Abstract
Fungal type I polyketides (PKs) are synthesized by PK synthases (PKSs) and include well known secondary metabolites such as the anticholesterol drug lovastatin and the potent natural carcinogen aflatoxin. Other type I PKs are known to be virulence factors for some plant pathogens and pigments such as melanin. In this study, a phylogenomic approach was used to investigate the origin and diversity of fungal genes encoding putative PKSs that are predicted to synthesize type I PKs. The resulting genealogy, constructed by using the highly conserved PKS ketosynthase (KS) domain, indicated that: (i) Species within subphylum Pezizomycotina (phylum Ascomycota) but not early diverging ascomycetes, like Saccharomyces cerevisiae (Saccharomycotina) or Schizosaccharomyces pombe (Taphrinomycotina), had large numbers (7–25) of PKS genes. (ii) Bacteria and fungi had separate groups of PKS genes; the few exceptions are the likely result of horizontal gene transfer from bacteria to various sublineages of fungi. (iii) The bulk of genes encoding fungal PKSs fell into eight groups. Four groups were predicted to synthesize variously reduced PKs, and four groups were predicted to make unreduced PKs. (iv) Species within different classes of Pezizomycotina shared the same groups of PKS genes. (v) Different fungal genomes shared few putative orthologous PKS genes, even between closely related genomes in the same class or genus. (vi) The discontinuous distributions of orthologous PKSs among fungal species can be explained by gene duplication, divergence, and gene loss; horizontal gene transfer among fungi does not need to be invoked.
Type I fungal polyketide (PK) synthases (PKSs) are closely related to fatty acid synthetases (FASs) (1). Both are multifunctional enzymes with the same ancestral enzymatic domain structure, namely ketoacyl synthase (KS), acyl transferase (AT), ketoreductase (KR), dehydratase (DH), enoyl reductase (ER), and acyl carrier protein [also known as a phosphopantetheine attachment site (PP) domains]. The KS, AT, and PP domains are essential for both FASs and PKSs, whereas the KR, DH, and ER domains are present in all FASs, but some or all are absent in various PKSs. KR, DH, and ER domains catalyze, in a stepwise fashion, reduction of a keto to a hydroxyl group, dehydration of the hydroxyl to an enoyl group, and reduction of the enoyl to an alkyl group, respectively. PKSs that lack some or all of these domains produce partially reduced or fully oxidized PKs.
Fungal type I PKSs, like FASs, are monomodular enzymes; most are iterative and use their active sites repeatedly to synthesize a PK, adding a two-carbon molecule (i.e., a CoA ester) to the growing chain with each condensation. The noniterative fungal PKSs perform only one condensation cycle and make a diketide. Each one that has been characterized so far is encoded by a gene that resides in a gene cluster (genes adjacent along one stretch of a chromosome), along with a PKS gene encoding an iterative PKS (2, 3). The products of an iterative and a noniterative PKS are joined to make a branched PK. The diversity of PKs is generated, in part, through the use of the three optional PKS reducing domains.
Access to genomes of saprobic and pathogenic members of the Ascomycota allowed us to address the following questions regarding distribution and evolution of Type I PKS genes: (i) How many PKS genes are there in these genomes? (ii) Do type I PKS genes from fungi and bacteria fall into the same or different groups? (iii) How many subgroups of type I PKS genes are found in the Ascomycota? (iv) Do species in the different classes of Ascomycota share the same or have different subgroups of type I PKS genes? (v) What is the frequency of putative orthologs among distantly related fungi, including those that evolved independently as plant pathogens and among closely related species? (vi) Which evolutionary process can account for the phylogenetic distribution of PKS genes: gene duplication, divergence, gene loss, horizontal gene transfer (HGT), or all of these?
To address these questions, we performed phylogenetic analyses on the amino acid sequences of KS domains encoded by all previously characterized fungal PKS genes and by all putative PKS genes extracted from genomic sequences of five taxonomically diverse fungal species in the Ascomycota, subphylum Pezizomycotina (4) [the saprobe Neurospora crassa (5); the maize pathogens Cochliobolus heterostrophus (Bipolaris maydis) (6, 7) and Gibberella moniliformis (Fusarium verticillioides) (8); the cereal pathogen Gibberella zeae (Fusarium graminearum) (8); and the cosmopolitan dicot pathogen Botryotinia fuckeliana (Botrytis cinerea) (8)]. We also searched for PKS genes in the genomes of three earlier diverging ascomycetes [Saccharomyces cerevisiae (9), the plant pest Eremothecium (Ashbya) gossypii (10), (both in the Saccharomycotina), and the yeast saprobe Schizosaccharomyces pombe (11) (Taphrinomycotina)].
Materials and Methods
Detailed treatments of all section below are provided as Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
DNA and Protein Genomic Databases. Four genomes were sequenced by Celera Genomics for the Torrey Mesa Research Institute (TMRI)/Syngenta: B. fuckeliana strain B05.10 (to ≈5× coverage), C. heterostrophus strain C4 (ATCC 48331) (to ≈5× coverage), G. moniliformis strain ATCC 38932 (FGSC 7600) (to ≈4× coverage), and G. zeae lineage 7 strain GZ3639 (12) (to ≈2× coverage). The shotgun sequence assemblies of these genomes were used for these studies, along with the genome sequences of N. crassa (5), S. cerevisiae (9), E. gossypii (10) (sequenced by Syngenta), and S. pombe (11). The protein databases for C. heterostrophus, B. fuckeliana, and G. moniliformis were compiled by using four gene models that were found to generate the largest numbers of predicted proteins: genemark.hmm (13) (Arabidopsis and Caenorhabditis models), fgenesh (14) (dicot model), and genscan (15) (Arabidopsis model). N. crassa, S. cerevisiae, and S. pombe protein databases are available at the National Center for Biotechnology Information.
