino acids important for biofilm formation are upstream of those removed in the GroEL1ΔC protein, or that removal of the last 18 residues may affect the folding of at least a part of GroEL1. Additionally, as this newly created strain was constructed for the overexpression of mycobacterial proteins, its growth in 7H9 expression medium was compared to the original expression strain M. smegmatis mc2155 (Figure 2B). We observed no significant differences in growth between the two strains, with both reaching an OD600 of between 2.5-3.0 after approximately 18 hours, at which time expression is usually induced.
Figure 2.
Biofilm formation and growth rates of M. smegmatis mc2 155 and M. smegmatis groEL1ΔC are comparable. (A) Both M. smegmatis mc2 155 (WT) and M. smegmatis groEL1ΔC strains are able to form biofilms after an incubation time of 7 days at 30°C. (B) Growth curve of M. smegmatis mc2155 (WT = black) and M. smegmatis groEL1ΔC strains (grey) in 7H9 expression medium. The arrow represents the typical time of induction in M. smegmatis.
GroEL1ΔC is absent during nickel affinity purification of proteins expressed in M. smegmatis groEL1ΔC
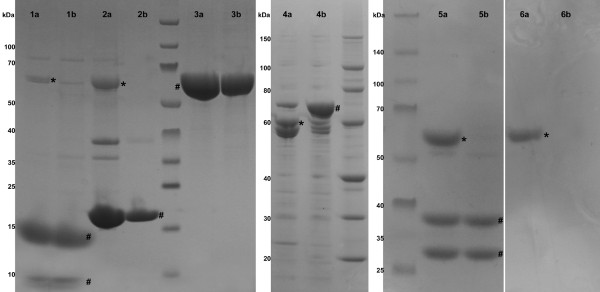
To demonstrate the absence of GroEL1ΔC as a contaminant when using the M. smegmatis groEL1ΔC expression strain, we determined the expression and purification efficiency of our strain in comparison to the wild type strain using five different constructs, representing a variety of different protein molecules, including the mycobacterial proteasome, the CFP10-ESAT6 complex, the AccD5-AccA3 dodecameric acyl-CoA carboxylase complex and the holo-acyl-carrier protein synthase (for details, see Table 3). Additionally, we also used the empty pMyNT vector, to check for GroEL1 binding in the absence of a his-tagged protein. All constructs were transformed into both M. smegmatis mc2155 and groEL1ΔC and the resulting transformants were cultured in 7H9 expression medium and induced by the addition of acetamide to a final concentration of 35 mM. Eighteen hours after induction, the cells were collected by centrifugation, lysed and the soluble protein fraction was passed over a nickel affinity column, with the elution fraction being analysed by SDS-PAGE (Figure 3). While GroEL1 was visible in samples purified from M. smegmatis mc2155 (Figure 3, lanes a), the protein was noticeably absent in five out of six protein samples isolated from the groEL1ΔC strain (Figure 3, lanes b). Due to the fact that AccD5 has a similar size to GroEL1, we were unable to determine its presence or absence in samples of the purified acyl-CoA carboxylase complex by SDS-PAGE. Therefore, samples isolated from gel (Figure 3) were analyzed by mass spectrometry (Additional file 1). While numerous peptides from both GroEL1 and AccD5 could be identified from gel slices deriving from the mc2155 strain, only AccD5 peptides could be detected in the sample obtained from the groEL1ΔC strain (Additional file 1). Likewise, MALDI-TOF mass spectrometry was performed on the other protein samples, verifying the absence of GroEL1 peptides in the protein samples derived from M. smegmatis groEL1ΔC (data not shown).
Table 3.
