Abstract
Gene duplications are one of the primary driving forces in the evolution of genomes and genetic systems. Gene duplicates account for 8–20% of the genes in eukaryotic genomes, and the rates of gene duplication are estimated at between 0.2% and 2% per gene per million years. Duplicate genes are believed to be a major mechanism for the establishment of new gene functions and the generation of evolutionary novelty, yet very little is known about the early stages of the evolution of duplicated gene pairs. It is unclear, for example, to what extent selection, rather than neutral genetic drift, drives the fixation and early evolution of duplicate loci. Analysis of recently duplicated genes in the Arabidopsis thaliana genome reveals significantly reduced species-wide levels of nucleotide polymorphisms in the progenitor and/or duplicate gene copies, suggesting that selective sweeps accompany the initial stages of the evolution of these duplicated gene pairs. Our results support recent theoretical work that indicates that fates of duplicate gene pairs may be determined in the initial phases of duplicate gene evolution and that positive selection plays a prominent role in the evolutionary dynamics of the very early histories of duplicate nuclear genes.
Gene duplications are one of the primary driving forces in the evolution of genomes and genetic systems (1, 2). Duplicate genes are believed to be a major mechanism for the establishment of new gene functions (3) and the generation of evolutionary novelty (4). Around 15% of genes in the human genome are believed to arise from duplication events, whereas gene duplicates account for 8–20% of the Drosophila melanogaster, Caenorhabditis elegans, and Saccharomyces cervisiae genomes (5, 6). The rates of gene duplication in these model species are estimated at between 0.2% and 2% per gene per million years (5, 6).
Most studies on the evolution of gene duplications examine the macroevolutionary patterns of gene diversification (7–9) and focus on the fates of duplicate loci long after their establishment and fixation within species. In contrast, we know very little about the initial stages of duplicate gene evolution. For example, it is unclear whether the process of evolutionary fixation of duplicate loci from a single individual to the entire species is governed by random genetic drift or through the action of positive selection acting on an adaptive phenotype associated with the gene duplication event (10, 11). We simply do not know which of these two evolutionary forces govern the critical early phases of duplicate gene evolution.
Theoretical studies suggest that the relative importance of these two evolutionary forces, neutral genetic drift and positive selection, differs depending on the ultimate functional fate of the duplicate gene pair (10). Gene duplication can lead to one of several functional relationships between duplicate gene copies, including (i) loss of gene function by pseudogene formation, (ii) the establishment of redundant loci (12), (iii) the evolutionary diversification of gene function by means of neofunctionalization, or (iv) the partitioning of ancestral gene function by a process of subfunctionalization (10, 13). Both pseudogenes and completely redundant unlinked genes are fixed by neutral genetic drift (11). Fixation of an unlinked duplicate gene by means of the subfunctionalization of ancestral functions is also believed to occur by genetic drift (10). Gene preservation by neofunctionalization or functional divergence, however, appears to be driven by selective advantage of the duplicate locus (11, 13). The precise mechanism of fixation of duplicated loci depends on several factors, including the relative levels of adaptive, neutral and deleterious mutations acting on duplicate gene pairs, the selection coefficients on duplicate loci, and, in some cases, the effective population size (10, 13).
We have undertaken a molecular population genetic study of three recently duplicated genes in the Arabidopsis thaliana genome to assess what evolutionary forces are associated with the initial phases of duplicate gene evolution. In at least two cases, there are significantly reduced levels of nucleotide polymorphism of the duplicate gene copies, which suggests that adaptive sweeps are associated with the fixation of these duplicated loci. Moreover, progenitor gene copies of two of the three duplicate pairs also show evidence of positive selection, demonstrating that adaptive forces may also govern the early evolutionary dynamics of progenitor loci. This study investigates the molecular evolutionary forces associated with the initial phases of gene duplications and suggests that positive selection characterizes the initial period of duplicate gene evolution. This work also confirms models that suggest that the ultimate fate of duplicate loci may be determined early in their evolutionary history (10).
