Abstract
There is considerable interest in understanding how cis-regulatory modifications drive morphological changes across species. Because developmental regulatory genes, including Hox genes, are remarkably conserved, their noncoding regulatory regions are likely sources for variations. Modifications of Hox cis-regulatory elements have potential to alter Hox gene expression and, hence, axial morphologies. In vertebrates, differences in the axial levels of Hox gene expression correlate with differences in the number and relative position of thoracic vertebrae. Variation in cis-regulatory elements of Hox genes can be identified by comparative sequence and reporter gene analyses in transgenic mouse embryos. Using these approaches, we show a remarkable divergence of the Hoxc8 early enhancers between mammals and fishes representing diverse axial morphologies. Extensive restructuring of the Hoxc8 early enhancer including nucleotide substitutions, inversion, and divergence result in distinct patterns of reporter gene expression along the embryonic axis. Our results provide an evolutionary perspective on how the enhancer elements are engineered and support the hypothesis that remodeling of Hox regulatory elements in different species has played a significant role in generating morphological diversity.
The Hox family of transcription factors is ideally suited for comparative regulatory analysis because of their role in specifying positional information during embryonic development of all metazoans studied to date. Genetic analyses have firmly established roles for Hox genes in specification of structures along the embryonic axes (1). Hox genes show remarkable conservation in their architectural organization and function across species. This raises the question of how a common genetic program has evolved to account for diverse morphologies. Several mechanisms, including cluster duplication, expansion in the number of genes, divergence of protein-coding sequences, changes in expression patterns, and alterations in cis-regulatory regions have been implicated in the generation of diversity in the metazoan body plan (reviewed in refs. 2–4). In vertebrates, differences in the spatial and temporal distribution of Hox gene expression correlate with differences in the number, relative position, and identities of axial skeletal elements among different species (5–8). Changes in the Hox expression patterns in the paraxial mesoderm are reflected in the transition from one type of vertebra to another (4, 9). For instance, comparative expression analyses reveal that the anterior boundary of Hoxc8 expression lies within the thoracic regions of mouse, chick, snake, and fish even though the number of thoracic vertebra vary extensively among these organisms (5–8). In the chick with a smaller thoracic region comprising seven vertebrae, the embryonic Hoxc8 expression domain is relatively smaller compared to the mouse (13 vertebrae) and the python (>100 vertebrae), possessing an increasingly expanded thoracic identity (7, 8). Among fishes, both zebrafish and Fugu appear to have eight to 10 vertebrae in the trunk region (10–12). The spatial domain of Hoxc8 expression extends anteriorly in zebrafish and is coincident with the presence of the rib-bearing vertebrae (13, 14). Mechanistically, an alteration in Hoxc8 expression may be brought about by divergence in transcriptional regulation machineries of different species. This may involve changes in cis-regulatory elements and transacting factors whose interactions determine the embryonic expression pattern of Hoxc8. Because a majority of transacting factors are likely to be conserved among vertebrates, variation in cis-regulatory elements of regulatory genes is likely to be the source of divergence.
Transcriptional regulation of Hoxc8 during mouse embryonic development is controlled by separate cis-regulatory modules (15–19). The early enhancer, located in the 5′ region of the Hoxc8 transcriptional unit, controls the initiation and establishment of Hoxc8 expression in the posterior regions of the neural tube and mesoderm (15, 17). The early enhancer has been delimited to a 200-bp fragment by progressive deletions (15, 17). Mutational analyses of reporter gene constructs in transgenic mouse embryos identified five distinct cis-acting elements (A–E) that are potential protein-binding sites for caudal, Hox, fork-head, and the high mobility group/box family of transcription factors (17). Different combinations of cis-elements determine expression in different regions of the embryo. Furthermore, mutations within these cis-elements result in posteriorization of reporter gene expression, indicating that the anterior extent of expression is determined by the combined action of transacting factors at these elements. A survey of Hoxc8 early enhancer sequences of ≈30 mammalian species belonging to different clades revealed a high degree of sequence similarity with all defined cis-acting elements being invariant (20). A major exception was found in all five extant species belonging to the baleen whale lineage, which carry a 4-bp deletion of a cis-acting element. When assayed in transgenic mouse embryos, the baleen whale Hoxc8 early enhancer carrying the 4-bp deletion of an element failed to direct reporter gene expression to the posterior mesoderm. This difference in the early enhancer activity may reflect modification of axial structures in baleen whales as a consequence of their secondary adaptation to aquatic life. The chicken Hoxc8 early enhancer, differing in a few nucleotides from the mouse enhancer, directed reporter gene expression to a more posterior and smaller domain in transgenic mouse embryos reflecting the chicken pattern of Hoxc8 expression, which in turn parallels the smaller thoracic region (7).