List of test proteins used to validate the groEL1ΔC expression strain
| ORF | Annotation | Description | Expressed ... | Mol. Mass (kDa) | |||
|---|---|---|---|---|---|---|---|
| Rv2109c Rv2110c |
PrcA PrcB |
α- and β-subunit of the mycobacterial proteasome (α7β7β7α7 subunit organisation) | Using native operon content, producing a 730 kDa multimeric complex | 26.8 30.3 |
|||
| Rv3285 Rv3280 |
AccA3 AccD5 |
α- and β-subunit from acyl-CoA carboxylase AccD5-AccA3 complex (α3β3β3α3 subunit organisation) | As monomeric proteins, mixed to form a acyl-CoA carboxylase complex of 740 kDa | 63.8 59.4 |
|||
| Rv3874 Rv3875 |
CFP10 ESAT6 |
Potential virulence factor CFP10-ESAT6 complex | Using native operon content, producing a heterodimeric (1:1) complex | 10.8 9.9 |
|||
| Rv2523c | ACPS | Holo-acyl-carrier protein synthase | As monomeric protein | 14 | |||
Figure 3.
GroEL1ΔC is absent from protein samples purified from M. smegmatis groEL1ΔC. SDS-PAGE analysis of protein samples isolated from cells expressing either CFP10-ESAT6 heterodimer (1), a holo-acyl-carrier protein synthase (ACPS) (2), the β- and α-subunit D5 (3) and A3 (4) of the acyl-CoA carboxylase AccD5-AccA3 complex, the PrcA-B complex (5) or the empty expression vector (empty pMyNT) (6). a, proteins expressed in M. smegmatis mc2155; b, proteins expressed in M. smegmatis groEL1ΔC. Purified proteins of interest are labelled with #. Visible presence of GroEL1 is depicted with *. Squares identify the bands excised for peptide mass fingerprinting. GroEL1 = 56 kDa.
Proteins purified from M. smegmatis groEL1ΔC behave identically to those purified from the wild type strain
M. smegmatis encodes three forms of the Hsp60 chaperone GroEL: Msmeg1583 (GroEL1), Msmeg0880 (GroEL2) and Msmeg1978. However, the precise molecular function of each protein remains unclear. Changing the last 18 amino acids of GroEL1 does not alter growth but does result in a strong defect in biofilm formation [3]. To confirm that the newly created recombinant version of GroEL1 has no effect on the correct folding and, ultimately, the function of the proteins expressed in M. smegmatis groEL1ΔC, a number of different proteins and protein complexes have been expressed and analysed.
In the previous section, we have shown that it is possible to express and purify potentially challenging protein complexes, such as the proteasome complex PrcA-B and the CFP10-ESAT6 complex, from the recombinant groEL1ΔC strain. These data imply that the proteins isolated from the groEL1ΔC strain are correctly folded, since we were able to observe all components after purification. In both examples, complex formation requires direct protein-protein interactions between subunits of the complex as only one subunit is his-tagged.
Taking our analysis one step further, we directly tested the structural and functional properties of proteins isolated from the groEL1ΔC strain. We used the five expression constructs described above and transformed them into both M. smegmatis mc2155 and groEL1ΔC. Proteins were expressed and purified using a nickel affinity column as described above. AccD5 and AccA3 protein samples were mixed in a 1:1 stoichiometry to form the high-molecular-weight AccD5-AccA3 complex. Size exclusion chromatography was performed on all samples as a final purification step.
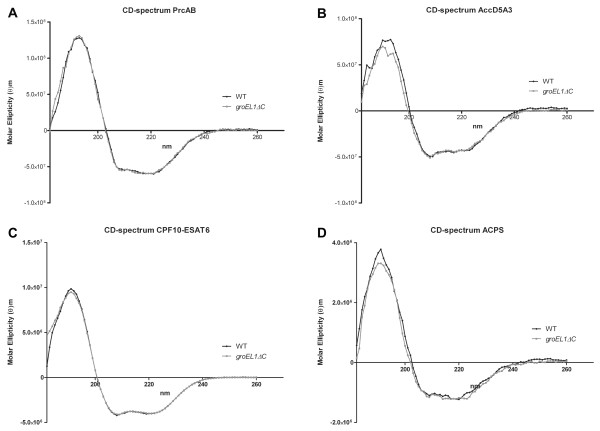
Circular dichroism (CD) spectroscopy is a powerful tool used to visualise the secondary structure properties of protein samples. We observed that the four protein samples isolated from groEL1ΔC gave virtually identical CD spectra to those purified from the wild type strain (Figure 4), implying that they are correctly folded. Furthermore, the CD spectra of the CFP10-ESAT6 complexes, showing a protein with high helical content, are comparable to those collected previously [12] and are in line with the X-ray structure, which consists of a four-helical bundle complex (PDB ID: 3FAV) [12].