PKS Gene Identification. To retrieve PKS genes or predicted proteins from each genome, we used the consensus sequence of the KS domain, the most highly conserved domain in type I PKSs and FASs (1), as a query in tblastn searches of the TMRI genome assemblies; in psi-blast searches of the TMRI protein databases of C. heterostrophus, B. fuckeliana, and G. moniliformis; and in psi-blast searches of the eukaryote protein database (www.ncbi.nlm.nih.gov/BLAST) for N. crassa, S. cerevisiae, and S. pombe. The psi-blast searches were also used to retrieve all previously published fungal PKS protein sequences from National Center for Biotechnology Information.
PKS Gene Annotation. The protein sequences of the KS domains obtained from tblastn and psi-blast searches were used as queries to retrieve the complete PKS protein sequences from the protein databases. The C. heterostrophus, B. fuckeliana, and G. moniliformis PKS-predicted sequences from each of the four protein databases were retrieved and aligned by clustalw by using the amino acid substitution matrix blosum62, implemented in megalign (DNASTAR, Madison, WI), and the protein predictions and corresponding DNA sequences were evaluated for their predicted introns. The predicted PKS proteins from N. crassa were also aligned to verify intron predictions. The predicted proteins were submitted to Hidden Markov Model searches, implemented in timelogic (TimeLogic, Carlsbad, CA), to determine their multidomain PKS protein structures.
KS-Domain Genealogy Construction and Evaluation. The predicted KS domains of all newly sequenced fungal genomes plus those from all previously published PKS protein sequences from ascomycete fungi, the homologous FASs from animals (1), and representative type I PKSs from bacteria (Table 1, which is published as supporting information on the PNAS web site) were aligned with clustalw (Table 2, which is published as supporting information on the PNAS web site) and phylogenetically analyzed with neighbor joining and maximum parsimony in paup4.0b8 (Sinauer, Sunderland, MA), by using the settings given in Supporting Materials and Methods. The resulting KS-domain genealogy was evaluated to rank major clades and subclades of PKS enzymes. The criteria used to categorize proteins in major clades and subclades were moderate bootstrap support, similar domain structure, and similar chemical characteristics of the resulting PK products for those PKSs that have been previously characterized. KS-domain genealogy was also used to predict putative phylogenetic orthologs that are expected to encode enzymes of identical or nearly identical biochemical function (16, 17). The criteria used to predict proteins as orthologs were bootstrap values >80%, identical predicted domain structure (or nearly so), and chemical characteristics similar to those of the resulting PK product for those proteins whose product has been characterized previously. Putative orthologs were further supported by a KS-domain genealogy that was consistent with the currently understood organismal phylogeny (4), applicable when three or more orthologs are present. Alternative phylogenetic hypotheses were tested for significance with a Kishino–Hasegawa test (18). The hypothesis underlying the constraint tree is rejected if the difference in logarithm likelihood compared to the unconstrained tree divided by the SD (Ln L/SD) is >1.96 (P ≤ 0.05) (19).
Results and Discussion
Species Within the Pezizomycotina Have Many PKS Genes. The four newly sequenced genomes have large numbers of putative PKS genes: 15 in G. moniliformis, 16 in G. zeae, 20 in B. fuckeliana, and 25 in C. heterostrophus (Table 3, which is published as supporting information on the PNAS web site). The PKS genes in G. zeae were represented as fragments, due to the small contig size and many gaps in the 2× coverage; however, a search of the publicly available G. zeae genome {albeit a different strain [PH-1 (NRRL 31084, but the same lineage, lineage 7)]} did not reveal additional PKS genes beyond those we identified in our 2× coverage. These numbers are greater than those of the saprobe N. crassa, which is thought to produce only one PK, namely PK-derived melanin, yet contains seven putative PKS genes, only one of which is a melanin-type PKS gene (5). By contrast, the three earlier-diverging ascomycetes (S. pombe, S. cerevisiae, and E. gossypii) lacked identifiable PKS genes, including those involved in pigment formation.