To identify significant divergence of the enhancer elements, we surveyed fish Hoxc8 early enhancer sequences. Mammals and fish diverged >450 million years ago, and two species of fish, zebrafish (Danio rerio) and Fugu (Fugu rubripes) diverged >150 million years ago (21, 22). Besides diverged morphologies, these two fishes also present differing genome sizes and duplicated Hox clusters. In this study, we report a remarkable divergence of the Hoxc8 early enhancers between mammals and fishes representing diverse axial morphologies. Extensive restructuring of the Hoxc8 early enhancer including nucleotide substitutions, inversion, and divergence result in distinct patterns of reporter gene expression along the embryonic axis. Our results provide an evolutionary perspective on how the enhancer elements are engineered and support the hypothesis that remodeling of Hox regulatory elements in different species has played a significant role in generating morphological diversity.
Materials and Methods
Standard subcloning procedures were followed to generate constructs for microinjection into mouse embryos. A reporter gene construct carrying a 399-bp fragment that contains the Hoxc8 early enhancer elements has been described (17). Alterations in the 399-bp reporter construct were introduced by an overlapping PCR strategy by using synthetic oligonucleotide primers containing appropriate changes in the nucleotide sequence. Three overlapping synthetic oligonucleotides were synthesized based on the Fugu (Fugu rubripes) enhancer sequence and used in successive PCR amplifications to replace the corresponding mouse enhancer (200 bp) region in the 399-bp reporter construct (23). The zebrafish (Danio rerio) Hoxc8 enhancer region was subcloned from a P1 artificial chromosome (PAC) (PAC17B1) containing genomic regions surrounding Hoxc8 (24). PAC DNA was digested with several restriction enzymes and electrophoresed, and fragments were transferred to nitrocellulose by using a Turboblotter system (Schleicher & Schuell). The blots were hybridized with radioactively labeled probe (mouse Hoxc8 early enhancer fragment) by using a random labeling kit. HindIII digestion of the PAC17B1 DNA yielded a 2-kb fragment that hybridized with the probe. Corresponding fragments were eluted from the gel, cloned into the pBSK+ (Stratagene) vector, and sequenced. The nucleotide sequences of two independent clones were aligned with the mouse and Fugu Hoxc9-Hoxc8 intergenic sequences to identify the Hoxc8 early enhancer region (DNAMAN). A PCR based strategy was used to replace 200 bp of the mouse enhancer with the corresponding zebrafish enhancer fragment in the reporter gene construct. The replacement constructs were generated according to a previously described strategy (7, 20). A 199-bp fish enhancer fragment containing the first nucleotide of element A at the 30th position was linked to the 5′ 200 bp of the mouse enhancer region, resulting in a replacement of the mouse enhancer elements with those of the fish elements. The 5′ 200-bp fragment, which is devoid of cis-acting elements, acts as a “stuffer fragment” and provides for a more robust reporter gene expression in transgenic mouse embryos (15, 17). The procedures for the preparation of DNA for microinjection, production of transgenic embryos, and detection of β-galactosidase activity assays have been described (15).