Materials and Methods
Database Identification of A. thaliana Duplicate Gene Pairs. The database of duplicated genes was obtained from a duplicate gene database of the A. thaliana genome, available at www.csi.uoregon.edu/projects/genetics/duplications/letters (5, 6). Putative pseudogenes, transposable elements, and members of large multigene families were removed from this database (5, 6). We limited our analyses to recent duplications with synonymous site divergence, Ks, <0.02. Gene pairs that were redundant or of unknown annotation, tandem or closely linked duplications (<20 intervening genes), triplicated loci, and those associated with transposable elements were also excluded from our analyses.
Isolation and Sequencing of Alleles. Genomic DNA was isolated from young leaves of 14–16 A. thaliana ecotypes (Table 3, which is published as supporting information on the PNAS web site) and one Arabidopsis lyrata accession by using the Plant DNeasy Mini kit (Qiagen, Valencia, CA). A. lyrata seed from a Karhumaki, Russia, population was provided by O. Savolainen (University of Oulu, Oulu, Finland) and Helmi Kuittinen (University of Barcelona, Barcelona). PCR primers were designed from the Col-0 genomic gene sequences by using primer3 (www-genome.wi.mit.edu/genome_software/other/primer3.html). PCR of A. thaliana and A. lyrata samples was performed with Taq DNA polymerase (Roche). Flanking regions from some of the gene duplicates were isolated from A. lyrata by using thermal isometric interlaced PCR (14). DNA fragments amplified from A. thaliana were purified with the QIAquick PCR Purification and Gel Extraction kits (Qiagen) and sequenced directly. Amplified A. lyrata products were cloned with the TOPO TA PCR Cloning Kit (Invitrogen), and plasmid DNA from five to six independent clones was sequenced. DNA sequencing was conducted at the North Carolina State University Genome Research Laboratory with a Prism 3700 96-capillary automated sequencer (Applied Biosystems). All polymorphisms were visually confirmed, and ambiguous polymorphisms were rechecked with PCR reamplification and sequencing. GenBank accession numbers for these genes are AY469987 to AY470073.
Expression Analyses. Total RNA was extracted from whole seedlings and floral bud (<1 mm) tissues of Col-0 plants with the RNeasy kit (Qiagen). Poly(A)+ RNA was isolated from DNase-treated (Ambion, Austin, TX) total RNA by using Oligotex spin columns (Qiagen). The cDNA was derived from poly(A)+ RNA with the Retroscript reverse transcription kit (Ambion). RT-PCR was conducted for each gene pair by using primers anchored in exons but designed to amplify across an intron. To distinguish between the expression of the gene duplicates, cleaved amplified polymorphic sequence (CAPS) markers (15) or derived CAPS (16) markers were designed. These markers were designed to allow for differential cleavage of each copy of the duplicate gene pair with specific restriction enzymes. The constitutively expressed gene EF1-α was used as a control for cDNA quality and to equalize loadings of RT-PCR products between seedling and floral reactions.
Molecular Population Genetic Data Analysis. Sequences were visually aligned against the A. thaliana sequence previously identified in the Arabidopsis whole genome sequence (17). The A. lyrata ortholog was used as the outgroup in the analyses. Interspecific nucleotide sequence divergence distances were estimated from silent sites with the Kimura 2-parameter model by using mega 2.1 software (www.megasoftware.net) (18). Polymorphism analyses were conducted by using dnasp 3.51(www.ub.es/dnasp) (19). Levels of silent site nucleotide diversity per site were estimated as π (20) and θW (21). Haplotype networks were constructed from substitution polymorphisms by using a maximum parsimony criterion in paup* 4.0 (22).
The Hudson–Kreitman–Aguade (HKA) test (23) was conducted by using silent site changes. The following loci were chosen as the neutral reference loci in these tests: AP1 (24), AP3 and PI (25), CAL (26), and F3H and FAH1 (27). The HKA multilocus test was conducted with silent site changes by using a program available from Jody Hey (Rutgers University, Piscataway, NJ). Individual HKA tests were also independently conducted for each of the loci against each of the neutral reference loci by using dnasp 3.51 (19). Probabilities of each of these pairwise tests were corrected by using Sime's method (28). The HKA test, which is comparative, appears to be the most robust test for selection in A. thaliana. Other tests of selection that rely on site frequency spectra are difficult to interpret given the predominantly selfing nature of this species (25).