Figure 4.
Proteins isolated from both strains give virtually identical CD spectra. CD spectra of the multimeric proteasome complex PrcA-B (A), the dodecameric acyl-CoA carboxylase AccD5-AccA3 complex (B), CFP10-ESAT6 heterodimer (C), and monomeric protein ACPS (D) expressed in M. smegmatis mc2155 (WT = black) and M. smegmatis groEL1ΔC (grey) are virtually identical. For A and B, a concentration between 170 and 200 nM was used while for C and D, concentrations were between 5 and 10 μM.
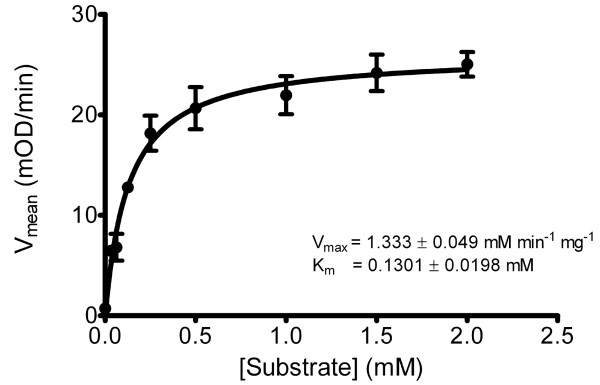
Additionally, we have demonstrated carboxylase activity of the acyl-CoA carboxylase AccD5-AccA3 complex, isolated from groEL1ΔC, using an enzyme-coupled reaction (Figure 5). Using propionyl-CoA as a substrate, AccD5-AccA3 showed carboxylase activity with a Km = 0.1301 ± 0.0198 mM and a Vmax = 1.333 ± 0.049 mM min-1 mg-1, data which are similar to the parameters determined using the AccD5-AccA3 complex isolated from E. coli [19], indicating that the AccD5-AccA3 complex isolated from groEL1ΔC is a functional carboxylase. Carboxylase activity requires the α-subunit of the carboxylase to be post-translationally biotinylated [19], implying that the subunits of this large megasynthase are folded correctly and, in the case of the α-subunit, correctly post-translationally modified, when isolated from groEL1ΔC.
Figure 5.
Kinetics of AccD5-AccA3 isolated from M. smegmatis groEL1ΔC. Carboxylation activity of the acyl-CoA carboxylase AccD5-AccA3 complex isolated from groEL1ΔC was measured using an enzyme-coupled reaction with propionyl-CoA as substrate, providing Km = 0.1469 mM and Vmax = 28.5 mOD/min.
Conclusions
We have developed an M. smegmatis expression strain that allows efficient expression and purification of mycobacterial proteins, multi-subunit protein complexes and post-translationally modified proteins while concomitantly removing the troublesome contaminant GroEL1 and consequently increasing the speed and efficiency of protein purification. The M. smegmatis groEL1ΔC strain is particularly suitable for laboratories performing in vitro activity assays and structural studies on mycobacterial proteins and protein complexes.
Abbreviations
PCR: Polymerase chain reaction; kDa: kilo Dalton; Hsp60: Heat shock protein 60; ADC: Albumine-dextrose-catalase; DMSO: dymethylsulfoxide; NiAc: Nickel affinity sepharose column; SDS-PAGE: sodium dodecyl sulfate polyacrylamide gel electroporesis; MALDI-TOF: matrix-assisted laser desorption/ioization reflection time-of-flight.