Results and Discussion
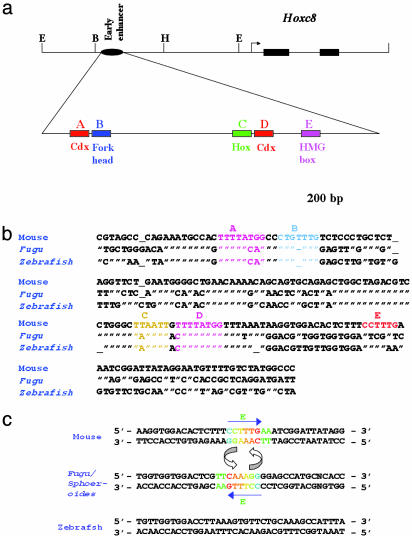
For cis-regulatory analysis, the zebrafish Hoxc8 early enhancer fragment was isolated from a PAC clone containing the Hoxc genomic region (24). A single fragment containing the Hoxc8 early enhancer region was identified by hybridization with a probe containing the mouse enhancer region. This fragment was cloned and sequenced. The Fugu Hoxc8 early enhancer sequence was obtained from the literature (23). A sequence comparison of the Hoxc8–Hoxc9 intergenic regions between mouse and fish show very little sequence similarity, outside the early enhancer region (not shown). Compared to the mouse enhancer both zebrafish and Fugu Hoxc8 early enhancers occupy similar intergenic positions between Hoxc8 and Hoxc9 genes and are recognizable because of their similar organization of the 5′ region of the enhancer containing elements A–D (Fig. 1). The two fish sequences differ considerably from the mouse sequence with only 47–55% sequence similarity over 188 nucleotides. The nucleotide similarity between the two fish enhancer sequences is ≈66%. Relative to mammalian sequences, many scattered changes are observed throughout the Hoxc8 enhancer region, with substitutions found in elements A–D. Most importantly, the 3′ region of the enhancer, immediately downstream of element D, has diverged significantly between mammalian and fish sequences. Both fish sequences fail to align with the mammalian sequences over a stretch of 56 bp downstream of element D. Although a few stretches of sequence similarity are evident between zebrafish and Fugu sequences in this region, overall the sequence similarity is <40%. At the point of divergence from the mouse sequence, the two fish enhancers share a 17-nt sequence of a high degree of similarity, suggesting that this sequence was part of other conserved blocks of sequences present before these two fishes diverged ≈150 million years ago. Furthermore, the zebrafish sequence lacks element E. Sequence comparisons of the zebrafish Hoxc8-Hoxc9 intergenic region, immediately flanking the early enhancer fail to identify additional sequences resembling early enhancer elements (not shown). In contrast, the Fugu enhancer sequence contains element E on the opposite strand as part of an 8-bp inversion between mouse and Fugu (5′-CCTTTGAA-3′–5′-TTCAAAGG-3′; Fig. 1c). A similar 8-bp inversion is also found in the Hoxc8 early enhancer region of Sphoeroides nephelus, a fish belonging to the same subfamily, Tetraodentinae, as Fugu (Fig. 1c).
Fig. 1.
Hoxc8 early enhancer. (a) Schematic of Hoxc8 genomic region showing the position of the early enhancer. E, EcoRI, B, BspEI, H, HindIII. Black boxes indicate two exons of Hoxc8. Black sphere is the position of the early enhancer located ≈3 kb upstream of the translational start site. Arrowhead indicates transcriptional start site. Cis-acting elements, A–E (colored boxes), and potential transcription factors interacting with these elements are indicated. (b) Nucleotide sequence of the Hoxc8 early enhancer. The colored sequences indicate cis-acting elements defined by mutational analysis in transgenic mouse embryos. Similarity with the mouse enhancer sequence is denoted by quotation marks, a dash denotes a missing base, and variant bases are indicated. (c) The colored sequence indicates inversion of the 8-bp sequence between the mouse and Fugu/Sphoeroides sequences. The sequence of the element E is underlined. Arrows indicate the direction of the element. Fugu and Sphoeroides sequences in this region are similar to each other, except for the base N, which is A-T in Fugu and G-C in Sphoeroides sequence.