Results and Discussion
Recently Duplicated Genes in the Arabidopsis Genome. The molecular evolutionary analysis of the early stages of duplicate gene evolution begins with the identification of recently duplicated gene pairs. In a survey of the A. thaliana genome, 3,712 gene pairs were identified, indicating that >29% of the genes in this plant genome are duplicated (5). Recently duplicated and unlinked genes were based on two main criteria: (i) a synonymous site divergence of <2%, which is ≈20% of the mean nucleotide divergence between A. thaliana and its closest relative, A. lyrata, and (ii) separation by at least 20 intervening genes. The second criterion circumvents any possible confounding effects of gene conversion events on the patterns of nucleotide polymorphisms for the duplicate gene pairs.
Only 68 valid gene pairs (<2%) of the total number of gene pairs in the A. thaliana genome had synonymous site divergence of <2%. Most of these duplications resulted in closely linked gene pairs (≈57%) or were associated with transposable elements (≈24%). Three gene pairs (≈4%) appeared to be part of a triplicated locus. Only four gene pairs (≈6%) were dispersed duplicate gene pairs, and, of these, one pair was found only in the Col-0 ecotype. This small number is not unusual given the low rate of gene duplication in eukaryotic genomes (5, 6).
The three dispersed duplicate gene pairs in our analysis (Fig. 1; see also Fig. 5, which is published as supporting information on the PNAS web site) include histidinol phosphate aminotransferase-like genes assigned the Arabidopsis annotated gene identification numbers AT1G71920 and AT5G10330. We refer to these as GD1-A and GD1-B (GD for gene duplicate). The other two duplicate genes include one pair encoding a predicted membrane-bound protein with six sodium dicarboxylic acid symporter domains (AT3G03700 and AT3G04440), which we will refer to as GD2-A and GD2-B, and a pair encoding an expressed protein of unknown function (AT1G05090 and AT4G20720), which we will refer to as GD3-A and GD3-B.
Fig. 1.
Recent duplicate genes in the A. thaliana genome. Thin arrows indicate translation start sites, and arrowheads depict positions of PCR primers. The genes are identified according to the identification number assigned by the Arabidopsis Information Resource. The chromosomes where these genes are located are also indicated.
PCR screening of 38 ecotypes indicate that all of the genes in these three duplicate gene pairs are either fixed or close to fixation in A. thaliana, with the gene being present at frequencies >97%. Analyses of flanking genes indicate that these duplicate loci were not generated by segmental duplication events and instead are the result of small-scale duplications of 2–5 kb. The boundaries of the all of the duplication events include <1 kb of duplicated flanking sequence (Fig. 5). Between 220 and 750 bps of sequence upstream of the translation start site is duplicated in all gene pairs, indicating that only a small portion of the promoter is conserved between duplicate loci.
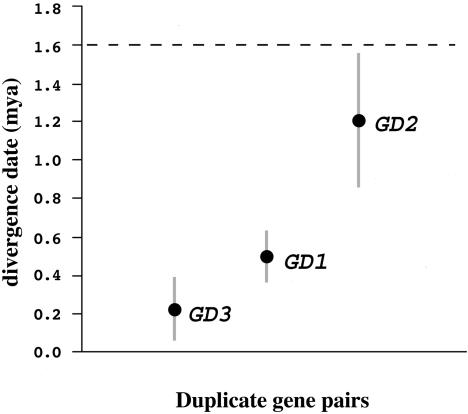
These three gene pairs are the products of duplications that occurred after the divergence of A. thaliana and the closely related species A. lyrata ≈5.2 million years ago (mya) (29). Based on a silent nucleotide site molecular clock for each gene calibrated with the A. thaliana/A. lyrata divergence date, we can estimate the date of duplication for these three gene pairs (Fig. 2). The GD1 and GD3 gene pairs were duplicated relatively recently, with duplications occurring 0.24 ± 0.16 mya and 0.50 ± 0.14 mya, respectively (Table 1). The GD2 gene duplication event occurred earlier, with molecular clock estimates suggesting a divergence date of 1.20 ± 0.35 mya between the pair (Table 1). The levels of silent site divergence between the duplicate gene pairs (0.5–1.8%) are the same order of magnitude as mean within-species nucleotide diversity levels for nuclear genes in A. thaliana (25).