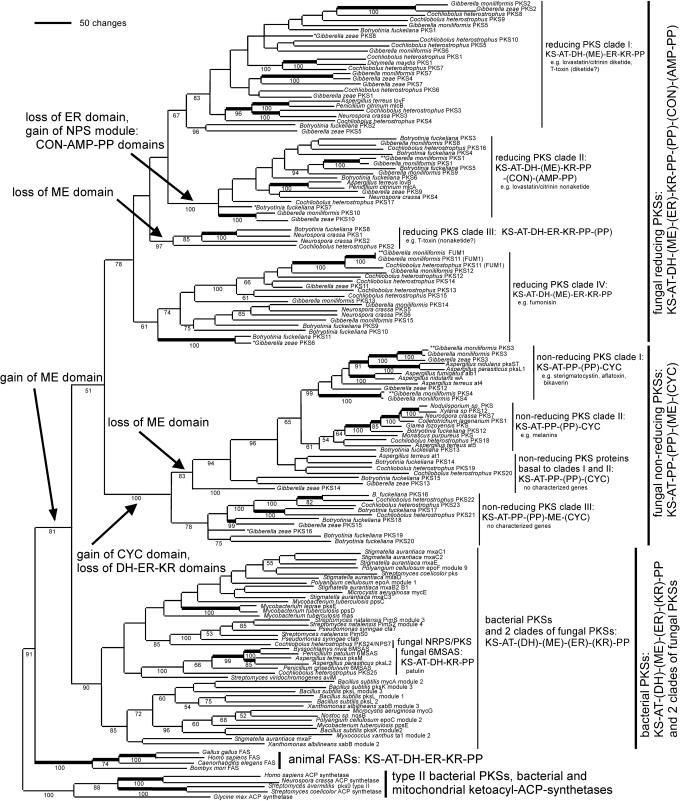
KS-Domain Genealogy and Prediction of PK Structure. KS-domain genealogy (Fig. 1) was used to infer the genealogy of type I PKSs. The predicted fungal and bacterial PKSs grouped in three main clades (20–22). Together, these three major clades grouped into a larger clade of type I PKSs, which was sister to the clade of FASs found in animals. All of the type I PKSs and FASs grouped into a larger clade, which was sister to the type II PKSs found in bacteria and to the bacterial and mitochondrial acyl-ACP synthetases (1). KS-domain genealogy suggested that the ancestral domain structure of type I PKSs was KS-AT-DH-ME-ER-KR-PP, because these domains were present, and in this order, in the PKS protein sequences in members of two of the three main clades of PKSs, one of which (the bacterial PKS clade) was in a basal position.
Fig. 1.
Genealogy of type I PKSs, inferred by maximum parsimony analysis of the KS domain. Major clades and subclades are indicated by vertical bars, each of which shares a common organization of domains (those in parentheses are variable in their presence/absence within that clade). Branch length indicates number of inferred amino acid changes. Numbers below branches indicate percentage bootstrap support for each clade. All branches present in a strict consensus of the maximum parsimony trees received bootstrap support. Bold branches indicate putative orthologs, which were inferred as described in Materials and Methods. Monophyletic gains and losses of domains are noted by arrows. Three G. moniliformis PKSs were previously represented by GenBank submissions; these are noted with double asterisks. The accession numbers for all sequences obtained from GenBank are given in Table 1. Three G. zeae and one B. fuckeliana PKSs represented by partial C-terminal fragments of the KS domain were mapped onto the tree that was based on an alignment that included only N-terminalfragments; these are marked with a single asterisk. Alignment was based on 4,862 amino acids from the KS domain; 462 characters were informative, 20 were uninformative, and 4 were constant. Parsimony was performed with 100 random additions. Shown is one of 18 most-parsimonious trees of 17,859 steps, consistency index (CI) = 0.2763, rescaled CI = 0.1491. The maximum parsimony trees generated by coding gaps as either 21st amino acid or as missing were not significantly different from each other or from the tree generated by neighbor joining.
Two of these three main clades of microbial type I PKSs were identified as exclusively fungal and correlated with the two largest structural classes of fungal PKs: reduced and unreduced (1). Each main fungal PKS clade was further divided into four groups, each with a typical domain structure, depending on the loss of ancestral domains, or the gain of novel domains (mapped onto Fig. 1 and listed in Table 3). The third major clade included all bacterial PKSs, and within this clade were nested two small, additional fungal clades, the first of which comprised the 6-methylsalicylic acid synthases, and the second of which was a nonribosomal peptide synthetase (NPS)/PKS hybrid gene found in C. heterostrophus.
Fungal PKSs Producing Reduced PKs. The first main fungal clade included the PKSs that synthesize variously reduced, and usually linear, PKs (Fig. 1). The characterized PKs serve as precursors to toxins that are active in animals [e.g., lovastatin (2), citrinin (3), and fumonisin (23)] and to toxins that are active in plants [e.g., T-toxin (6), and PM-toxin (24)]. These reduced PKs frequently are synthesized from CoA thioesterified carboxylic acids other than acetyl and malonyl CoA, and the extent of keto-group processing varies from one condensation cycle to the next (1). In addition, one or more sites may be methylated by a ME domain (3, 25). However, many of the predicted PKSs in the PKS clade that produces reduced PKs have highly divergent and presumably nonfunctional ME domains.
The clade of PKSs that synthesize reduced PKs was subdivided into four subclades (I—IV), each of which had a typical domain structure (Fig. 1). Reducing PKS subclade I included Aspergillus terreus LovF and its ortholog Penicillium citrinum MlcB, which synthesize the diketide portion of lovastatin (2) and citrinin (3), respectively, and C. heterostrophus PKS1 and its ortholog Didymella (Mycosphaerella) maydis PKS1, which are necessary for the synthesis of structurally similar linear PKs, T-toxin (6), and PM-toxin (26), respectively.