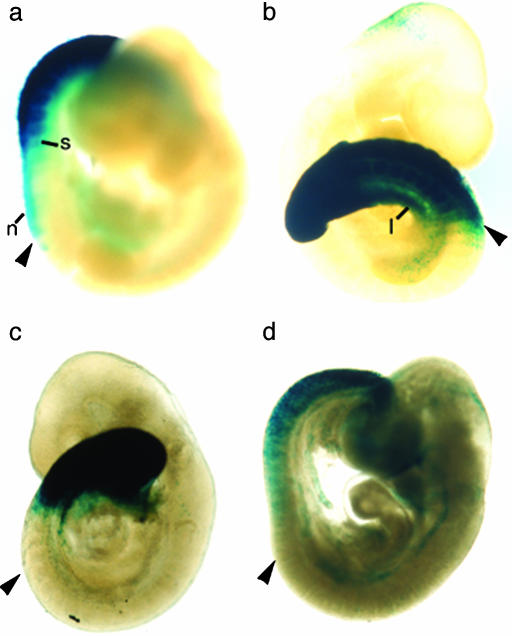
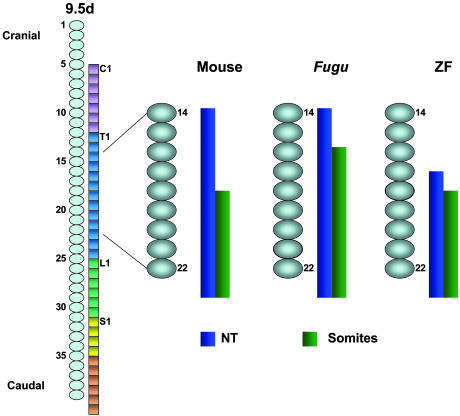
Because the mouse, Fugu, and zebrafish Hoxc8 early enhancer sequences contain significant restructuring of the cis-acting elements, their enhancer activities were assayed in transgenic mouse embryos. The three enhancers directed reporter gene expression to the posterior neural tube and mesoderm, but each had distinct rostral boundaries (Fig. 2). Whereas the mouse (Fig. 2a) and the Fugu (Fig. 2b) enhancers directed reporter gene expression to the same rostral levels in the neural tube (14th somite), the zebrafish (Fig. 2c) enhancer directed the expression to a more posterior level (17th somite). The posterior shift observed in the neural tube expression directed by the zebrafish enhancer is consistent with the posteriorizing effect seen by mutating element E in the mouse enhancer (19). In contrast, the mouse and zebrafish enhancers directed reporter gene expression to the 18th somite, but the Fugu enhancer directed somite expression to more rostral levels at the 16th somite. In the lateral plate mesoderm, the Fugu and zebrafish enhancers directed expression to more anterior levels (15th and 18th somites, respectively) compared to the mouse enhancer (20th somite). Thus, both fish enhancers directed reporter gene expression in the mesoderm to more anterior levels than the mouse enhancer. The rostral boundaries of Hox gene expression in the neural tube and somites are characteristically staggered. The somite expression is posterior to that of neural tube expression by several somite levels. The difference in the rostral boundary is specific to each species: five somite levels in the mouse and three somite levels in the zebrafish (14). The reporter gene expression driven by heterospecific Hoxc8 early enhancers may also reflect this characteristic stagger in neural tube and somite (Fig. 3). Changes in rostral boundaries of expression have also been reported for a zebrafish Hoxd11 enhancer assayed in transgenic mice (25). Targeting the zebrafish enhancer to the endogenous mouse genomic locus resulted in an anterior shift of the sacrum. These results are consistent with the idea that regulatory changes affecting anterior boundaries of Hox gene expression also affect anatomical changes in different species (25).
Fig. 2.