Fig. 2.
Divergence date estimates for duplicate gene pairs. The thin line shows the standard errors around the estimates. The dashed line is positioned at 4Ne years (= generations) given an estimate of Ne for A. thaliana at 4 × 105. The duplication events that gave rise to each duplicate pair occurred after the species split between A. lyrata and A. thaliana, which occurred ≈5.2 mya (29).
Table 1. Molecular evolutionary parameters for duplicate gene pairs.
| Duplicate gene pair | Gene annotation number* | Ks† | Divergence time, mya | n | Length, bp | π‡ | θW‡ |
|---|---|---|---|---|---|---|---|
| GD1 | 0.014 ± 0.004 | 0.50 ± 0.14 | |||||
| A | AT1G71920 | 16 | 966 | 0.004 | 0.006 | ||
| B | AT5G10330 | 16 | 955 | 0.001 | 0.003 | ||
| GD2 | 0.018 ± 0.005 | 1.20 ± 0.035 | |||||
| A | AT3G03700 | 16 (15)§ | 893 | 0.001 (0.000)§ | 0.002 (0.000)§ | ||
| B | AT3G04440 | 15 | 893 | 0.002 | 0.003 | ||
| GD3 | 0.005 ± 0.004 | 0.24 ± 0.16 | |||||
| A | AT1G05090 | 16 | 1064 | 0.002 | 0.004 | ||
| B | AT4G20720 | 14¶ | 1064 | 0.001 | 0.002 |
Gene identification number designated by the Arabidopsis Information Resource.
Silent site divergence between duplicate gene pairs (substitutions per site).
Nucleotide polymorphism levels based on silent sites.
Figure in parentheses based on exclusion of Ita-0 ecotype.
Unable to amplify in two ecotypes.
Identifying the Progenitor and Duplicate Copies. Approximately 0.8–1.0 kb was sequenced for each gene pair for 14–16 A. thaliana ecotypes and one A. lyrata accession. Haplotype networks for each duplicate gene pair, rooted by using the A. lyrata ortholog as an outgroup sequence (Fig. 3), permit identification of the progenitor and duplicate genes for two of these gene pairs. Inspection of the networks indicates that alleles from the B copies of GD1 and GD3 form monophyletic groups derived from the A-copy alleles of these loci (Fig. 3 A and C). GD1-B and GD3-B thus appear to be duplicated from the progenitor A copies.
Fig. 3.
Maximum parsimony haplotype networks for GD1 (A), GD2 (B), and GD3 (C) gene duplicate pairs. All trees are rooted with the appropriate A. lyrata orthologues. Circles and squares indicate A and B gene copy haplotypes, respectively. The diamonds depict the A. lyrata haplotypes as the outgroup sequence. Short lines represent mutational changes corresponding to substitution polymorphisms, and black circles indicate inferred intermediate haplotypes. Numbers of substitutions between the A. lyrata and A. thaliana haplotypes are shown. The consistency index for each tree = 1.00, and the number of equally parsimonious trees are 1 for GD1, 26 for GD2, and 3 for GD3. In the last two cases, there are minor differences in the placement of specific haplotypes. These differences do not change the relationship of A and B haplotypes with each other.