Reducing PKS subclade II was characterized by enzymes that have lost the ER domain (Fig. 1); PKs synthesized by PKSs in this clade are predicted to either lack reduced alkyl groups or to contain alkyl groups whose reduction is completed by the product of an external ER domain-containing gene [e.g., A. terreus lovC (25), P. citrinum mlcG (3)]. The PKSs of this clade were also found to have either a condensation (CON) domain typical of NPSs (25) or an entire NPS module consisting of a CON domain, an adenylation (AMP) domain, and a PP domain (Table 3 and Fig. 1). Genealogies of the AMP and CON domains, along with those from characterized fungal and bacterial NPS proteins, grouped the AMP domains with those that adenylate amino acids that are subsequently N-methylated and grouped the CON domains with those that condense N-methylated amino acids (analysis not shown). These data suggest that the PKSs in this subclade gained an N-methylated amino acid-type NPS module (described in refs. 27–30). However, none of these PKSs had an N-methylation domain that is found between the AMP and CON domains of N-methylated amino acid type NPS modules. Many of these PKSs also lacked AMP and PP domains; these PKSs were scattered throughout this subclade, suggesting that the AMP and PP domains have been lost repeatedly, including those from the only two characterized proteins, A. terreus LovB and its ortholog P. citrinum MlcA, which synthesize the cyclic nonaketide portion of lovastatin and citrinin, respectively (3, 25). Each cyclic PK is derived from the action of the CON domain on a methylated keto group. This mode of action suggests that the loss of the AMP and PP domains was involved in a change of function of the CON domain from acting on methylated amino acids to acting on methylated keto groups. Uncharacterized PKSs with the same domain structure are predicted to make cyclic PKs. PKSs that have lost the entire C-terminal NPS module, consisting of the CON, AMP, and PP domains, are also scattered throughout this clade. The inferred polyphyletic losses of the CON, AMP, and PP domains were tested by constraint topologies forcing subsets of sequences into clades, which were subjected to parsimony analyses, and the likelihoods of the resulting genealogies were compared to the likelihood of the maximum parsimony tree found without these topological constraints. The Kishino–Hasegawa tests (18) rejected the alternative hypothesis of a monophyletic origin of the four PKSs that lack the entire NPS-like module via a sequential loss of first the AMP-PP domains and then the CON domain [P < 0.0001) (19)] or single loss of all three domains (P = 0.0302).
PKSs in reducing subclade III all lacked a ME domain, and two of the four PKSs had an additional PP domain. The only characterized protein is C. heterostrophus PKS2, which, along with PKS1, is required for synthesis of T-toxin (S. E. Baker, personal communication).
Reducing PKS subclade IV, like subclades I and II, included PKS enzymes that may or may not have a conserved ME domain. The only characterized PKS in this subclade is G. moniliformis FUM1, which makes the linear PK precursor of the toxin fumonisin (31, 32).
The PKSs that lack conserved and putatively functional ME domains are scattered throughout reducing PKS clades I, III, and IV and comprise reducing PKS clade II. The alternative hypothesis of a monophyletic loss of ME domains in the reducing PKS clade was rejected by the Kishino–Hasegawa test (P < 0.0001).
Fungal PKSs Producing Unreduced PKs. The second main fungal clade included fungal PKSs that synthesize unreduced, and usually cyclic, i.e., aromatic, PKs that are precursors to toxins [e.g., sterigmatocystin (33) and aflatoxin (34)] and pigments [e.g., melanin (35), bikaverin (36), and green spore pigment (37)]. All PKSs within this clade lacked ER, DH, and KR domains, which is interpreted as a single loss of these reducing domains, compared to the inferred ancestral domain structure of type I PKSs (Fig. 1, arrows). The nonreducing fungal PKSs are predicted to synthesize PKs in which the keto groups are either not reduced or may be reduced by enzymes other than a PKS. Unreduced PKs are typically synthesized from acetyl- and malonyl-CoA. PKS proteins with an additional PP domain were scattered throughout the nonreducing PKS clade, as was the case for reducing PKS subclade III. The functional significance of these duplicated PP domains is not known.
A Claisen-type cyclase (38) domain was present at the C terminus of all PKS proteins in sister subclades I (toxins and nonmelanin pigments) and II (melanin). The PKSs that make melanin precursors are encoded by the most widely distributed fungal PKS genes and have been characterized in Colletotrichum lagenarium (PKS1) (35), Glarea sp., Nodulisporium sp. (39), and C. heterostrophus (PKS18) (B. Robbertse and S. Baker, personal communication). N. crassa PKS7 maps to the same region as the per locus; per mutants have nonmelanized perithecia and ascospores (40). The melanin-type PKS gene is not ubiquitous, because the two Gibberella genomes lacked a gene within unreduced PKS clade I, which correlates with a lack of melanized structures in their life cycles.
All PKSs basal to nonreducing PKS subclades I and II are uncharacterized. These basal PKSs occur both with and without the Claisen-type cyclase (CYC domain); the PKSs without CYC are predicted to make unreduced noncyclic PKs. We propose calling this clade the nonreducing PKSs, because this character is invariable, whereas not all PKSs in this clade are likely to make aromatic (cyclic) PKs. Several of these basal PKSs formed a subclade (nonreducing PKS subclade III) characterized by a ME domain located after the PP domain [KS-AT-PP-(PP)-ME-(CYC)], the apparent result of a domain rearrangement. The other PKSs basal to nonreducing subclades I and II do not have ME domains; we hypothesize that the genes encoding these PKSs diverged after the loss of the ME domain that was retained in nonreducing PKS subclade III (Fig. 1, arrow).