Authors' contributions
EN and CP designed the study. EN made the groEL1ΔC strain, tested its functionality and growth, expressed and purified all proteins described except the AccD5-AccA3 complex and wrote the manuscript. CW carried out the CD measurements, provided technical assistance and participated in writing the manuscript. MA carried out all experiments concerning AccD5-AccA3. CP provided expression constructs. ME participated in testing the strain's growth and feasibility. MW organized the funding, supervised the work and helped revising the manuscript. All authors read and approved the final manuscript.
Supplementary Material
GroEL1 is absent from an AccD5 protein sample derived from M. smegmatis groEL1ΔC. Results of peptide mass fingerprinting analysis of samples excised from SDS-PAGE gel (Figure 3, boxes). Shown in red are the peptides that could be identified. (a) Sample derived from M. smegmatis mc2155. (b) Sample derived from M. smegmatis groEL1ΔC.
Contributor Information
Elke E Noens, Email: e.noens@embl-hamburg.de.
Chris Williams, Email: c.williams@embl-hamburg.de.
Madhankumar Anandhakrishnan, Email: m.anandhakrishnan@embl-hamburg.de.
Christian Poulsen, Email: poulsen@scripps.edu.
Matthias T Ehebauer, Email: m.ehebauer@embl-hamburg.de.
Matthias Wilmanns, Email: matthias.wilmanns@embl-hamburg.de.
Acknowledgements and Funding
We thank Arie Geerlof for the pMyNT expression vector, Young-Hwa Song for her contribution in the early stages of the work, the Proteomics Core Facility of EMBL Heidelberg for performing peptide mass fingerprinting, the Mandelkow Lab (Max Planck Institute for Structural Molecular Biology, Hamburg, Germany) for access to their circular dichroism spectroscope. CW is funded by a Rubicon post-doctoral fellowship (825.08.023) from the Netherlands organization for scientific research (NWO). ME is funded by an EMBO long-term fellowship (ALTF-7272008). The project has been supported by grants awarded to MW from BMBF (Pathogenomik Plus PTJ-BIO 0313801L), from the European Commission Framework VII (NATT, 222965 and SystemTB, 241587) and from the DFG (SPP1170, WI 1058/6-3).
References
- Goldstone RM, Moreland NJ, Bashiri G, Baker EN, Lott JS. A new Gateway® vector and expression protocol for fast and efficient recombinant protein expression in Mycobacterium smegmatis. Protein Expr Purif. 2008;57:81–87. doi: 10.1016/j.pep.2007.08.015. [DOI] [PubMed] [Google Scholar]
- Poulsen C, Ahkter Y, Jeon AH, Schmitt-Ulms G, Meyer HE, Stühler K, Wilmanns M, Song YH. Proteome-wide identification of mycobacterial pupylation targets. Mol Syst Biol. 2010;6:386–394. doi: 10.1038/msb.2010.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojha A, Anand M, Bhatt A, Kremer L, Jacobs WR Jr, Hatfull GF. GroEL1: a dedicated chaperone involved in mycolic acid biosynthesis during biofilm formation in Mycobacteria. Cell. 2005;123:861–873. doi: 10.1016/j.cell.2005.09.012. [DOI] [PubMed] [Google Scholar]
- Rinke de Wit TF, Bekelie S, Osland A, Miko TL, Hermans PW, van Soolingen D, Drijfhout JW, Schöningh R, Janson AA, Thole JE. Mycobacteria contain two groEL genes: the second Mycobacterium leprae groEL gene is arranged in an operon with groES. Mol Microbiol. 1992;6(14):1995–2007. doi: 10.1111/j.1365-2958.1992.tb01372.x. [DOI] [PubMed] [Google Scholar]
- Rao T, Lund PA. Differential expression of the multiple chaperonins of Mycobacterium smegmatis. FEMS Micobiol Lett. 2010;310:24–31. doi: 10.1111/j.1574-6968.2010.02039.x. [DOI] [PubMed] [Google Scholar]
