Abstract
To elucidate the mechanism of transcription by cellular RNA polymerases (RNAPs), high resolution X-ray crystal structures together with structure-guided biochemical, biophysical and genetics studies are essential. The recently-solved X-ray crystal structures of archaeal RNA polymerase (RNAP) allow a structural comparison of the transcription machinery among all three domains of life. The archaea were once thought of closely related to bacteria, but they are now considered to be more closely related to the eukaryote at the molecular level than bacteria. According to these structures, the archaeal transcription apparatus, which includes RNAP and general transcription factors, is similar to the eukaryotic transcription machinery. Yet, the transcription regulators, activators and repressors, encoded by archaeal genomes are closely related to bacterial factors. Therefore, archaeal transcription appears to possess an intriguing hybrid of eukaryotic-type transcription apparatus and bacterial-like regulatory mechanisms. Elucidating the transcription mechanism in archaea, which possesses a combination of bacterial and eukaryotic transcription mechanisms that are commonly regarded as separate and mutually exclusive, can provide data that will bring basic transcription mechanisms across all three domains of life.
INTRODUCTION
The last decade marked a revolution in understanding of the molecular detail of cellular RNA polymerases (RNAPs) from bacteria, archaea and eukaryotes and their respective transcription mechanisms. The structure of bacterial Thermus aquaticus RNAP core enzyme, the first high-resolution X-ray crystal structure of any cellular RNAP revealed the structure-function relationship of the university conserved core part of cellular RNAP (Zhang et al., 1999, Darst, 2001). The structural studies of T. aquaticus holoenzyme and holoenzyme-fork junction DNA complex have explained how the promoter recognition σ factor associates with the core enzyme and how the bacterial promoter DNA is recognized by holoenzyme to form the “transcription ready” promoter open complex (Murakami and Darst, 2003, Murakami et al., 2002a, Murakami et al., 2002b). The Saccharomyces cerevisiae RNAP II (Pol II) structures that include 10 subunits form (Cramer et al., 2001), 12 subunits form (Armache et al., 2005), complexed with a general transcription factor TFIIB (Kostrewa et al., 2009, Liu et al., 2010) and the transcription elongation form (Gnatt et al., 2001), have shown how the Pol II looks like and how it forms the promoter complex and how it transcribes RNA. These structures boosted our understanding of the structural basis of eukaryotic transcription. The structure of Sulfolobus solfataricus RNAP, the first X-ray crystal structure of archaeal RNAP solved recently, has completed the suite of cellular RNAPs of all three domains of life (Hirata et al., 2008b).
Archaea was discovered as one of three branches of life, and since then, interest has grown for many reasons (Woese and Fox, 1977, Pace, 1997). The archaeal transcription system has been characterized as a hybrid of eukaryotic and bacterial transcription systems (Bell and Jackson, 1998). The archaeal basal transcription apparatus is very similar to that of eukaryote (Zillig et al., 1978, Langer et al., 1995), but its transcriptional regulatory factors are similar to those of bacteria (Brinkman et al., 2003, Ouhammouch, 2004). In this review article, we will compare the architectural features of cellular RNAPs from three domains of life. We will then discuss the mechanism of transcription regulation in archaea, which are informed by high-resolution structure as well as biochemistry and genetics experiments.
Archaeal RNA Polymerase
The development of molecular biology has shown that molecular criteria like DNA sequence comparison of ribosomal RNA genes reveals more precise evolutionary relationships among the organisms than the classical morphological or cytological criteria. The classification of organisms into the “three domains” - bacteria, archaea and eukaryote - is based on the molecular criteria and widely accepted (Woese et al., 1990). Interestingly, archaea are so-called prokaryotic cells in terms of cytological features, but are thought to have common ancestry with eukaryotes considering its molecular features. Particularly, proteins involved in gene maintenance and expression are similar to those of eukaryotes. It has been shown that the basal transcription machinery including RNAP in archaea is closely related to the transcription machinery found in eukaryotes based on their subunit compositions (Rowlands et al., 1994, Langer et al., 1995) (Table 1). In the purified archaeal RNAP, the protein complex consists of 11~13 subunits depending on the species (Zillig et al., 1978, Huet et al., 1983) and it became clear, based on DNA cloning of each subunit coding gene, that the amino acid sequence similarities between archaeal and eukaryotic RNAPs are closer (Huet et al., 1983, Langer et al., 1995). Finally, in 2008, it became possible to compare the X-ray crystal structures of RNAP from the three domains of life and to begin to understand their evolutionary relationships (Zhang et al., 1999, Cramer et al., 2001, Hirata et al., 2008b).
Table 1.
Subunit composition of cellular RNAPs
| Eukaryotic Pol II | Archaeal RNAP | Bacterial RNAP | |
|---|---|---|---|
| Class I subunit | Rpb1 | A′+A″ | β ′ |
| Rpb2 | B | β | |
| Rpb3 | D | α I | |
| Rpb6 | K | ω | |
| Rpb11 | L | α II | |
|
| |||
| Class II subunit | Rpb4 | F | |
| Rpb5 | H | ||
| Rpb7 | E | ||
| Rpb8 | G | ||
| Rpb10 | N | ||
| Rpb12 | P | ||
|
| |||
| Class III subunit | Rpb91 | ||
| Gdown12 | Rpo132 | δ 2 | |
found only in the eukaryotic Pol II
found only in certain species and these subunits are not orthologs
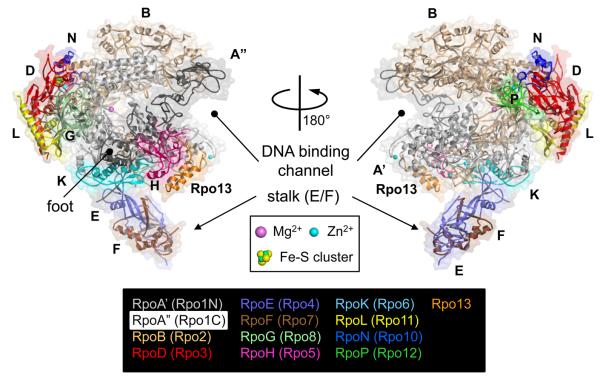
The X-ray crystal structures of archaeal RNAPs have been determined from two Sulfolobus species, Sulfolobus solfataricus (Hirata et al., 2008b, Hirata and Murakami, 2009) and Sulfolobus shibatae (Korkhin et al., 2009) and the cryo-electron microscopy structure has been determined from Pyrococcus furiosus (Kusser et al., 2008). Two X-ray crystal structures are very similar and are composed of 13 subunits with a molecular weight of about 380 kDa. The overall shape of Sulfolobus solfataricus RNAP resembles a ‘crab claw’ that includes 11 subunits with a protruding stalk that consists of E and F subunits (Figure 1). Based on a structural comparison among the archaeal, bacterial and eukaryotic RNAPs, it was determined that the dimensions of the double-stranded DNA binding channel and the architecture of active site of archaeal RNAP are highly conserved. In archaea, the largest subunit is divided into two polypeptides, A′ and A″ subunits, which are encoded by separate genes in an operon (Langer et al., 1995). Sequence alignments reveal that archaeal A′ and A″ correspond to the N-terminal two-thirds and the C-terminal one-third of the Rpb1 subunit of Pol II (Table 1), respectively, and that the junction between A′ and A″ is positioned at the ‘foot’ domain (Cramer et al., 2001). The C- and N-termini of A′ and A″ forms a four α-helix bundle domain, composed of 1 α-helix from C-terminus of A′ and 3 α-helices from N-terminus of A″ (Figure 1) (Hirata et al., 2008b).
Figure 1.
The 13-subunit Sulfolobus solfataricus RNAP structure (PDB: 3HKZ). The α-carbon backbone is shown as cartoon model, along with the transparent molecular surface. Each subunit is denoted by a unique color and labeled. Two sets of nomenclatures, one for traditional and another is based on the eukaryotic terminology proposed by Korkhin et al., are shown. Various structural features discussed in the text are also labeled. Color version of the figure is available online.
Several archaeal RNAP subunits form stable subcomplexes. For examples, the D and L subunits form a heterodimer and functions as a platform for the assembly of subunits (A′, A″ and B) that comprise the active site (Goede et al., 2006). The D/L subcomplex is located on the opposite side of the opening of the claw. Interestingly, the D subunit contains a 4Fe-4S cluster binding motif that has been structurally characterized by the atomic resolution X-ray crystal structure of D/L subcomplex in addition to the entire Sulfolobus solfataricus RNAP structure (Hirata et al., 2008b, Hirata and Murakami, 2009). Three Cys residues (C183, C203 and C209) are ligands to the 3Fe-4S cluster and one additional C206 is positioned near the Fe-S cluster, suggesting that the cluster may exist as a 4Fe-4S in vivo. The Fe-S cluster is located ~45 Å from the enzyme active site, which suggests a structural role rather than a catalytic one. Furthermore, mutagenesis study has shown that the Fe-S cluster plays a role in supporting the structural integrity of the D subunit and that it is essential in the formation of D/L subcomplex (Hirata et al., 2008b). Interestingly, the 4Fe–4S cluster-binding domain is not conserved in all archaeal RNAPs. In fact, amino acid residues holding the Fe–S cluster in the D subunit characterize a specific evolutionary lineage of archaea (Hirata and Murakami, 2009).