Other Fungal and Bacterial PKSs. The third main fungal PKS clade comprised the 6-methylsalicylic acid-type (i.e., simple aromatic) of PKSs that make 6-methylsalicylic acid, a precursor to toxins [e.g., patulin (41)]. These toxins have been characterized from Aspergillus spp. (27, 42), Byssochlamys nivea, and Penicillium spp. (27). The C. heterostrophus genome had a PKS in this clade, but the other Pezizomycotina genomes did not. This fungal clade was nested within the large clade that comprised all bacterial type I PKSs. A second fungal clade was also nested within this bacterial clade and was represented by a single member, C. heterostrophus PKS24. PKS24 is a hybrid NPS/PKS gene, predicted to encode an enzyme that synthesizes a partly reduced PK decorated with a single amino acid.
Origin of PKS Genes in the Ascomycota. Fungal genome sequences available were limited to the three subphyla of Ascomycota, i.e., Taphrinomycotina, Saccharomycotina, and Pezizomycotina, as well as to one of the three subphyla of Basidiomycota, Hymenomycetes (Phanerochaete chrysosporium, www.jgi.doe.gov/programs/whiterot.htm). PKS genes were found only in the genomes of Pezizomycotina. It is possible that these genes have been lost in the non-Pezizomycotina taxa or that PKS genes came to the Pezizomycotina via HGT, perhaps from the bacterial PKS genes that form the sister clade to the main fungal clade. However, until fungal genomes from other Basidiomycota and from Zygomycota and Chytridiomycota are sequenced, this question will remain unresolved.
Diversification of PKS Genes Among Species in the Pezizomycotina. The diversity of PKS genes and PKs found in Pezizomycotina led to speculation, including our own (6, 43), that HGT is involved in generating and maintaining this diversity (ref. 44 and references therein). However, other discontinuous genome events such as gene duplication and differential gene loss are alternative explanations to HGT for the distributions of PKS genes (45).
We have data for genomes of species within three classes (Sordariomycetes, Dothideomycetes, and Leotiomycetes) of the Pezizomycotina (Table 3). For the fungal-reducing PKSs, all three classes are represented in each of the four subclades. For the fungal-nonreducing PKSs, all three classes are represented, but not in each of the four subclades. We also have data from a fourth class, Eurotiomycetes, in the form of many PKS proteins in GenBank from Penicillium and Aspergillus species (Table 1); these PKSs were found in two of the four fungal-reducing PKS subclades and three of the four fungal-nonreducing PKS subclades. This phylogenomic distribution (Fig. 1) is consistent with the hypothesis that all eight types of PKS gene were present in the common ancestor of the four classes of the Pezizomycotina, before the radiation of Pezizomycotina 300–700 million years ago (46).
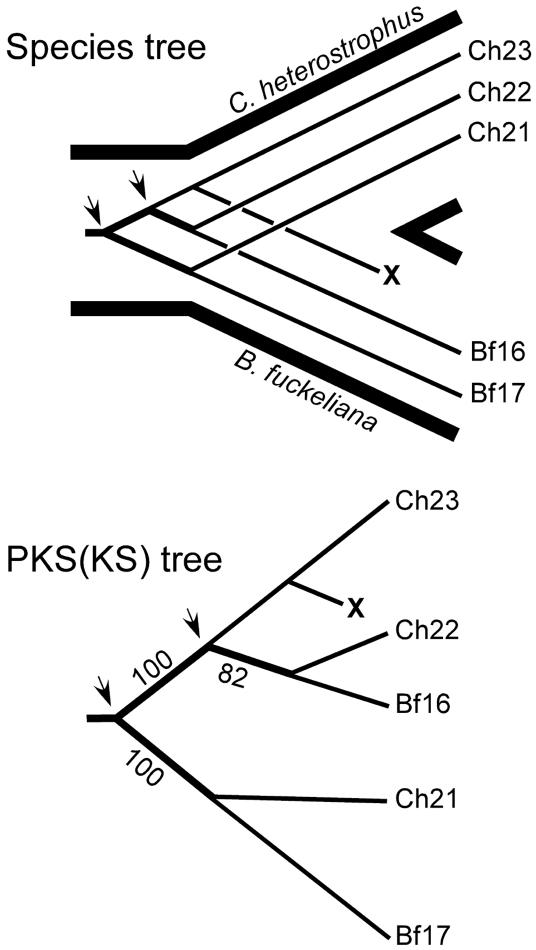
Evidence for gene duplication and gene loss was observed; for example, in nonreducing PKS subclade III, a clade with many strongly supported internal branches (Fig. 2). Using only the genomes of C. heterostrophus and B. fuckeliana, two gene duplications in an ancestor of these two fungi would account for the present distribution of three pairs of paralogs, ChPKS21/BfPKS17, ChPKS22/BfPKS16, and ChPKS23, provided that the counterpart of C. heterostrophus PKS23 was lost in the lineage leading to B. fuckeliana. HGT does not need to be invoked.
Fig. 2.
Species tree (Upper) depicts the inferred gene duplication and gene losses necessary to account for the distribution of these PKS proteins present in fungal nonreducing PKS subclade III (Lower). Arrows indicate the two gene duplication events, and X indicates the extinction of the ortholog of C. heterostrophus PKS23 in the lineage leading to B. fuckeliana.
It would seem possible to distinguish between gene duplication and loss and HGT by comparing the time of divergence of PKS genes to that of genes taken to represent the organismal divergence. Similar divergence times for the two types of gene would favor gene duplication. More recent divergence for the PKS genes would favor HGT. Although rates of nucleotide substitution have been estimated for fungal tubulin genes (47), substitution rates for PKS genes are not known and will require data sets with large numbers of orthologous PKS genes.