- Basu D, Khare G, Singh S, Tyagi A, Khosla S, Mande SC. A novel nucleoid-associated protein of Mycobacterium tuberculosis is a sequence homolog of GroEL. Nucleic Acids Res. 2009;37(15):4944–4954. doi: 10.1093/nar/gkp502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring harbor: Cold Spring harbor laboratory press; 1989. [Google Scholar]
- Recht J, Kolter R. Glycopeptidolipid acetylation affects sliding motility and biofilm formation in Mycobacterium smegmatis. J Bacteriol. 2001;183:5718–5724. doi: 10.1128/JB.183.19.5718-5724.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kessel JC, Hatfull GF. Recombineering in Mycobacterium tuberculosis. Nat Methods. 2007;4(2):147–152. doi: 10.1038/nmeth996. [DOI] [PubMed] [Google Scholar]
- Bardarov S, Bardarov S Jr, Pavelka Ms Jr, Sambandamurthy V, Larsen M, Tufariello J, Chan J, Hatfull G, Jacobs WR Jr. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology. 2002;148:3007–3017. doi: 10.1099/00221287-148-10-3007. [DOI] [PubMed] [Google Scholar]
- Piuri M, Hatfull GF. A peptidoglycan hydrolase motif within the mycobacteriophage TM4 tape measure protein promotes efficient infection of stationary phase cells. Mol Microbiology. 2006;62:1569–1585. doi: 10.1111/j.1365-2958.2006.05473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen C, Holton S, Geerlof A, Wilmanns M, Song YH. Stoichiometric protein complex formation and over-expression using the prokaryotic native operon structure. FEBS Lett. 2010;584:669–674. doi: 10.1016/j.febslet.2009.12.057. [DOI] [PubMed] [Google Scholar]
- Daugelat S, Kowall J, Matthow J, Bumann D, Winter R, Hurwitz R, Kaufmann SH. The RD1 proteins of Mycobacterium tuberculosis: expression in Mycobacterium smegmatis and biochemical characterization. Microbes Infect. 2003;5:1082–1095. doi: 10.1016/S1286-4579(03)00205-3. [DOI] [PubMed] [Google Scholar]
- Diacovich L, Peiru S, Kurth D, Rodriguez E, Podesta F, Khosla C, Gramajo H. Kinetic and Structural Analysis of a New Group of Acyl-CoA Carboxylases Found in Streptomyces coelicolor A3(2) J Biol Chem. 2002;277:31228–31236. doi: 10.1074/jbc.M203263200. [DOI] [PubMed] [Google Scholar]
- Xu Z, Horwich AL, Sigler PB. The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex. Nature. 1997;388(6644):741–50. doi: 10.1038/41944. [DOI] [PubMed] [Google Scholar]
- Fukami TA, Yohda M, Taguchi H, Yoshida M, Miki K. Crystal structure of chaperonin-60 from Paracoccus denitrificans. J Mol Biol. 2001;312(3):501–9. doi: 10.1006/jmbi.2001.4961. [DOI] [PubMed] [Google Scholar]
- Kumar CM, Khare G, Srikanth CV, Tyagi AK, Sardesai AA, Mande SC. Facilitated oligomerization of mycobacterial GroEL: Evidence for phosphorylation-mediated oligomerization. J Bacteriol. 2009;191:6525–6538. doi: 10.1128/JB.00652-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson SGE, Sharp PM. Codon usage in the Mycobacterium tuberculosis complex. Microbiology. 1996;142:915–925. doi: 10.1099/00221287-142-4-915. [DOI] [PubMed] [Google Scholar]
- Gago G, Kurth D, Diacovich L, Tsai SC, Gramajo H. Biochemical and structural characterization of an essential acyl Coenzyme A carboxylase from Mycobacterium tuberculosis. J Bacteriol. 2006;188:477–486. doi: 10.1128/JB.188.2.477-486.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GroEL1 is absent from an AccD5 protein sample derived from M. smegmatis groEL1ΔC. Results of peptide mass fingerprinting analysis of samples excised from SDS-PAGE gel (Figure 3, boxes). Shown in red are the peptides that could be identified. (a) Sample derived from M. smegmatis mc2155. (b) Sample derived from M. smegmatis groEL1ΔC.