The E and F subunits form a stalk-like E/F heterodimer that binds to the core part of RNAP. It may modulate the position of the clamp of RNAP, thereby making the Sulfolobus solfataricus RNAP adopt a closed clamp conformation. The E/F subcomplex is also known to be involved in both transcription initiation and elongation; it stimulates DNA melting and interacts with the newly synthesized RNA transcript through the RNA-binding motifs in the E and F subunits (Naji et al., 2007, Werner, 2007). The RNA-protein interaction has been demonstrated by studies on its Pol II counterpart, Rpb7/Rpb4 subcomplex in vitro (Meka et al., 2005, Ujvari and Luse, 2006), however, the interaction between RNA and this subcomplex has not been confirmed in vivo. The genetic study of hyperthermophilic archaeon, Thermococcus kodakarensis, has shown that the F subunit coding gene rpoF is not essential but ΔrpoF cells shows temperature sensitive phenotype (Hirata et al., 2008a). RNAP preparations purified from ΔrpoF cells lacked subunit F and also subunit E and a transcription factor TFE that co-purifies with RNAP from wild-type cells, but in vitro, this mutant RNAP exhibited no differences from wild-type RNAP in promoter-dependent transcription, abortive transcript synthesis, transcript elongation or termination. The RpoE/F subcomplex could be a binding platform of the TFE. This is consistent with TFE stimulation of archaeal RNAP activity requiring subunit E (Naji et al., 2007, Ouhammouch et al., 2004) and with reports of stimulatory transcription factor interactions with the homologous complexes in Pol I, II and III. The extensions formed in Pol I by A43 plus A14, and in Pol III by C25 plus C17 interact with polymerase-specific transcription initiation factors that recruit Pol I and Pol III to the appropriate promoters (Peyroche et al., 2000, Kassavetis et al., 2001). Given these observations, it seems most likely that the extensions formed by the archaeal subunits E plus F, and eukaryotic subunits A43 plus A14 (Pol I), Rpb4 plus Rpb7 (Pol II) and C25 plus C17 (Pol III) provide targets for some transcription factor binding, and so facilitate RNAP recruitment and transcription factor activation of the transcription machinery embodied in the core structures of these enzymes.
Structural Comparison of RNA Polymerases from Three Domains of Life
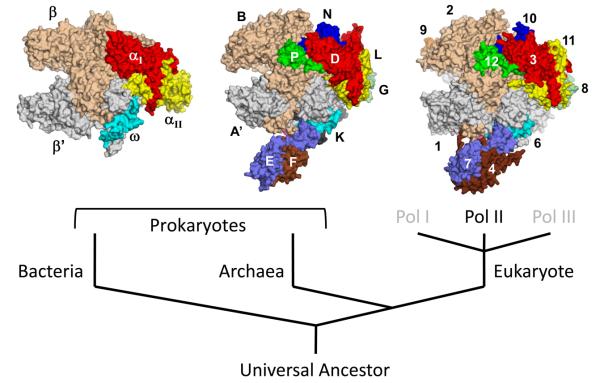
The overall shape of bacterial, archaeal, and eukaryotic RNAPs is look like a crab claw (Figure 2). The largest 2~3 subunits form the most part of each claw-arm, generating a cleft for double-stranded DNA binding (Cramer et al., 2001, Hirata et al., 2008b, Zhang et al., 1999). The enzyme active site is located at the bottom of the cleft that coordinates a catalytic metal Mg2+ by three invariant Asp residues found in the absolutely-conserved NADFDGD motif. The architecture around the cleft including the active site is highly conserved among all cellular RNAPs, which suggests that the catalytic mechanism of RNA synthesis from bacteria to human is conserved (Darst, 2001). But there are significant differences on the surfaces of these structures. The most significant difference is that archaeal RNAP and all three types of eukaryotic RNAPs have a protruding stalk-like structure that is absent from bacterial RNAP (Figure 2) (Armache et al., 2005, Hirata et al., 2008b, Kuhn et al., 2007, Jasiak et al., 2006).
Figure 2.
Surface representations of cellular RNAP structures from bacteria (left, Thermus aquaticus core enzyme), archaea (center, Sulfolobus solfataricus RNAP), and eukaryote (right, Saccharomyces cerevisiae Pol II). Each subunit is denoted by a unique color and labeled. Orthologous subunits are depicted with the same color. Color version of the figure is available online.
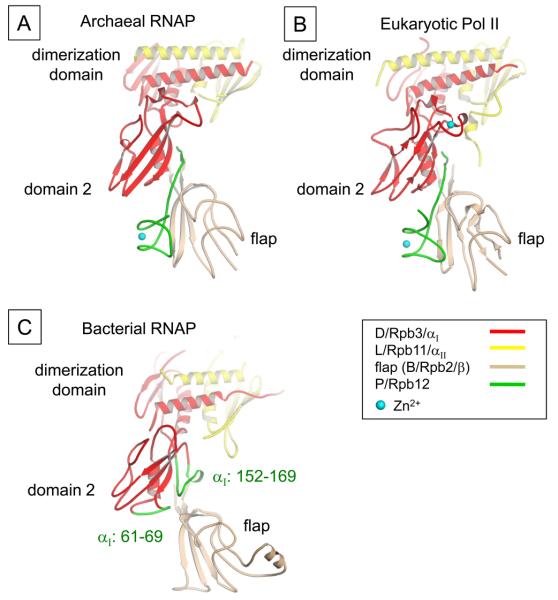
Based on structural comparison, the subunits of cellular RNAPs can be characterized into 3 classes (Table 1). Class I subunits are conserved in all 3 domains, class II subunits are shared between archaeal and eukaryotic RNAPs, class III subunits are unique to each domain and except Rpb9, others are found in certain species. All 5 subunits of the bacterial RNAP core enzyme are class I indicating they all play essential roles in cellular RNAP functions. Structural similarities calculated by secondary structure matching algorithm have shown that the structures of class I subunits of eukaryote are more similar with archaeal subunits than those of bacteria (Hirata et al., 2008b). Especially, the second largest Rpb2 subunit of S. cerevisiae Pol II shows about 90 % structural similarity with the B subunit of S. solfataricus RNAP whereas 65 % with the β subunit of T. aquaticus RNAP (Hirata et al., 2008b). The differences in the structures of class I subunits of eukaryote and bacteria come from the extensive interactions with small subunits like class II and class III on the periphery (Cramer et al., 2001). Class II subunits decorate the outside of class I subunits and are involved in the proper folding and assembly of RNAP (Werner and Weinzierl, 2002). The individual structures of class II subunits as well as their interactions with other subunits are highly conserved between archaea and eukaryote (Hirata et al., 2008b). For E, N and P subunits, the 3 smallest class II subunits in S. solfataricus RNAP, the structural similarities with the counterparts of S. cerevisiae Pol II are higher than 90 %. Although there is no class II subunit in bacteria RNAP, there are several motifs generated from class I subunits, which are located at the equivalent positions to the motifs from class II subunits of archaeal and eukaryotic RNAPs (Table 1). For example, the C-terminal tail of P subunit in archaeal RNAP (Rpb12 in Pol II) occupies the space between domain 2 of archaeal D subunit (Rpb3 in Pol II) and flap domain of B subunit (Rpb2 in Pol II) (Figures 3A and B). P subunit is essential subunit for the assembly of archaeal RNAP (Werner and Weinzierl, 2002). In bacterial RNAP, two loops in αI subunit are present at the corresponding space of C-terminal tail of P subunit (Figure 3C). These loops likely help the assembly of αI subunit and β subunit as C-terminal tail of P subunit does in archaeal RNAP.
Figure 3.
Interface of the D and P subunits and flap domain. (A) Ribbon representation of the archaeal domain 2 (D subunit), P subunit and flap domain (B subunit) is shown (color coding of ribbons is indicated). A gap between domain 2 and flap domain is filled by P subunit, which forms a β-addition motif (in which a strand from one subunit is added to a β sheet of another). (B) Ribbon representation of the eukaryotic Pol II domain 2 (Rpb3), Rpb12 and flap domain (Rpb2) is shown. The orientation is the same as shown in the A. C) Ribbon representation of the domain 2 (αI subunit) and flap domain (β subunit) is shown. Bacterial αI subunit residues 61-69 and 152-169 are present to the equivalent position of the C-terminal tail of P subunit, which buttresses αI domain 2 and β flap interaction. Color version of the figure is available online.
The Rpb9 of Pol II belongs to the class III subunit that is found only in Pol II (Table 1 and Figure 4C). The Rpb9 plays an important role in maintaining transcriptional fidelity by mediating the intrinsic nuclease activity of Pol II (Nesser et al., 2006) and also it is involved in the repair of damage in the DNA of an actively transcribed gene, termed transcription-coupled repair (TCR) (Li and Smerdon, 2002). Although Rpb9 is highly conserved in eukaryotic organisms, yeast null mutants of RPB9 have only a limited growth defect (Woychik et al., 1991), whereas it is required for viability in higher organisms like Drosophila (Harrison et al., 1992) showing again its intriguing position in evolution.
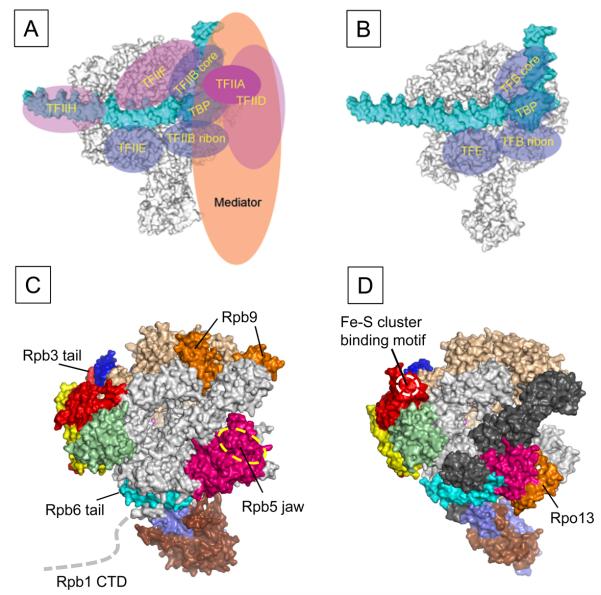
Figure 4.
Comparison of the transcription machinery between eukaryote and archaea. (Top) Comparison of the eukaryotic Pol II PIC (A) and the archaeal PIC (B). RNAP (gray) and promoter DNA (cyan) are represented as surface and general transcription factors are represented as ellipses. The common general transcription factors between eukaryote and archaea are colored transparent blue. (Bottom) Structural differences between eukaryotic Pol II (C) and archaeal RNAP (D) are highlighted and labeled. Subunit color-code is the same as in Figure 2. Color version of the figure is available online.
The δ subunit of bacterial RNAP, the Rpo13 of archaeal RNAP and Gdown1 of eukaryotic Pol II belong to the class III subunit. RNAP isolated from several Gram (+) bacteria, including Bacillus subtilis, contains an additional δ subunit (~20 kDa) in the core enzyme. The δ subunit consists with the N-terminal domain and negatively charged C-terminal tail. The NMR structure of the N-terminal domain has been determined (Motackova et al., 2010), but its binding site on RNAP is unknown. In vitro transcription assays have shown that the δ subunit either increases or decreases activities of transcription depending on promoters, and it may potentially influence the isomerization between the closed complex and the transcription-competent open complex (Lopez de Saro et al., 1995).