The N. crassa genome is remarkable for its lack of duplicated genes (5), due presumably to repeat-induced point mutation that alters nucleotides in both copies of duplicated genes and renders them nonfunctional. If HGT were the main method of generating PKS gene diversity, N. crassa might be expected to have as many PKS genes as the other fungi. However, N. crassa has by far the fewest PKS genes, suggesting that gene duplication may be more important than HGT for generating PKS gene diversity.
Origins of Fungal PKS Genes in the Bacterial PKS Gene Clade. We found two PKS genes in C. heterostrophus, each sister to different PKS genes from bacteria. Several branches uniting the fungal and bacterial PKS genes in the bacterial PKS clade were so well supported that it seems certain that these fungal sequences belong in the bacterial clade. Kishino–Hasegawa tests rejected the alternative hypothesis that these sequences are fungal in origin [C. heterostrophus PKS24 (P = 0.0001) and C. heterostrophus PKS25 and the other members of the MSAS clade (P < 0.0001)]. These fungi may have acquired these PKS genes by one or more HGT events from bacteria.
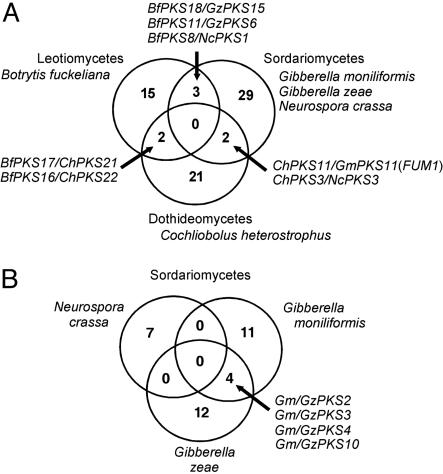
Orthologs. Gene duplication and loss have left few putative orthologous matches among fungal PKS genes, and in no case were orthologous genes found for members of all of the three classes (Sordariomycetes, Dothideomycetes, and Leotiomycetes) for which we have genomic sequence (Fig. 3A). Few orthologs were observed among members of the Sordiariomycetes, even within a genus (Fig. 3B); the N. crassa PKS genes have no orthologs in G. zeae or G. moniliformis, and only four PKS genes of 25 are orthologous between G. zeae and G. moniliformis.
Fig. 3.
Venn diagrams showing predicted PKS gene orthologs shared between and among taxa, inferred as described in Materials and Methods. (A) Among the classes Leotiomycetes (represented by B. fuckeliana), Dothideomycetes (C. heterostrophus), and Sordariomycetes (N. crassa, G. moniliformis, and G. zeae). (B) Among the Sordariomycetes: N. crassa, G. moniliformis, and G. zeae.
Orthologous PKS genes found among closely or distantly related species reflect examples of nearly identical PKs being produced by different fungi. For example, citrinin and lovastatin are nearly identical compounds, made by closely related P. citrinum (3, 24) and A. terreus (25), respectively. Both citrinin and lovastatin require two PKSs for their synthesis; A. terreus LovF and P. citrinum MlcB genes are orthologs, and LovB and MlcA genes are orthologs. We note that both pairs of proteins receive strong bootstrap support (Fig. 1) and have identical domain structures, which is why we used these criteria to predict orthologs among the uncharacterized proteins. Another example of orthologs that fit these criteria, C. heterostrophus PKS1 (6) and D. maydis PKS1 (26), synthesize nearly identical compounds (T- and PM-toxins, respectively), and the biological activities of these toxins on the fungal host, maize, are identical (24). In a third example, Dothistroma pini makes dothistromin with products of genes that are orthologous to those of the aflatoxin biosynthetic pathway found in Aspergillus spp. (48).
PKS Diversity in Ascomycota. Given that a fungal species has as many as 25 PKS genes, our small sampling of fungal genomes predicts that an astonishingly large number of PKs are produced by species of Pezizomycotina. The unknown PKs may be toxins and pigments, or they may be involved in developmental processes that require small molecule signaling, such as growth and hyphal fusion, aerial hyphae formation, conidiation, and sexual reproduction. For example, in Dictyostelium discoideum, a type II PKS produces the 12-carbon PK precursor for a diffusible signaling molecule (DIF-1, a chlorinated alkyl phenone), which induces the differentiation of prestalk-O cells (49).
This inventory and categorization of all type I PKS genes in the genomes of five species within the Pezizomycotina is a starting point in our understanding of the diversity of PKs generated by these multidomain enzymes. A better understanding of the function of these compounds requires the characterization of PKs, including their modification and decoration by enzymes that are part of the biochemical pathway [some or all of which may be found in gene clusters (50, 51)], and the analysis of their roles in the biology of these fungi (52).