Expression patterns of reporter genes in transgenic embryos. Expression of the β-galactosidase gene is detected in the posterior regions of transgenic embryos staged between days 9.0 and 9.5. Shown are mouse enhancer (a), Fugu enhancer (b), zebrafish enhancer (c), and mouse enhancer carrying the 8-bp inversion of the sequence flanking element E (d). At least two independent transgenic embryos were examined for reporter gene expression for each construct. The expression boundary varied at the most by one somite level among independent transgenic embryos. Arrowhead indicates the position of the 14th somite. n, neural tube; s, somites; l, lateral plate mesoderm.
Fig. 3.
Schematic representation of anterior boundary of reporter gene expression in the neural tube and somites along the anteroposterior embryonic axis. Colored spheres indicate somites, and square boxes indicate vertebrae. NT, neural tube, C, cervical, T, thoracic, L, lumbar, S, sacral, ZF, zebrafish.
Mutating element E in the mouse enhancer results in lack of mesoderm expression (19). Because the zebrafish enhancer lacks element E, we postulated that other elements present in the fish enhancer contribute to mesoderm expression. Because element E is on the opposite strand in the Fugu enhancer, we engineered the same 8-bp inversion in the mouse Hoxc8 early enhancer. The mouse enhancer carrying the inversion failed to direct expression to the mesoderm (Fig. 2d). The rostral boundary of expression in the neural tube was posteriorized to the 16th somite. This result is similar to mutating mouse elements D or E (17). Thus, while having element E in the opposite strand, the Fugu enhancer is able to direct reporter gene expression to the mesoderm. In contrast, the introduction of the same inversion into the mouse enhancer leads to a loss of mesoderm expression. This inversion may have resulted in disruption of adjacent sequences in the mouse enhancer that are critical for mesoderm expression. Thus, the diverged enhancer sequence may contain additional cis-acting elements that contribute to the mouse Hoxc8 early enhancer activity.
These studies enable us to trace the evolutionary history of the Hoxc8 early enhancer. A survey of Hoxc8 enhancers from other fish lineages and other vertebrates will provide valuable insight on the evolutionary restructuring of the enhancer. One could argue that the existence of duplicated Hox clusters in fish have made divergence of regulatory elements possible because of relaxation of stabilizing selection due to redundancy. Although there is no direct evidence for duplication of Hoxc8 or of its regulatory elements in fish, it is tempting to postulate that because of an increase in the number of Hox genes potentially involved in axial specification, the Hoxc8 early enhancer is more open to modification and selection. Duplication, deletion, inversion, and other modifications have been postulated to contribute to cis-regulatory evolution (4). Here we provide evidence for an inversion of a cis-acting element that alters enhancer activity. The remarkable plasticity shown by the Hoxc8 early enhancer also suggests that the latter may be a critical regulatory region where significant changes affecting axial patterning can be introduced. Other paralogous Hox enhancers involved in thoracic specification may also integrate parallel changes similar to the ones reported here in the Hoxc8 early enhancer. Thus, modifications occurring in a number of Hox enhancers together contribute to changes in axial specification across species. Our studies support the notion that many subtle changes occurring at cis-regulatory elements may act as general developmental and evolutionary mechanisms that determine diversity.
Acknowledgments
We thank Henry Belting, Kevin Bentley, Ann Burke, Sangwei Lu, Cindy McKinney, Tsutomu Miyake, Laura Polanec, David Stock, Dan Seufert, Günter Wagner, and Ken Weiss for their comments on the manuscript. C.S.S. wishes to acknowledge the continued interest, support, and encouragement provided by Frank Ruddle. This work was supported by faculty start-up funds to C.S.S.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: PAC, P1 artificial chromosome.
References
- 1.Krumlauf, R. (1994) Cell 78, 191–201. [DOI] [PubMed] [Google Scholar]
- 2.Holland, P. W. (1999) Nature 402, C41–C44. [DOI] [PubMed] [Google Scholar]
- 3.Ruddle, F. H., Amemiya, C. T., Carr, J. L., Kim, C. B., Ledje, C., Shashikant, C. S. & Wagner, G. P. (1999) Ann. N.Y. Acad. Sci. 870, 238–248. [DOI] [PubMed] [Google Scholar]
- 4.Carroll, S. B., Grenier, J. K. & Weatherbee, S. D. (2001) From DNA to Diversity: Molecular Genetics and the Evolution of Animal Design (Blackwell, Oxford), p. 214.