The Rpo13 has been identified as a new subunit of archaeal RNAP and Rpo13, which is only found in the order Sulfolobales, is located at a groove between the H subunit and the clamp head domain of the A′ subunit (Figures 1 and 4D) (Korkhin et al., 2009). It has a helix-turn-helix motif and has been suggested to bind DNA. The role of Rpo13 in archaeal RNAP is an open question. (1) Rpo13 might have been introduced into the RNAP of last archaeal common ancestor to meet the evolutionary need of some subgroup of archaea. In this case its function would be a new one that does not exist in the subunits of eukaryotic RNAPs. (2) Rpo13 might function as a ‘built-in’ transcription factor like A49 and A34.5 of eukaryotic Pol I and C37 and C53 of eukaryotic Pol III that are related to TFIIF (Kuhn et al., 2007, Jasiak et al., 2006). (3) Rpo13 might play a similar role with the Rpb5 jaw domain of Pol II since they occupy similar locations in RNAPs (Figures 4C and D) and both were suggested to have DNA binding activities.
The Pol II isolated from metazoan cells contains an additional tightly associated polypeptide Gdown1, which is about 43 kDa molecular weight (Hu et al., 2006). In vitro transcription assay provided some evidences that the Gdown1 has a functional interaction with mediator complex for responding activator-dependent transcription. The structure of Gdown1 and its binding sites on the Pol II are unknown.
Comparison of Transcription Pre-Initiation Complexes from Three Domains of Life
When gene is expressed, the transcription machinery including general transcription factors and RNAP are recruited to promoter DNA to form pre-initiation complex (PIC) (Hampsey, 1998, Hahn, 2004). Significant conformational changes of PIC form the transcription competent open complex that include the unwinding of DNA around the transcription start site and the positioning single-stranded template DNA near the RNAP active site. In bacteria, σ factor binds to core enzyme to form holoenzyme, which recognizes promoter DNA at around -35 and -10 from the transcription start site and makes a closed RNAP-promoter complex (Murakami and Darst, 2003). In the case of Pol II transcription, 6 general transcription factors, TFIIA, TFIIB, TFIID, TFIIE, TFIIF and TFIIH, mediator complex and Pol II are recruited to the promoter DNA and form the PIC (Hampsey, 1998, Hahn, 2004) (Figure 4A). The archaeal PIC is similar to the eukaryotic Pol II transcription system that contains TBP, TFB and TFE, which are orthologs of TBP, TFIIB and TFIIEα, respectively, are recruited with RNAP to the promoter DNA (Figure 4B) (Bell and Jackson, 1998). Only the structure of bacterial RNAP-promoter complex has been determined by X-ray crystallography (Murakami et al., 2002a). However, the topological arrangements of archaeal transcription apparatus including general transcription factors (TBP and TFB) and RNAP on promoter DNA were well determined by high-resolution DNA photo-crosslink experiments (Renfrow et al., 2004, Bartlett et al., 2004), and the arrangements of these proteins on promoter DNA are in good agreements with their counterparts in the eukaryotic Pol II transcription system (Chen and Hahn, 2004, Kim et al., 2000, Chen et al., 2004, Forget et al., 2004). Although archaeal PIC is much simpler than eukaryotic one, all the components of archaeal basal transcription machinery are highly related to the eukaryotic counterparts suggesting that the archaeal and eukaryotic transcription machines have come from the same origin. Tight conservation but much simpler and robust archaeal transcription system has been used for understanding the basic mechanism of Pol II transcription. For example, Cramer and Thomm groups have used archaeal in vitro transcription system to characterize the structure and function relationship of the yeast TFIIB in transcription initiation (Kostrewa et al., 2009). Based on newly determined the Pol II-TFIIB complex X-ray crystal structure, they expected the DNA opening activity of the B-linker domain of TFIIB and have shown the same region of archaeal TFB has the activity.
Compared with the Pol II transcription system, another advantage of the archaeal transcription system is that the active archaeal RNAP can be conveniently reconstituted from its individual subunits in vitro (Werner and Weinzierl, 2002, Naji et al., 2007). With the reconstituted archaeal RNAP, it has been shown in vitro that the interaction between the B-linker domain of TFB and the coiled-coil region of clamp domain of RNAP is required for the DNA opening activity of TFB, which was nicely complement to the observation from the X-ray crystal structure of Pol II-TFIIB complex (Kostrewa et al., 2009).
Given that RNAP is the center of transcription machinery, structural comparisons between archaeal and eukaryotic RNAPs would give the insight for understanding the transcription machinery of two systems. The structural differences between archaeal and eukaryotic RNAPs can be regarded as simple additions of polypeptides to the archaeal RNAP rather than changes to the core structure (Hirata et al., 2008b) (Figures 4C and D). Differences between archaeal and eukaryotic RNAPs found by X-ray crystal structures are Rpb9 and the N-terminal domain of Rpb5. In the X-ray crystal structure of S. cerevisiae Pol II, several regions are disordered that include the C-terminal hepta-peptide repeats of Rpb1 (Pol II CTD), Rpb3 C-terminal tail, the Rpb6 N-terminal tail and the Rpb12 N-terminal region. Interestingly, these flexible regions do not exist in the archaeal RNAP amino acid sequence. Therefore, it is tempting to speculate that these flexible regions in Pol II have been gained during the evolution for interacting with the eukaryotic specific transcription factors (Figure 4A). Further comparisons between archaeal and eukaryotic transcription apparatus could give the clues of how archaea and eukaryotes have developed their unique transcription system from their common ancestry during the evolution.
Transcription Regulation in Archaea
One striking feature of the archaea is that they possess a high degree of similarity to eukaryotes in the proteins utilized for gene expression. This includes proteins involved in coordinating DNA replication, translation and transcription, and it is reflective of the common ancestry shared by the two groups (Woese et al., 1990). The focus of second part of review will be to summarize the key proteins and the steps involved in establishing archaeal transcription initiation and the transcription regulation.
Components of Archaeal Pre-Initiation Complex
Despite their capacity for de novo transcription without a primer, multi-subunit cellular RNAPs are incapable of sequence-specific promoter recognition. These RNAPs require the aid of additional general transcription factors to properly position them at near the transcription start site before engaging in transcription. The first of the two required archaeal GTFs is an ortholog to the eukaryotic TBP, which is a highly conserved protein found in all known species of archaea and eukaryote, but not in bacteria. TBP orthologs are essential for the transcription of virtually all genes in these two domains (Thomm, 2007, Pugh, 2000). TBP binds promoter DNA along the minor groove of an AT-rich TATA-box sequence located approximately 25 bp upstream of the transcription start site and causes a sharp DNA bend (Kosa et al., 1997, Kim and Burley, 1994, Nikolov et al., 1996). The distortion of DNA backbone is thought to assist in the recruitment of other general transcription factors to the promoter. Archaeal TBPs and the C-terminal domain of the eukaryotic TBPs (TBPc) are approximately 180 amino acids in length, and consist of two domains which are direct imperfect repeats. Archaeal TBPs and the eukaryotic TBPc have approximately 30~40 % amino acid sequence identity (Soppa, 1999) and have close structural similarity (Kosa et al., 1997, Bell et al., 1999b, Nikolov et al., 1996, Kim and Burley, 1994). Although their structures are very similar, eukaryotic TBPs generally have an overall basic charge whereas archaeal TBPs tend to be acidic (Thomsen et al., 2001). The eukaryotic TBPs have additional N-terminal domain that is absent in the archaeal molecule (Soppa, 1999, Thomsen et al., 2001).
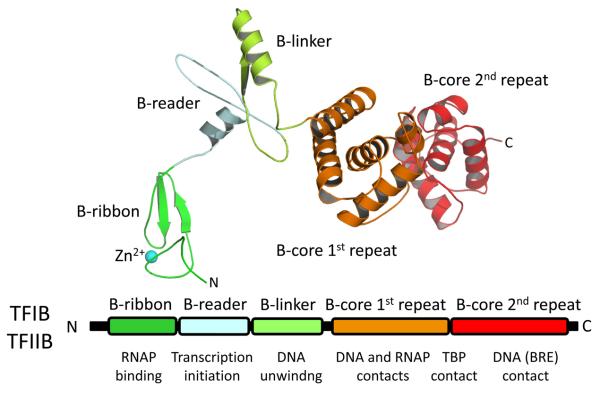
The second required archaeal GTF is TFB which is orthologous to eukaryotic TFIIB (Figure 5). TFB is highly conserved among different species (50~60 % amino acid similarity), but sequence similarity between archaeal TFBs and eukaryal TFIIBs is only approximately 20~30 % (Soppa, 1999). TFB interacts with a DNA sequences known as the B-factor Recognition Element (BRE), which is a purine rich (within non-template strand) sequence found immediately upstream of the TATA-box. The interaction between TFB and BRE appears to be necessary to determine direction of transcription (Bell et al., 1999b, Littlefield et al., 1999). The C-terminal of TFB has two domains, each consisting of 5 or 6 α-helices (Figure 5). The X-ray crystal structure of DNA-TBP-TFB complex showed that both domains are involved in interaction with DNA and TBP (Korkhin et al., 2001). The last two α-helices in the second repeat form a helix-turn-helix motif, which recognizes the DNA sequence of BRE (Littlefield et al., 1999). The structure of archaeal TFB N-terminal domain including Zn2+ binding site has been determined by NMR (Zhu et al., 1996). Later, X-ray structure of the yeast Pol II-TBP-TFIIB complex provided a nearly entire structure of TFIIB and positions of each TFIIB domain on the Pol II for understanding their functions (Kostrewa et al., 2009). The N-terminal domain of TFB contains several motifs that play important roles in RNAP binding (B-ribbon and B-linker), DNA unwinding (B-linker) and transcription start site selection (B-reader). Previous in vitro transcription analyses showed that once 9-12 bases of the nascent transcripts are formed, TFB is released from the transcription initiation complex and recycled for the next round of transcription (Xie and Reeve, 2004, Spitalny and Thomm, 2003). TFB release from RNAP is also necessary to open the channel for nascent RNA exit from RNAP (Kostrewa et al., 2009).