Supplementary Material
Acknowledgments
We thank Steven Briggs, President and Chief Executive Officer of TMRI, for his helmsmanship. Intermediate genome assembly of raw shotgun sequences was performed with in-house software (Don Hutchinson, TMRI). The gene predictions were created by Darrell Ricke (TMRI). Preliminary analyses of PKSs in C. heterostrophus were conducted by Barbara Robbertse and Scott E. Baker (TMRI). S.K. was supported by a collaborative agreement between University of California, Berkeley, and TMRI/Syngenta. The C. heterostrophus, B. fuckeliana, G. moniliformis, and G. zeae genome sequences were provided by Celera Genomics (Rockville, MD) for TMRI/Syngenta. The genome of E. gossypii was sequenced by SBI Technology/Syngenta (Research Triangle Park, NC). The genome of P. chrysosporium was made available by the U.S. Department of Energy (http://genome.jgi-psf.org/whiterot1/whiterot1.home.html).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PK, polyketide; PKS, PK synthase; KS, ketoacyl synthase; AT, acyl transferase; DH, dehydratase; NPS, nonribosomal peptide synthetase; ME, methyl transferase; ER, enoyl reductase; KR, ketoreductase; PP, phosphopantetheine attachment site (acyl carrier protein of PKSs and thiolation of NPSs); AMP, adenylation; CON, condensation; HGT, horizontal gene transfer; TMRI, Torrey Mesa Research Institute; FAS, fatty acid synthase.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AY495643–AY495666 (C. heterostrophus PKS2–25), AY495606–AY495625 (B. fuckeliana PKS1–20), AY495591–AY495605 (G. moniliformis PKS1–15), AY495626–AY495641 (G. zeae PKS1–16), and AY495642 (PKS1 from Didymella maydis)].
References
- 1.Hopwood, D. A. (1997) Chem. Rev. 97, 2465–2497. [DOI] [PubMed] [Google Scholar]
- 2.Hendrickson, L., Davis, C. R., Roach, C., Nguyen, D. K., McAda, P. C. & Reeves, C. D. (1999) Chem. Biol. 6, 429–439. [DOI] [PubMed] [Google Scholar]
- 3.Abe, Y, Suzuki, T., Mizvno, T., Ono, C., Iwamoto, K., Hosobuchi, M. & Yoshikawa, H. (2002) Mol. Genet. Genomics 267, 636–646. [DOI] [PubMed] [Google Scholar]
- 4.Berbee, M. L. (2001) Physiol. Mol. Plant Pathol. 59, 165–187. [Google Scholar]
- 5.Galagan, J. E., Calvo, S. E., Borkovich, K. A., Selker, E. U., Read, N. D., Jaffe, D., FitzHugh, W., Ma, L. J., Smirnov, S., Purcell, S., et al. (2003) Nature 442, 859–868. [DOI] [PubMed] [Google Scholar]
- 6.Yang, G., Rose, M. S., Turgeon, B. G. & Yoder, O. C. (1996) Plant Cell 8, 2139–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoder, O. C. & Turgeon, B. G. (2001) Curr. Opin. Plant Biol. 4, 315–321. [DOI] [PubMed] [Google Scholar]
- 8.Agrios, G. N. (1997) Plant Pathology (Academic, San Diego).
- 9.Goffeau, A., Barrell, B. G., Bussey, H., Davis, R. W., Dujon, B., Feldmann, H., Galibert, F., Hoheisel, J. D., Jacq, C., Johnston, M., et al. (1996) Science 274, 546–567. [DOI] [PubMed] [Google Scholar]
- 10.Brachat, S., Dietrich, F. S., Voegeli, S., Zhang, Z., Stuart, L., Lerch, A., Gates, K., Gaffney, T. & Philippsen, P. (2003) Genome Biol. 4, R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wood, V., Gwilliam, R., Rajandream, M. A., Lyne, M., Lyne, R., Stewart, A., Sgouros, J., Peat, N., Hayles, J., Baker, S., et al. (2002) Nature 415, 871–880. [DOI] [PubMed] [Google Scholar]
- 12.Bowden, R. L. & Leslie, J. F. (1999) Phytopathology 89, 182–188. [DOI] [PubMed] [Google Scholar]
- 13.Lukashin, A. V. & Borodovsky, M. (1998) Nucleic Acids Res. 26, 1107–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salamov, A. A. & Solovyev, V. (2000) Genome Res. 10, 516522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burge, C. & Karlin, S. (1997) J. Mol. Biol. 268, 78–94. [DOI] [PubMed] [Google Scholar]
- 16.Thornton, J. W. & DeSalle, R. (2000) Annu. Rev. Genomics Hum. Genet. 1, 41–73. [DOI] [PubMed] [Google Scholar]
- 17.Eisen, J. A. (1998) Genome Res. 8, 163–167. [DOI] [PubMed] [Google Scholar]
- 18.Kishino, H. & Hasegawa, M. (1989) J. Mol. Evol. 29, 170–179. [DOI] [PubMed] [Google Scholar]
- 19.Felsenstein, F. (1990) phylip (Phylogeny Inference Package) (Univ. of Washington, Seattle), Ver. 3.3.