- 5.Gaunt, S. J. (1994) Int. J. Dev. Biol. 38, 549–552. [PubMed] [Google Scholar]
- 6.Burke, A. C., Nelson, C. E., Morgan, B. A. & Tabin, C. (1995) Development (Cambridge, U.K.) 121, 333–346. [DOI] [PubMed] [Google Scholar]
- 7.Belting, H. G., Shashikant, C. S. & Ruddle, F. H. (1998) Proc. Natl. Acad. Sci. USA 95, 2355–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohn, M. J. & Tickle, C. (1999) Nature 399, 474–479. [DOI] [PubMed] [Google Scholar]
- 9.Burke, A. C. (2000) Curr. Top. Dev. Biol. 47, 155–181. [PubMed] [Google Scholar]
- 10.Morin-Kensicki, E. M., Melancon, E. & Eisen, J. S. (2002) Development (Cambridge, U.K.) 129, 3851–3860. [DOI] [PubMed] [Google Scholar]
- 11.Sanger, T. J. & McCune, A. R. (2002) Zool. J. Linn. Soc. London 135, 529–546. [Google Scholar]
- 12.Tyler, J. C. (1980) Osteology, Phylogeny, and Higher Classification of the Fishes of the Order Plectognathi (Tetraodontiformes) (National Oceanic and Atmospheric Administration, Seattle), NOAA technical report NMFS circular, Vol. 434.
- 13.Prince, V. E., Joly, L., Ekker, M. & Ho, R. K. (1998) Development (Cambridge, U.K.) 125, 407–420. [DOI] [PubMed] [Google Scholar]
- 14.Prince, V. E., Price, A. L. & Ho, R. K. (1998) Dev. Genes Evol. 208, 517–522. [DOI] [PubMed] [Google Scholar]
- 15.Shashikant, C. S., Bieberich, C. J., Belting, H. G., Borbely, M. A., Wang, J. C. H. & Ruddle, F. H. (1995) Development (Cambridge, U.K.) 121, 4339–4347. [DOI] [PubMed] [Google Scholar]
- 16.Bradshaw, M. S., Shashikant, C. S., Bollekens, J. A., Belting, H. G. & Ruddle, F. H. (1996) Proc. Natl. Acad. Sci. USA 93, 2426–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shashikant, C. S. & Ruddle, F. H. (1996) Proc. Natl. Acad. Sci. USA 93, 12364–12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanson, R. D. (1999) Proc. Natl. Acad. Sci. USA 96, 14372–14377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milne, T. A. (2002) Mol. Cell 10, 1107–1117. [DOI] [PubMed] [Google Scholar]
- 20.Shashikant, C. S., Kim, C. B., Borbely, M. A., Wang, W. C & Ruddle, F. H. (1998) Proc. Natl. Acad. Sci. USA 95, 15446–15451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prince, V. (2002) Dev. Biol. 249, 1–15. [DOI] [PubMed] [Google Scholar]
- 22.Prince, V. E. & Pickett, F. B. (2002) Nat. Rev. Genet. 3, 827–837. [DOI] [PubMed] [Google Scholar]
- 23.Aparicio, S., Hawker, K., Cottage, A., Mikawa, Y., Zuo, L., Venkatesh, B., Chen., E, Krumlauf, R. & Brenner, S. (1997) Nat. Genet. 16, 79–83. [DOI] [PubMed] [Google Scholar]
- 24.Amores, A. Force, A., Yan, Y. L., Joly, L., Amemiya, C., Fritz, A., Ho, R. K., Langeland J., Prince, V., Wang, Y. L., et al. (1998) Science 282, 1711–1714. [DOI] [PubMed] [Google Scholar]
- 25.Gerard, M., Zakany, J. & Duboule, D. (1997) Dev. Biol. 190, 32–40. [DOI] [PubMed] [Google Scholar]