Figure 5.
Schematic of TFB/TFIIB sequence architecture. The black bar represents the primary sequence. The conserved regions are labeled and their functions are indicated. The structure of TFIIB is also shown. Each conserved region has the same color as in the primary sequence. Color version of the figure is available online.
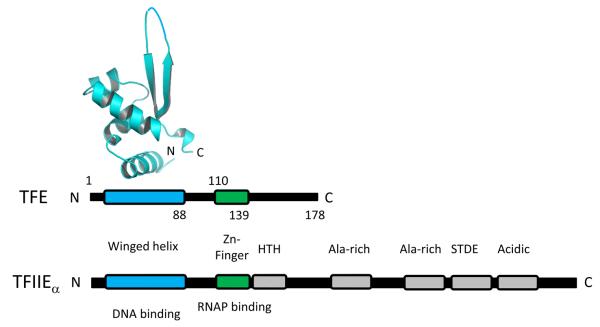
In addition to having TBP and TFIIB orthologs, archaea possesses less well characterized orthologs to the N-terminal domain of the α-subunit of eukaryotic TFIIE designated TFE (Figure 6) (Bell et al., 2001, Thomm, 2007). Transcription from strong promoters is apparently unaffected by TFE (Bell et al., 2001). It has been suggested that TFE is involved in stabilizing the RNAP open complex formation by enhancing DNA melting and DNA loading, and this activity is dependent upon the E subunit of RNAP (Naji et al., 2007, Thomm et al., 2009). Accordingly, TFE is co-purified with RNAP but the mutant RNAP preparations purified from ΔrpoF cells lacked subunit F and also subunit E and TFE (Hirata et al., 2008a). It has also been demonstrated that unlike TFIIE, TFE remains associated with RNAP during elongation (Grunberg et al., 2007).
Figure 6.
Schematic of TFE/TFIIEα sequence architecture. The black bar represents the primary sequence. The conserved regions are labeled and their functions are indicated. The X-ray crystal structure of the N-terminal winged helix domain of TFE (Sulfolobus solfataricus TFE, residues 1–88, PDB: 1Q1H) is also shown. Color version of the figure is available online.
The TFS is the ortholog of C-terminal domain of the eukaryotic transcription elongation factor TFIIS, which plays a role in transcription proofreading by RNA hydrolysis (Langer and Zillig, 1993, Hausner et al., 2000, Thomm, 2007). There have been no identified archaeal orthologs to eukaryotic TFIIA, TFIIF or TFIIH nor have there been any identified archaeal homologs to any of the myriad of eukaryotic TBP associated factors (TAF) which are found in TFIID (Thomm, 2007). Although no archaeal homologs to any eukaryotic TAFs have been identified, one protein, designated TBP-interacting protein 26 (TIP26) has been identified in T. kodakarensis KOD1 which can bind TBP and inhibit it from binding DNA (Matsuda et al., 2001, Yamamoto et al., 2006, Matsuda et al., 1999). Although TIP26 homologs are not widespread in the archaea, its finding gives rise to the possibility that other proteins with analogous functions might exist in other archaeal species.
Transcription Regulation in the Archaea
In order to effectively survive in a competitive environment, organisms need to balance production of gene required for cell activities, while avoiding extraneous production of unnecessary proteins. The regulation of gene expression could potentially take place at any stage from transcription initiation to protein degradation. Regulation of transcription initiation is one of the important steps governing gene expression. Therefore, organisms have developed a variety of methods to achieve effective outcomes. Some archaeal transcription regulation mechanisms have been shown to be more closely related to bacterial than eukaryotic systems (Bell, 2005, Geiduschek and Ouhammouch, 2005), whereas others are more closely related to those in eukaryotes. Three examples of currently proposed models of transcription regulation in archaea are described below.
The first example is the utilization of bacterial-type transcription activators and repressors for transcription regulation. Although archaeal transcription machinery consists of a eukaryotic-type RNAP and GTFs, most archaeal genomes encode homologues to bacterial-type transcription regulators (Bell, 2005, Bell and Jackson, 2001, Geiduschek and Ouhammouch, 2005, Vierke et al., 2003). The mechanism of repressing transcription in these cases entails binding to DNA in the promoter by a transcription factor leading to the blocking of TFB and/or TBP bindings to DNA (e.g. Sulfolobus solfataricus Lrs-14 (Fiorentino et al., 2003)) or inhibiting RNAP recruitment to the promoter (e.g. Pyrococcus furiosus LrpA (Brinkman et al., 2002, Dahlke and Thomm, 2002), Archaeoglobus fulgidus MDR1 (Bell et al., 1999a)). Some bacterial Lrp (Leucine responsive regulator protein)-like factors in archaea are proposed to function as transcription activators, which enhance the formation of PIC (Ouhammouch, 2004). Examples of currently identified activators are Sulfolobus solfataricus LysM (Brinkman et al., 2002) and Methanocaldococcus janaschii Ptr2 (Brinkman et al., 2003, Ouhammouch et al., 2003). The second example involves DNA packaging proteins, which is analogous to transcription regulation in eukaryotes; the nucleosome structure and histone modification play key roles in gene regulation. Eukaryotic histones possess positively charged N-terminal tails, and specific residues within the tails are covalently modified by acetylation, phospholylation, metylation and ubiquitination (reviewed in (Wu and Grunstein, 2000)). Acetylation of histone tails, for example, allows the access of transcription factors to DNA to promote transcription initiation. Archaeal species from Euryarchaeota kingdom possess one to six different histones forming both heterodimers and homodimers. For example, two histone proteins in Methanothermus fervidus, HMfA and HMfB, exist as either a heterodimer (HMfA+HMfB) or a homodimer ((HMfA)2 or (HMfB)2) in vivo (Pereira and Reeve, 1998). Interestingly, alternative dimer forms have different affinities to DNA sequences. Therefore, combinations of histone proteins could potentially control accessibility of transcription factors to DNA promoters in the archaea (reviewed in (Pereira and Reeve, 1998, Reeve, 2003)). A recent in vitro transcription experiments revealed that the M. jannaschii histones inhibit transcription from promoter and the M. jannaschii transcription activator Ptr2 competes for DNA binding with histones in effect counteracting the repressive effect of histones (Wilkinson et al., 2010). Identification of archaeal histones raises the possibility of nucleosome structure presence in Euryarchaeota and a potential transcription regulation mechanism via its modification (Reeve, 2003). However, the archaeal histones lack the N- and C-terminal tails, which are the targets for modifications in eukaryotes. In addition, homologues to histone modification factors found in eukaryotes have not been identified in any archaeal genomes (Reeve, 2003).
Alba (Acetylation lowers binding affinity) is another double-stranded DNA binding protein identified in the archaea, which can be reversibly acetylated and deacetylated by modification enzymes (Bell et al., 2002). It has been proposed that acetylation and deacetylation of Alba could influence transcription activity due to the lower DNA binding affinity exhibited by acetylated Alba compared with deacetylated form (Bell et al., 2002). Also, repressed transcription activity in vitro on DNA templates in the presence of acetylated Alba had been reported (Bell et al., 2002).
The third proposed model involves the multiplicity of GTFs that include TBP and TFB. Most higher eukaryotes are known to carry multiple orthologs of both TBP and TFIIB with divergent functional roles from these GTFs. These include at least four TBP orthologs known as TBP-related factors (TRF) with a variety of different functional roles mostly during embryonic development (Crowley et al., 1993, Dantonel et al., 1999, Persengiev et al., 2003), and two different TFIIB orthologs known as TFIIB-related factors (Brf) in the Pol III transcription system (Ferrari et al., 2004, Paule and White, 2000, Saxena et al., 2005). The presence of multiple TBP and TFIIB homologs is not unique to eukaryotes. Not long after the discovery of archaeal TBP and TFB, it was discovered that some archaeal species possessed more than one gene for TBP and/or TFB consistent with the possibility that archaeal species might utilize these alternative GTF orthologs to differentially regulate gene expression. The sequencing of an extrachromosomal megaplasmid in the extreme halophile, Halobacterium sp. NRC-1 revealed four open reading frames (orfs) encoding for putative TBPs located on this megaplasmid (Ng et al., 1998). Later, the complete genome sequencing of this organism revealed two additional TBP orthologs and seven different TFB orthologs (Ng et al., 2000, Baliga et al., 2000). Subsequent genome sequencing projects revealed multiple TFB orthologs (Kawarabayasi et al., 1999, Thompson et al., 1999, Lecompte et al., 2001, Fukui et al., 2005, She et al., 2001, Kawarabayasi et al., 1998, Facciotti et al., 2007, Coker and DasSarma, 2007) or multiple TBP orthologs (Deppenmeier et al., 2002, Galagan et al., 2002, Maeder et al., 2006) in several other archaeal species. It is now apparent that the presence of multiple archaeal GTF orthologs is not rare. The Institute for Genomic Research (TIGR) currently lists the completed genomes for 42 archaeal species on its database, and 29 of these species have at least two identified TBPs or TFBs (Table 2). Among those species with multiple GTF orthologs, there is a tendency towards multiple TFBs in the halophiles, hyperthermophiles and thermoacidophiles, whereas there is a tendency towards multiple TBPs in the methanogens. Whether or not this trend has any functional significance is unclear. However, in light of the important roles for TBP and TFB in DNA binding and DNA opening respectively, this trend could be an indication that for the halophiles and hyperthermophiles the DNA opening step is the major target for gene regulation whereas for the methanogens, the DNA binding step is the primary target for regulation.
Table 2.