- 20.Bingle, L. E. H., Simpson, T. J. & Lazarus, C. M. (1999) Fungal Genet. Biol. 26, 209–223. [DOI] [PubMed] [Google Scholar]
- 21.Nicholson, T. P., Rudd, B. A. M., Dawson, M., Lazarus, C. M., Simpson, T. J. & Cox, R. J. (2001) Chem. Biol. 8, 157–178. [DOI] [PubMed] [Google Scholar]
- 22.Miao, V., Coeffet-LeGal, M.-F., Brown, D., Sinnemann, S., Donaldson, G. & Davies, J. (2001) Trends Biotechnol. 19, 349–355. [DOI] [PubMed] [Google Scholar]
- 23.Nelson, P. E., Desjardins, A. E. & Plattner, R. D. (1993) Annu. Rev. Phytopathol. 31, 233–252. [DOI] [PubMed] [Google Scholar]
- 24.Yoder, O. C. (1973) Phytopathology 63, 1361–1366. [Google Scholar]
- 25.Kennedy, J., Auclair, K., Kendrew, S. G., Park, C., Vederas, J. C. & Hutchinson, C. R. (1999) Science 284, 1368–1372. [DOI] [PubMed] [Google Scholar]
- 26.Yun, S.-H., Turgeon, B. G. & Yoder, O. C. (1998) Physiol. Mol. Plant Path. 52, 53–66. [Google Scholar]
- 27.Smith, D. J., Earl, A. J. & Turner, G. (1990) EMBO J. 9, 2743–2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacCabe, A. P., van Liempt, H., Palissa, H., Unkles, S. E., Riach, M. B., Pfeifer, E., von Dohren, H. & Kinghorn, J. R. (1991) J. Biol. Chem. 266, 12646–12654. [PubMed] [Google Scholar]
- 29.Haese, A., Schubert, M., Herrmann, M. & Zocher, R. (1993) Mol. Microbiol. 7, 905–914. [DOI] [PubMed] [Google Scholar]
- 30.Weber, G., Schorgendorfer, K., Schneider-Scherzer, E. & Leitner, E. (1994) Curr. Genet. 26, 120–125. [DOI] [PubMed] [Google Scholar]
- 31.Proctor, R. H., Desjardins, A. E., Plattner, R. D. & Hohn, T. M. (1999) Fungal Genet. Biol. 27, 100–112. [DOI] [PubMed] [Google Scholar]
- 32.Proctor, R. H., Brown, D. W., Plattner, R. D. & Desjardins, A. E. (2003) Fungal Genet. Biol. 38, 237–249. [DOI] [PubMed] [Google Scholar]
- 33.Yu, J. H. & Leonard, T. J. (1995) J. Bacteriol. 177, 4792–4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng, G. H. & Leonard, T. J. (1995) J. Bacteriol. 177, 6246–6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perpetua, N. S., Kubo, Y. Yakano, Y. & Furusawa, I. (1996) Mol. Plant–Microbe Interact. 9, 232–239. [DOI] [PubMed] [Google Scholar]
- 36.Linnemannstons, P., Schulte, J., del Mar Prado, M., Proctor, R. H., Avalos, J. & Tudzynski, B. (2002) Fungal Genet. Biol. 37, 134–148. [DOI] [PubMed] [Google Scholar]
- 37.Mayorga, M. E. & Timberlake, W. E. (1992) Mol. Gen. Genet. 235, 205–212. [DOI] [PubMed] [Google Scholar]
- 38.Watanabe, A., Fujii, I., Tsai, H., Chang, Y. C., Kwon-Chung, K. J. & Ebizuka, Y. (2000) FEMS Microbiol. Lett. 192, 39–44. [DOI] [PubMed] [Google Scholar]
- 39.Fulton, T. R., Ibrahim, N., Losada, M. C., Grzegorski, D. & Tkacz, J. S. (1999) Mol. Gen. Genet. 262, 714–720. [DOI] [PubMed] [Google Scholar]
- 40.Howe, H. B., Jr., & Benson, E. W. (1974) Mol. Gen. Genet. 131, 79–83. [DOI] [PubMed] [Google Scholar]
- 41.Beck, J., Ripka, S., Siegner, A., Schiltz, E. & Schweizer, E. (1990) Eur. J. Biochem. 192, 487–498. [DOI] [PubMed] [Google Scholar]
- 42.Pazoutova, S, Linka, M., Storkova, S. & Schwab, H. (1997) Folia Microbiol. 43, 419–430. [DOI] [PubMed] [Google Scholar]
- 43.Rose, M. S., Yun, S. H., Asvarak, T., Lu, S.-W., Yoder, O. C. & Turgeon, B. G. (2003) Mol. Plant–Microbe Interact. 15, 883–893. [DOI] [PubMed] [Google Scholar]
- 44.Walton, J. D. (2000) Fung. Genet. Biol. 30, 167–171. [DOI] [PubMed] [Google Scholar]
- 45.Kurland, C. G., Canback, G. & Berg, O. G. (2003) Proc. Natl. Acad. Sci. USA 100, 9658–9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heckman, D. S., Geiser, D. M., Eidell, B. R., Stauffer, R. L., Kardos, N. L. & Hedges, S. B. (2001) Science 298, 1129–1133. [DOI] [PubMed] [Google Scholar]
- 47.Kasuga, T., White, T. J. & Taylor, J. W. (2002) Mol. Biol. Evol. 19, 2318–2324. [DOI] [PubMed] [Google Scholar]
- 48.Bradshaw, R. E., Bhatnagar, D., Ganley, R. J., Gillman, C. J., Monahan, B. J. & Seconi, J. M. (2002) Appl. Environ. Microb. 68, 2885–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson, C. R. & Kay, R. R. (2000) Mol. Cell. 6, 1509–1504. [DOI] [PubMed] [Google Scholar]
- 50.Keller, N. P. & Hohn, T. M. (1997) Fungal Genet. Biol. 21, 17–29. [PubMed] [Google Scholar]
- 51.Bentley, R. & Bennett, J. (1999) Annu. Rev. Microbiol. 53, 411–446. [DOI] [PubMed] [Google Scholar]
- 52.Wolpert, T. J., Dunkle, L. D. & Cuiffetti, L. M. (2002) Annu. Rev. Phytopathol. 40, 251–285. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.