Archaeal species with multiple annotated TBP or TFB genes
| Organism | Putative TBPs | Putative TFBs |
|---|---|---|
| Aeropyrum pernix K1 | 1 | 2 |
| Candidatus methanoregula boonei 6A8 | 3 | 1 |
| Haloarcula marismortui ATCC 43049 | 1 | 9 |
| Halobacterium sp. NRC-1 | 6 | 7 |
| Haloquadratum walsbyi DSM 16790 | 2 | 9 |
| Hyperthermus butylicus DSM 5456 | 1 | 2 |
| Metallosphaera sedula DSM 5348 | 1 | 2 |
| Methanococcus maripaludis C5 | 2 | 1 |
| Methanocorpusculum labreanum Z | 2 | 1 |
| Methanoculleus marisnigri JR1 | 2 | 2 |
| Methanosarcina acetivorans C2A | 3 | 1 |
| Methanosarcina barkeri fusaro | 2 | 1 |
| Methanosarcina mazei Goe1 | 3 | 1 |
| Methanospirillum hungatei JF-1 | 2 | 1 |
| Natronomonas pharaonis sp | 1 | 9 |
| Picrophilus torridus DSM 9790 | 1 | 2 |
| Pyrobaculum aerophilum IM2 | 1 | 3 |
| Pyrobaculum arsenaticum DSM 13514 | 1 | 2 |
| Pyrobaculum islandicum DSM 4184 | 1 | 2 |
| Pyrococcus furiosus DSM 3638 | 1 | 2 |
| Pyrococcus horikoshii shinkaj OT3 | 1 | 2 |
| Sulfolobus acidocaldarius DSM 639 | 1 | 2 |
| Sulfolobus solfataricus P2 | 1 | 2 |
| Sulfolobus tokodaii strain 7 | 1 | 2 |
| Thermococcus kodakarensis KOD1 | 1 | 2 |
| Thermofilum pendens Hrk 5 | 1 | 2 |
| Thermoplasma acidophilum DSM 1728 | 1 | 3 |
| Thermoplasma volcanium GSS1 | 1 | 3 |
The majority of the experimental investigations into the function of multiple archaeal GTFs have been carried out in Halobacterium sp. NRC-1. This has been due to the large number of unique GTF homologs (six TBPs and seven TFBs which is the most for any archaeal species) (Table 2) (Ng et al., 2000, Baliga et al., 2000), and because of the availability of facile genetic tools. These investigations have largely concluded that the individual GTF isomers in Halobacterium sp. NRC-1 function to differentially regulate gene expression. Genetic analyses have established that at least 10 of its 13 GTF orthologs (4 TBPs and 6 TFBs) can be individually deleted under standard laboratory growth conditions without hindering cell viability (Coker and DasSarma, 2007, Facciotti et al., 2007) although no investigations into the consequences of deleting more than one GTF have been reported. One of the two TBPs that could not be deleted was tbpE, which is located on the main chromosome of Halobacterium sp. NRC-1. Proteomics analysis and quantitative RT-PCR established that the tbpE is more robustly expressed than the other TBP genes, and it was suggested that TBPe is the primary TBP utilized during growth. It was proposed that the additional TBPs provide added fitness based on defective growth of strains lacking the additional TBP genes (Goo et al., 2003, Teufel et al., 2008). It is unclear if one TFB ortholog functions as the primary TFB during growth, although tfbB is the only TFB that could not be knocked out (Coker and DasSarma, 2007, Facciotti et al., 2007). In a global analysis, a preliminary TFB regulatory network was deduced based on integrated data from ChIP-chip analysis, transcriptomic data and in vivo protein-protein interactions between the various TBP and TFB orthologs (Facciotti et al., 2007). In a separate report, one TBP, tbpD and one TFB, tfbA were shown to coordinately regulate roughly 10 % of the Halobacterim sp. NRC-1 genome including several heat shock response genes suggesting that preferential pairings of TBP-TFB may be exploited to direct gene expression. Mutant strains with either tbpD or tfbA deleted elicited defective growth in response to heat shock (Kaur et al., 2006). TFBb was shown to preferentially bind the heat shock inducible promoter Phsp5 in vitro during incubation at 50 °C, but not during incubation at 37 °C. Furthermore, TFBg binding at Phsp5 was not detected at either temperature consistent with a specific role for TFBb in the regulation of hsp5 during Halobacterium sp. NRC-1 heat shock response (Lu et al., 2008). Consistent with these observations for Halobacterium salinarum, TFB orthologs from Haloferax volcanii, P. furiosus and S. solfataricus have been implicated in the heat shock response or UV irradiation response as well (Thompson et al., 1999, Shockley et al., 2003, Paytubi and White, 2009). This suggests that the use of alternative TFB orthologs to direct the regulation of stress response genes may be a common feature in archaeal species.
A functional assessment of the two TFB orthologs in the hyperthermophile, T. kodakarensis suggested the possibility of functional redundancy for multiple TFBs (Santangelo et al., 2007). It was determined that either of the two TFB orthologs can be individually deleted without hindering growth under optimal growth conditions. Furthermore, both TFB orthologs could support in vitro transcription equally well from several different promoters showing no difference in TSS selection (Hirata et al., 2008a). In contrast, two TFB orthologs in P. furiosus (TFB1 and TFB2) showed substantial difference in their transcription activities in vitro. The TFB2, which lacks B-finger motif, is less active compared with TFB1 for all tested promoters (Micorescu et al., 2008).
To date, there has been only one reported experimental investigation into the functional roles of multiple TBP orthologs in organisms with only a single TFB which was carried out in the mesophilic methanogen, Methanosarcina acetivorans (Reichlen et al., 2010). As is the case for Halobacterium sp. NRC-1 TBPe, one of the M. acetivorans TBP orthologs, TBP1 appears to function in a dominant role to the two other TBPs, TBP2 and TBP3. However, these alternative TBPs did appear to be important for optimal growth when cells were cultured under nutrient limiting conditions or when forced to adjust their metabolic pathway from an energetically rich to an energetically poor substrate. These results suggested a possible role in for the alternative TBP orthologs transcriptional regulation specific to these conditions. However, specific targets for the alternative TBPs were not identified (Reichlen et al., 2010).
ACKNOWLEDGEMENTS
We thank Dr. Rieko Yajima for critical reading of the manuscript.
This work was supported by NIH grant GM087350.
Footnotes
A DECLARATION OF INTEREST SECTION
The authors report no declarations of interest.
REFERENCES
- ARMACHE KJ, MITTERWEGER S, MEINHART A, CRAMER P. Structures of complete RNA polymerase II and its subcomplex, Rpb4/7. J Biol Chem. 2005;280:7131–4. doi: 10.1074/jbc.M413038200. [DOI] [PubMed] [Google Scholar]
- BALIGA NS, GOO YA, NG WV, HOOD L, DANIELS CJ, DASSARMA S. Is gene expression in Halobacterium NRC-1 regulated by multiple TBP and TFB transcription factors? Mol Microbiol. 2000;36:1184–5. doi: 10.1046/j.1365-2958.2000.01916.x. [DOI] [PubMed] [Google Scholar]
- BARTLETT MS, THOMM M, GEIDUSCHEK EP. Topography of the euryarchaeal transcription initiation complex. J Biol Chem. 2004;279:5894–903. doi: 10.1074/jbc.M311429200. [DOI] [PubMed] [Google Scholar]
- BELL SD. Archaeal transcriptional regulation--variation on a bacterial theme? Trends Microbiol. 2005;13:262–5. doi: 10.1016/j.tim.2005.03.015. [DOI] [PubMed] [Google Scholar]
- BELL SD, BOTTING CH, WARDLEWORTH BN, JACKSON SP, WHITE MF. The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science. 2002;296:148–51. doi: 10.1126/science.1070506. [DOI] [PubMed] [Google Scholar]
- BELL SD, BRINKMAN AB, VAN DER OOST J, JACKSON SP. The archaeal TFIIEalpha homologue facilitates transcription initiation by enhancing TATA-box recognition. EMBO Rep. 2001;2:133–8. doi: 10.1093/embo-reports/kve021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BELL SD, CAIRNS SS, ROBSON RL, JACKSON SP. Transcriptional regulation of an archaeal operon in vivo and in vitro. Mol Cell. 1999a;4:971–82. doi: 10.1016/s1097-2765(00)80226-9. [DOI] [PubMed] [Google Scholar]
- BELL SD, JACKSON SP. Transcription and translation in Archaea: a mosaic of eukaryal and bacterial features. Trends Microbiol. 1998;6:222–8. doi: 10.1016/s0966-842x(98)01281-5. [DOI] [PubMed] [Google Scholar]
- BELL SD, JACKSON SP. Mechanism and regulation of transcription in archaea. Curr Opin Microbiol. 2001;4:208–13. doi: 10.1016/s1369-5274(00)00190-9. [DOI] [PubMed] [Google Scholar]
- BELL SD, KOSA PL, SIGLER PB, JACKSON SP. Orientation of the transcription preinitiation complex in archaea. Proc Natl Acad Sci U S A. 1999b;96:13662–7. doi: 10.1073/pnas.96.24.13662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRINKMAN AB, BELL SD, LEBBINK RJ, DE VOS WM, VAN DER OOST J. The Sulfolobus solfataricus Lrp-like protein LysM regulates lysine biosynthesis in response to lysine availability. J Biol Chem. 2002;277:29537–49. doi: 10.1074/jbc.M203528200. [DOI] [PubMed] [Google Scholar]
- BRINKMAN AB, ETTEMA TJ, DE VOS WM, VAN DER OOST J. The Lrp family of transcriptional regulators. Mol Microbiol. 2003;48:287–94. doi: 10.1046/j.1365-2958.2003.03442.x. [DOI] [PubMed] [Google Scholar]
- CHEN BS, MANDAL SS, HAMPSEY M. High-resolution protein-DNA contacts for the yeast RNA polymerase II general transcription machinery. Biochemistry. 2004;43:12741–9. doi: 10.1021/bi048993r. [DOI] [PubMed] [Google Scholar]
- CHEN HT, HAHN S. Mapping the location of TFIIB within the RNA polymerase II transcription preinitiation complex: a model for the structure of the PIC. Cell. 2004;119:169–80. doi: 10.1016/j.cell.2004.09.028. [DOI] [PubMed] [Google Scholar]
- COKER JA, DASSARMA S. Genetic and transcriptomic analysis of transcription factor genes in the model halophilic Archaeon: coordinate action of TbpD and TfbA. BMC Genet. 2007;8:61. doi: 10.1186/1471-2156-8-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CRAMER P, BUSHNELL DA, KORNBERG RD. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2001;292:1863–76. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- CROWLEY TE, HOEY T, LIU JK, JAN YN, JAN LY, TJIAN R. A new factor related to TATA-binding protein has highly restricted expression patterns in Drosophila. Nature. 1993;361:557–61. doi: 10.1038/361557a0. [DOI] [PubMed] [Google Scholar]
- DAHLKE I, THOMM M. A Pyrococcus homolog of the leucine-responsive regulatory protein, LrpA, inhibits transcription by abrogating RNA polymerase recruitment. Nucleic Acids Res. 2002;30:701–10. doi: 10.1093/nar/30.3.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DANTONEL JC, WURTZ JM, POCH O, MORAS D, TORA L. The TBP-like factor: an alternative transcription factor in metazoa? Trends Biochem Sci. 1999;24:335–9. doi: 10.1016/s0968-0004(99)01436-x. [DOI] [PubMed] [Google Scholar]
- DARST SA. Bacterial RNA polymerase. Curr Opin Struct Biol. 2001;11:155–62. doi: 10.1016/s0959-440x(00)00185-8. [DOI] [PubMed] [Google Scholar]
- DEPPENMEIER U, JOHANN A, HARTSCH T, MERKL R, SCHMITZ RA, MARTINEZ-ARIAS R, HENNE A, WIEZER A, BAUMER S, JACOBI C, BRUGGEMANN H, LIENARD T, CHRISTMANN A, BOMEKE M, STECKEL S, BHATTACHARYYA A, LYKIDIS A, OVERBEEK R, KLENK HP, GUNSALUS RP, FRITZ HJ, GOTTSCHALK G. The genome of Methanosarcina mazei: evidence for lateral gene transfer between bacteria and archaea. J Mol Microbiol Biotechnol. 2002;4:453–61. [PubMed] [Google Scholar]
- FACCIOTTI MT, REISS DJ, PAN M, KAUR A, VUTHOORI M, BONNEAU R, SHANNON P, SRIVASTAVA A, DONOHOE SM, HOOD LE, BALIGA NS. General transcription factor specified global gene regulation in archaea. Proc Natl Acad Sci U S A. 2007;104:4630–5. doi: 10.1073/pnas.0611663104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FERRARI R, RIVETTI C, ACKER J, DIECI G. Distinct roles of transcription factors TFIIIB and TFIIIC in RNA polymerase III transcription reinitiation. Proc Natl Acad Sci U S A. 2004;101:13442–7. doi: 10.1073/pnas.0403851101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FIORENTINO G, CANNIO R, ROSSI M, BARTOLUCCI S. Transcriptional regulation of the gene encoding an alcohol dehydrogenase in the archaeon Sulfolobus solfataricus involves multiple factors and control elements. J Bacteriol. 2003;185:3926–34. doi: 10.1128/JB.185.13.3926-3934.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FORGET D, LANGELIER MF, THERIEN C, TRINH V, COULOMBE B. Photo-cross-linking of a purified preinitiation complex reveals central roles for the RNA polymerase II mobile clamp and TFIIE in initiation mechanisms. Mol Cell Biol. 2004;24:1122–31. doi: 10.1128/MCB.24.3.1122-1131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FUKUI T, ATOMI H, KANAI T, MATSUMI R, FUJIWARA S, IMANAKA T. Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Res. 2005;15:352–63. doi: 10.1101/gr.3003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GALAGAN JE, NUSBAUM C, ROY A, ENDRIZZI MG, MACDONALD P, FITZHUGH W, CALVO S, ENGELS R, SMIRNOV S, ATNOOR D, BROWN A, ALLEN N, NAYLOR J, STANGE-THOMANN N, DEARELLANO K, JOHNSON R, LINTON L, MCEWAN P, MCKERNAN K, TALAMAS J, TIRRELL A, YE W, ZIMMER A, BARBER RD, CANN I, GRAHAM DE, GRAHAME DA, GUSS AM, HEDDERICH R, INGRAM-SMITH C, KUETTNER HC, KRZYCKI JA, LEIGH JA, LI W, LIU J, MUKHOPADHYAY B, REEVE JN, SMITH K, SPRINGER TA, UMAYAM LA, WHITE O, WHITE RH, CONWAY DE MACARIO E, FERRY JG, JARRELL KF, JING H, MACARIO AJ, PAULSEN I, PRITCHETT M, SOWERS KR, SWANSON RV, ZINDER SH, LANDER E, METCALF WW, BIRREN B. The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 2002;12:532–42. doi: 10.1101/gr.223902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GEIDUSCHEK EP, OUHAMMOUCH M. Archaeal transcription and its regulators. Mol Microbiol. 2005;56:1397–407. doi: 10.1111/j.1365-2958.2005.04627.x. [DOI] [PubMed] [Google Scholar]
- GNATT AL, CRAMER P, FU J, BUSHNELL DA, KORNBERG RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001;292:1876–82. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- GOEDE B, NAJI S, VON KAMPEN O, ILG K, THOMM M. Protein-protein interactions in the archaeal transcriptional machinery: binding studies of isolated RNA polymerase subunits and transcription factors. J Biol Chem. 2006;281:30581–92. doi: 10.1074/jbc.M605209200. [DOI] [PubMed] [Google Scholar]
- GOO YA, YI EC, BALIGA NS, TAO WA, PAN M, AEBERSOLD R, GOODLETT DR, HOOD L, NG WV. Proteomic analysis of an extreme halophilic archaeon, Halobacterium sp. NRC-1. Mol Cell Proteomics. 2003;2:506–24. doi: 10.1074/mcp.M300044-MCP200. [DOI] [PubMed] [Google Scholar]
- GRUNBERG S, BARTLETT MS, NAJI S, THOMM M. Transcription factor E is a part of transcription elongation complexes. J Biol Chem. 2007;282:35482–90. doi: 10.1074/jbc.M707371200. [DOI] [PubMed] [Google Scholar]
- HAHN S. Structure and mechanism of the RNA polymerase II transcription machinery. Nat Struct Mol Biol. 2004;11:394–403. doi: 10.1038/nsmb763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAMPSEY M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol Mol Biol Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HARRISON DA, MORTIN MA, CORCES VG. The RNA polymerase II 15-kilodalton subunit is essential for viability in Drosophila melanogaster. Mol Cell Biol. 1992;12:928–35. doi: 10.1128/mcb.12.3.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAUSNER W, LANGE U, MUSFELDT M. Transcription factor S, a cleavage induction factor of the archaeal RNA polymerase. J Biol Chem. 2000;275:12393–9. doi: 10.1074/jbc.275.17.12393. [DOI] [PubMed] [Google Scholar]
- HIRATA A, KANAI T, SANTANGELO TJ, TAJIRI M, MANABE K, REEVE JN, IMANAKA T, MURAKAMI KS. Archaeal RNA polymerase subunits E and F are not required for transcription in vitro, but a Thermococcus kodakarensis mutant lacking subunit F is temperature-sensitive. Mol Microbiol. 2008a;70:623–33. doi: 10.1111/j.1365-2958.2008.06430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HIRATA A, KLEIN BJ, MURAKAMI KS. The X-ray crystal structure of RNA polymerase from Archaea. Nature. 2008b;452:248. doi: 10.1038/nature06530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HIRATA A, MURAKAMI KS. Archaeal RNA polymerase. Curr Opin Struct Biol. 2009;19:724–31. doi: 10.1016/j.sbi.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HU X, MALIK S, NEGROIU CC, HUBBARD K, VELALAR CN, HAMPTON B, GROSU D, CATALANO J, ROEDER RG, GNATT A. A Mediator-responsive form of metazoan RNA polymerase II. Proc Natl Acad Sci U S A. 2006;103:9506–11. doi: 10.1073/pnas.0603702103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUET J, SCHNABEL R, SENTENAC A, ZILLIG W. Archaebacteria and eukaryotes possess DNA-dependent RNA polymerases of a common type. Embo J. 1983;2:1291–4. doi: 10.1002/j.1460-2075.1983.tb01583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JASIAK AJ, ARMACHE KJ, MARTENS B, JANSEN RP, CRAMER P. Structural biology of RNA polymerase III: subcomplex C17/25 X-ray structure and 11 subunit enzyme model. Mol Cell. 2006;23:71–81. doi: 10.1016/j.molcel.2006.05.013. [DOI] [PubMed] [Google Scholar]
- KASSAVETIS GA, LETTS GA, GEIDUSCHEK EP. The RNA polymerase III transcription initiation factor TFIIIB participates in two steps of promoter opening. Embo J. 2001;20:2823–34. doi: 10.1093/emboj/20.11.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAUR A, PAN M, MEISLIN M, FACCIOTTI MT, EL-GEWELEY R, BALIGA NS. A systems view of haloarchaeal strategies to withstand stress from transition metals. Genome Res. 2006 doi: 10.1101/gr.5189606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAWARABAYASI Y, HINO Y, HORIKAWA H, YAMAZAKI S, HAIKAWA Y, JIN-NO K, TAKAHASHI M, SEKINE M, BABA S, ANKAI A, KOSUGI H, HOSOYAMA A, FUKUI S, NAGAI Y, NISHIJIMA K, NAKAZAWA H, TAKAMIYA M, MASUDA S, FUNAHASHI T, TANAKA T, KUDOH Y, YAMAZAKI J, KUSHIDA N, OGUCHI A, KIKUCHI H, et al. Complete genome sequence of an aerobic hyper-thermophilic crenarchaeon, Aeropyrum pernix K1. DNA Res. 1999;6:83–101. doi: 10.1093/dnares/6.2.83. 145-52. [DOI] [PubMed] [Google Scholar]
- KAWARABAYASI Y, SAWADA M, HORIKAWA H, HAIKAWA Y, HINO Y, YAMAMOTO S, SEKINE M, BABA S, KOSUGI H, HOSOYAMA A, NAGAI Y, SAKAI M, OGURA K, OTSUKA R, NAKAZAWA H, TAKAMIYA M, OHFUKU Y, FUNAHASHI T, TANAKA T, KUDOH Y, YAMAZAKI J, KUSHIDA N, OGUCHI A, AOKI K, KIKUCHI H. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3 (supplement) DNA Res. 1998;5:147–55. doi: 10.1093/dnares/5.2.147. [DOI] [PubMed] [Google Scholar]
- KIM JL, BURLEY SK. 1.9 A resolution refined structure of TBP recognizing the minor groove of TATAAAAG. Nat Struct Biol. 1994;1:638–53. doi: 10.1038/nsb0994-638. [DOI] [PubMed] [Google Scholar]
- KIM TK, EBRIGHT RH, REINBERG D. Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science. 2000;288:1418–22. doi: 10.1126/science.288.5470.1418. [DOI] [PubMed] [Google Scholar]
- KORKHIN Y, LITTLEFIELD O, NELSON PJ, BELL SD, SIGLER PB. Preparation of components of archaeal transcription preinitiation complex. Methods Enzymol. 2001;334:227–39. doi: 10.1016/s0076-6879(01)34471-3. [DOI] [PubMed] [Google Scholar]
- KORKHIN Y, UNLIGIL UM, LITTLEFIELD O, NELSON PJ, STUART DI, SIGLER PB, BELL SD, ABRESCIA NG. Evolution of Complex RNA Polymerases: The Complete Archaeal RNA Polymerase Structure. PLoS Biol. 2009;7:e102. doi: 10.1371/journal.pbio.1000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOSA PF, GHOSH G, DEDECKER BS, SIGLER PB. The 2.1-A crystal structure of an archaeal preinitiation complex: TATA-box-binding protein/transcription factor (II)B core/TATA-box. Proc Natl Acad Sci U S A. 1997;94:6042–7. doi: 10.1073/pnas.94.12.6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOSTREWA D, ZELLER ME, ARMACHE KJ, SEIZL M, LEIKE K, THOMM M, CRAMER P. RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature. 2009;462:323–30. doi: 10.1038/nature08548. [DOI] [PubMed] [Google Scholar]
- KUHN CD, GEIGER SR, BAUMLI S, GARTMANN M, GERBER J, JENNEBACH S, MIELKE T, TSCHOCHNER H, BECKMANN R, CRAMER P. Functional architecture of RNA polymerase I. Cell. 2007;131:1260–72. doi: 10.1016/j.cell.2007.10.051. [DOI] [PubMed] [Google Scholar]
- KUSSER AG, BERTERO MG, NAJI S, BECKER T, THOMM M, BECKMANN R, CRAMER P. Structure of an archaeal RNA polymerase. J Mol Biol. 2008;376:303–7. doi: 10.1016/j.jmb.2007.08.066. [DOI] [PubMed] [Google Scholar]
- LANGER D, HAIN J, THURIAUX P, ZILLIG W. Transcription in archaea: similarity to that in eucarya. Proc Natl Acad Sci U S A. 1995;92:5768–72. doi: 10.1073/pnas.92.13.5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LANGER D, ZILLIG W. Putative tfIIs gene of Sulfolobus acidocaldarius encoding an archaeal transcription elongation factor is situated directly downstream of the gene for a small subunit of DNA-dependent RNA polymerase. Nucleic Acids Res. 1993;21:2251. doi: 10.1093/nar/21.9.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LECOMPTE O, RIPP R, PUZOS-BARBE V, DUPRAT S, HEILIG R, DIETRICH J, THIERRY JC, POCH O. Genome evolution at the genus level: comparison of three complete genomes of hyperthermophilic archaea. Genome Res. 2001;11:981–93. doi: 10.1101/gr.165301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI S, SMERDON MJ. Rpb4 and Rpb9 mediate subpathways of transcription-coupled DNA repair in Saccharomyces cerevisiae. Embo J. 2002;21:5921–9. doi: 10.1093/emboj/cdf589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LITTLEFIELD O, KORKHIN Y, SIGLER PB. The structural basis for the oriented assembly of a TBP/TFB/promoter complex. Proc Natl Acad Sci U S A. 1999;96:13668–73. doi: 10.1073/pnas.96.24.13668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIU X, BUSHNELL DA, WANG D, CALERO G, KORNBERG RD. Structure of an RNA polymerase II-TFIIB complex and the transcription initiation mechanism. Science. 2010;327:206–9. doi: 10.1126/science.1182015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOPEZ DE SARO FJ, WOODY AY, HELMANN JD. Structural analysis of the Bacillus subtilis delta factor: a protein polyanion which displaces RNA from RNA polymerase. J Mol Biol. 1995;252:189–202. doi: 10.1006/jmbi.1995.0487. [DOI] [PubMed] [Google Scholar]
- LU Q, HAN J, ZHOU L, COKER JA, DASSARMA P, DASSARMA S, XIANG H. Dissection of the regulatory mechanism of a heat-shock responsive promoter in Haloarchaea: a new paradigm for general transcription factor directed archaeal gene regulation. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkn152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAEDER DL, ANDERSON I, BRETTIN TS, BRUCE DC, GILNA P, HAN CS, LAPIDUS A, METCALF WW, SAUNDERS E, TAPIA R, SOWERS KR. The Methanosarcina barkeri genome: comparative analysis with Methanosarcina acetivorans and Methanosarcina mazei reveals extensive rearrangement within methanosarcinal genomes. J Bacteriol. 2006;188:7922–31. doi: 10.1128/JB.00810-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MATSUDA T, FUJIKAWA M, HARUKI M, TANG XF, EZAKI S, IMANAKA T, MORIKAWA M, KANAYA S. Interaction of TIP26 from a hyperthermophilic archaeon with TFB/TBP/DNA ternary complex. Extremophiles. 2001;5:177–82. doi: 10.1007/s007920100193. [DOI] [PubMed] [Google Scholar]
- MATSUDA T, MORIKAWA M, HARUKI M, HIGASHIBATA H, IMANAKA T, KANAYA S. Isolation of TBP-interacting protein (TIP) from a hyperthermophilic archaeon that inhibits the binding of TBP to TATA-DNA. FEBS Lett. 1999;457:38–42. doi: 10.1016/s0014-5793(99)01005-4. [DOI] [PubMed] [Google Scholar]
- MEKA H, WERNER F, CORDELL SC, ONESTI S, BRICK P. Crystal structure and RNA binding of the Rpb4/Rpb7 subunits of human RNA polymerase II. Nucleic Acids Res. 2005;33:6435–44. doi: 10.1093/nar/gki945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MICORESCU M, GRUNBERG S, FRANKE A, CRAMER P, THOMM M, BARTLETT M. Archaeal transcription: function of an alternative transcription factor B from Pyrococcus furiosus. J Bacteriol. 2008;190:157–67. doi: 10.1128/JB.01498-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MOTACKOVA V, SANDEROVA H, ZIDEK L, NOVACEK J, PADRTA P, SVENKOVA A, KORELUSOVA J, JONAK J, KRASNY L, SKLENAR V. Solution structure of the N-terminal domain of Bacillus subtilis delta subunit of RNA polymerase and its classification based on structural homologs. Proteins. 2010;78:1807–10. doi: 10.1002/prot.22708. [DOI] [PubMed] [Google Scholar]
- MURAKAMI KS, DARST SA. Bacterial RNA polymerases: the wholo story. Curr Opin Struct Biol. 2003;13:31–9. doi: 10.1016/s0959-440x(02)00005-2. [DOI] [PubMed] [Google Scholar]
- MURAKAMI KS, MASUDA S, CAMPBELL EA, MUZZIN O, DARST SA. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science. 2002a;296:1285–90. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- MURAKAMI KS, MASUDA S, DARST SA. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 A resolution. Science. 2002b;296:1280–4. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- NAJI S, GRUNBERG S, THOMM M. The RPB7 orthologue E' is required for transcriptional activity of a reconstituted archaeal core enzyme at low temperatures and stimulates open complex formation. J Biol Chem. 2007;282:11047–57. doi: 10.1074/jbc.M611674200. [DOI] [PubMed] [Google Scholar]
- NESSER NK, PETERSON DO, HAWLEY DK. RNA polymerase II subunit Rpb9 is important for transcriptional fidelity in vivo. Proc Natl Acad Sci U S A. 2006;103:3268–73. doi: 10.1073/pnas.0511330103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NG WV, CIUFO SA, SMITH TM, BUMGARNER RE, BASKIN D, FAUST J, HALL B, LORETZ C, SETO J, SLAGEL J, HOOD L, DASSARMA S. Snapshot of a large dynamic replicon in a halophilic archaeon: megaplasmid or minichromosome? Genome Res. 1998;8:1131–41. doi: 10.1101/gr.8.11.1131. [DOI] [PubMed] [Google Scholar]
- NG WV, KENNEDY SP, MAHAIRAS GG, BERQUIST B, PAN M, SHUKLA HD, LASKY SR, BALIGA NS, THORSSON V, SBROGNA J, SWARTZELL S, WEIR D, HALL J, DAHL TA, WELTI R, GOO YA, LEITHAUSER B, KELLER K, CRUZ R, DANSON MJ, HOUGH DW, MADDOCKS DG, JABLONSKI PE, KREBS MP, ANGEVINE CM, DALE H, ISENBARGER TA, PECK RF, POHLSCHRODER M, SPUDICH JL, JUNG KW, ALAM M, FREITAS T, HOU S, DANIELS CJ, DENNIS PP, OMER AD, EBHARDT H, LOWE TM, LIANG P, RILEY M, HOOD L, DASSARMA S. Genome sequence of Halobacterium species NRC-1. Proc Natl Acad Sci U S A. 2000;97:12176–81. doi: 10.1073/pnas.190337797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIKOLOV DB, CHEN H, HALAY ED, HOFFMAN A, ROEDER RG, BURLEY SK. Crystal structure of a human TATA box-binding protein/TATA element complex. Proc Natl Acad Sci U S A. 1996;93:4862–7. doi: 10.1073/pnas.93.10.4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OUHAMMOUCH M. Transcriptional regulation in Archaea. Curr Opin Genet Dev. 2004;14:133–8. doi: 10.1016/j.gde.2004.01.002. [DOI] [PubMed] [Google Scholar]
- OUHAMMOUCH M, DEWHURST RE, HAUSNER W, THOMM M, GEIDUSCHEK EP. Activation of archaeal transcription by recruitment of the TATA-binding protein. Proc Natl Acad Sci U S A. 2003;100:5097–102. doi: 10.1073/pnas.0837150100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OUHAMMOUCH M, WERNER F, WEINZIERL RO, GEIDUSCHEK EP. A fully recombinant system for activator-dependent archaeal transcription. J Biol Chem. 2004;279:51719–21. doi: 10.1074/jbc.C400446200. [DOI] [PubMed] [Google Scholar]
- PACE NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–40. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- PAULE MR, WHITE RJ. Survey and summary: transcription by RNA polymerases I and III. Nucleic Acids Res. 2000;28:1283–98. doi: 10.1093/nar/28.6.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAYTUBI S, WHITE MF. The crenarchaeal DNA damage-inducible transcription factor B paralogue TFB3 is a general activator of transcription. Mol Microbiol. 2009;72:1487–99. doi: 10.1111/j.1365-2958.2009.06737.x. [DOI] [PubMed] [Google Scholar]
- PEREIRA SL, REEVE JN. Histones and nucleosomes in Archaea and Eukarya: a comparative analysis. Extremophiles. 1998;2:141–8. doi: 10.1007/s007920050053. [DOI] [PubMed] [Google Scholar]
- PERSENGIEV SP, ZHU X, DIXIT BL, MASTON GA, KITTLER EL, GREEN MR. TRF3, a TATA-box-binding protein-related factor, is vertebrate-specific and widely expressed. Proc Natl Acad Sci U S A. 2003;100:14887–91. doi: 10.1073/pnas.2036440100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PEYROCHE G, MILKEREIT P, BISCHLER N, TSCHOCHNER H, SCHULTZ P, SENTENAC A, CARLES C, RIVA M. The recruitment of RNA polymerase I on rDNA is mediated by the interaction of the A43 subunit with Rrn3. Embo J. 2000;19:5473–82. doi: 10.1093/emboj/19.20.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PUGH BF. Control of gene expression through regulation of the TATA-binding protein. Gene. 2000;255:1–14. doi: 10.1016/s0378-1119(00)00288-2. [DOI] [PubMed] [Google Scholar]
- REEVE JN. Archaeal chromatin and transcription. Mol Microbiol. 2003;48:587–98. doi: 10.1046/j.1365-2958.2003.03439.x. [DOI] [PubMed] [Google Scholar]
- REICHLEN MJ, MURAKAMI KS, FERRY JG. Functional analysis of the three TATA binding protein homologs in Methanosarcina acetivorans. J Bacteriol. 2010;192:1511–7. doi: 10.1128/JB.01165-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RENFROW MB, NARYSHKIN N, LEWIS LM, CHEN HT, EBRIGHT RH, SCOTT RA. Transcription factor B contacts promoter DNA near the transcription start site of the archaeal transcription initiation complex. J Biol Chem. 2004;279:2825–31. doi: 10.1074/jbc.M311433200. [DOI] [PubMed] [Google Scholar]
- ROWLANDS T, BAUMANN P, JACKSON SP. The TATA-binding protein: a general transcription factor in eukaryotes and archaebacteria. Science. 1994;264:1326–9. doi: 10.1126/science.8191287. [DOI] [PubMed] [Google Scholar]
- SANTANGELO TJ, CUBONOVA L, JAMES CL, REEVE JN. TFB1 or TFB2 Is Sufficient for Thermococcus kodakaraensis Viability and for Basal Transcription in Vitro. J Mol Biol. 2007;367:344–57. doi: 10.1016/j.jmb.2006.12.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAXENA A, MA B, SCHRAMM L, HERNANDEZ N. Structure-function analysis of the human TFIIB-related factor II protein reveals an essential role for the C-terminal domain in RNA polymerase III transcription. Mol Cell Biol. 2005;25:9406–18. doi: 10.1128/MCB.25.21.9406-9418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHE Q, SINGH RK, CONFALONIERI F, ZIVANOVIC Y, ALLARD G, AWAYEZ MJ, CHAN-WEIHER CC, CLAUSEN IG, CURTIS BA, DE MOORS A, ERAUSO G, FLETCHER C, GORDON PM, HEIKAMP-DE JONG I, JEFFRIES AC, KOZERA CJ, MEDINA N, PENG X, THI-NGOC HP, REDDER P, SCHENK ME, THERIAULT C, TOLSTRUP N, CHARLEBOIS RL, DOOLITTLE WF, DUGUET M, GAASTERLAND T, GARRETT RA, RAGAN MA, SENSEN CW, VAN DER OOST J. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc Natl Acad Sci U S A. 2001;98:7835–40. doi: 10.1073/pnas.141222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHOCKLEY KR, WARD DE, CHHABRA SR, CONNERS SB, MONTERO CI, KELLY RM. Heat shock response by the hyperthermophilic archaeon Pyrococcus furiosus. Appl Environ Microbiol. 2003;69:2365–71. doi: 10.1128/AEM.69.4.2365-2371.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SOPPA J. Transcription initiation in Archaea: facts, factors and future aspects. Mol Microbiol. 1999;31:1295–305. doi: 10.1046/j.1365-2958.1999.01273.x. [DOI] [PubMed] [Google Scholar]
- SPITALNY P, THOMM M. Analysis of the open region and of DNA-protein contacts of archaeal RNA polymerase transcription complexes during transition from initiation to elongation. J Biol Chem. 2003;278:30497–505. doi: 10.1074/jbc.M303633200. [DOI] [PubMed] [Google Scholar]
- TEUFEL K, BLEIHOLDER A, GRIESBACH T, PFEIFER F. Variations in the multiple tbp genes in different Halobacterium salinarum strains and their expression during growth. Arch Microbiol. 2008;190:309–18. doi: 10.1007/s00203-008-0383-5. [DOI] [PubMed] [Google Scholar]
- THOMM M. Transcription: Mechanism and Regulation. In: CAVICCHIOLI R, editor. Archaea: Molecular and Cellular Biology. ASM Press; Washington, DC: 2007. [Google Scholar]
- THOMM M, REICH C, GRUNBERG S, NAJI S. Mutational studies of archaeal RNA polymerase and analysis of hybrid RNA polymerases. Biochem Soc Trans. 2009;37:18–22. doi: 10.1042/BST0370018. [DOI] [PubMed] [Google Scholar]
- THOMPSON DK, PALMER JR, DANIELS CJ. Expression and heat-responsive regulation of a TFIIB homologue from the archaeon Haloferax volcanii. Mol Microbiol. 1999;33:1081–92. doi: 10.1046/j.1365-2958.1999.01551.x. [DOI] [PubMed] [Google Scholar]
- THOMSEN J, DE BIASE A, KACZANOWSKI S, MACARIO AJ, THOMM M, ZIELENKIEWICZ P, MACCOLL R, CONWAY DE MACARIO E. The basal transcription factors TBP and TFB from the mesophilic archaeon Methanosarcina mazeii: structure and conformational changes upon interaction with stress-gene promoters. J Mol Biol. 2001;309:589–603. doi: 10.1006/jmbi.2001.4705. [DOI] [PubMed] [Google Scholar]
- UJVARI A, LUSE DS. RNA emerging from the active site of RNA polymerase II interacts with the Rpb7 subunit. Nat Struct Mol Biol. 2006;13:49–54. doi: 10.1038/nsmb1026. [DOI] [PubMed] [Google Scholar]
- VIERKE G, ENGELMANN A, HEBBELN C, THOMM M. A novel archaeal transcriptional regulator of heat shock response. J Biol Chem. 2003;278:18–26. doi: 10.1074/jbc.M209250200. [DOI] [PubMed] [Google Scholar]
- WERNER F. Structure and function of archaeal RNA polymerases. Mol Microbiol. 2007;65:1395–404. doi: 10.1111/j.1365-2958.2007.05876.x. [DOI] [PubMed] [Google Scholar]
- WERNER F, WEINZIERL RO. A recombinant RNA polymerase II-like enzyme capable of promoter-specific transcription. Mol Cell. 2002;10:635–46. doi: 10.1016/s1097-2765(02)00629-9. [DOI] [PubMed] [Google Scholar]
- WILKINSON SP, OUHAMMOUCH M, GEIDUSCHEK EP. Transcriptional activation in the context of repression mediated by archaeal histones. Proc Natl Acad Sci U S A. 2010;107:6777–81. doi: 10.1073/pnas.1002360107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WOESE CR, FOX GE. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci U S A. 1977;74:5088–90. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WOESE CR, KANDLER O, WHEELIS ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990;87:4576–9. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WOYCHIK NA, LANE WS, YOUNG RA. Yeast RNA polymerase II subunit RPB9 is essential for growth at temperature extremes. J Biol Chem. 1991;266:19053–5. [PubMed] [Google Scholar]
- WU J, GRUNSTEIN M. 25 years after the nucleosome model: chromatin modifications. Trends Biochem Sci. 2000;25:619–23. doi: 10.1016/s0968-0004(00)01718-7. [DOI] [PubMed] [Google Scholar]
- XIE Y, REEVE JN. Transcription by Methanothermobacter thermautotrophicus RNA polymerase in vitro releases archaeal transcription factor B but not TATA-box binding protein from the template DNA. J Bacteriol. 2004;186:6306–10. doi: 10.1128/JB.186.18.6306-6310.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YAMAMOTO T, MATSUDA T, INOUE T, MATSUMURA H, MORIKAWA M, KANAYA S, KAI Y. Crystal structure of TBP-interacting protein (Tk-TIP26) and implications for its inhibition mechanism of the interaction between TBP and TATA-DNA. Protein Sci. 2006;15:152–61. doi: 10.1110/ps.051788906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG G, CAMPBELL EA, MINAKHIN L, RICHTER C, SEVERINOV K, DARST SA. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 A resolution. Cell. 1999;98:811–24. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
- ZHU W, ZENG Q, COLANGELO CM, LEWIS M, SUMMERS MF, SCOTT RA. The N-terminal domain of TFIIB from Pyrococcus furiosus forms a zinc ribbon. Nat Struct Biol. 1996;3:122–4. doi: 10.1038/nsb0296-122. [DOI] [PubMed] [Google Scholar]
- ZILLIG W, STETTER KO, TOBIEN M. DNA-dependent RNA polymerase from Halobacterium halobium. Eur J Biochem. 1978;91:193–9. doi: 10.1111/j.1432-1033.1978.tb20951.x. [DOI] [PubMed] [Google Scholar]