The haplotype network is uninformative as to which gene is the progenitor and which is the copy for GD2, the oldest duplicate gene pair in our study (Fig. 3C). However, examination of the structure of the GD2-A and GD2-B copies suggests GD2-B is the duplicate locus (Fig. 5B). Immediately downstream of GD2-A is a gene encoding a putative polyribonucleotide nucleotidyltransferase (Arabidopsis Information Resource annotation no. AT3G03710). This gene is only partially duplicated in the 3′ region of GD2-B. The most parsimonious explanation for this is that GD2-A is the progenitor gene, and the duplication event resulting in the GD2-B copy included only part of the downstream flanking gene. Furthermore, the 5′ flanking region of the single-copy GD2 from A. lyrata can be aligned only with the 5′ flanking region of GD2-A (data not shown).
Expression Analysis of Gene Duplicates. The expression of these gene duplicates was examined to determine whether the duplicate gene copies continue to be transcribed and thus remain functional. Expression for these genes was assayed by using RT-PCR analysis on vegetative (seedlings) and reproductive (floral) tissues of A. thaliana. GD1 is the only gene pair with completely divergent expression patterns between duplicate copies (Fig. 4). GD1-B is expressed in both vegetative and reproductive tissues, although no transcripts were detected from the progenitor, GD1-A. Moreover, GD1-A is also not found in extant A. thaliana EST databases (www.arabidopsis.org). The structure of the GD1-A gene, however, does not have any features that suggest that it is a pseudogene, suggesting that it may be expressed either at very low levels or in tissues and/or developmental conditions that have not been tested.
Fig. 4.
Expression analyses of duplicate genes. The RT-PCR for gene duplicates are shown for floral and seedling tissues and genomic DNA positive controls. The specific restriction digests used as CAPS and/or dCAPS markers and the negative control (-RT) are indicated. (A) GD1 is expressed in both reproductive (floral) and vegetative (seedling) tissue. GD1-A and GD1-B share a common DdeI restriction site in intron 6, but GD1-A has an additional unique DdeI site in exon 7, producing three bands when genomic PCR product is digested. DdeI does not cut RT-PCR products from either floral or seedling cDNA, indicating that GD1-A is absent from these cDNA pools. RsaI cuts GD1-B but not GD1-A. RT-PCR product from floral and seedling DNA is completely digested, indicating that only GD1-B is expressed in these tissues. (B) GD2 is expressed in floral tissue, but only weakly in seedling tissue (data not shown). Both the unprocessed (higher-molecular-weight band) and processed mRNAs (white arrowheads) are expressed. GD2-A and GD2-B both share a common BccI restriction site in intron 2, but GD2-A has an additional unique BccI site in exon 1, producing three bands when genomic PCR product is digested. A BccI digest of floral RT-PCR product digests the unprocessed amplicons but not the processed products. This indicates that GD2-A is present in the unprocessed floral mRNA pool but not the processed floral mRNA pool, whereas GD2-B is present in both floral pools. VspI specifically cuts GD2-B in intron 1. A VspI digest of floral RT-PCR product cuts the unprocessed RT-PCR product, thus confirming that GD2-B is present in the unprocessed mRNA pool. (C) GD3 is expressed in both flowers and seedlings. RsaI cuts GD3-A specifically but not GD3-B. RT-PCR product digested with RsaI produces both cut and uncut product, indicating that GD3-A and GD3-B share the same general expression pattern in these tissues.
Both duplicates of GD2 and GD3 are expressed in vegetative and reproductive tissues (Fig. 4), although the vegetative RT-PCR product for GD2 is very weak (data not shown). Interestingly, both putatively processed and unprocessed mRNA of GD2 are expressed in floral tissues (Fig. 4) and weakly in seedlings (data not shown). Both GD2-A and GD2-B express the unprocessed mRNA, identifiable by the presence of an intervening intron. The processed mRNA that lacks this intron is found only for the duplicated GD2-B copy (Fig. 4).
Reduced Variation of Two Recently Duplicated Genes Associated with Selective Sweeps. What evolutionary forces drive the fixation and early evolution of duplicate genes? This question can be addressed by examining the levels and patterns of nucleotide polymorphisms for duplicate gene pairs, provided that the duplication was sufficiently recent to retain the signature of any possible selective events that may accompany the fixation process. The mean time to fixation of neutral alleles is 4Ne generations (30). The effective population size of A. thaliana is ≈4 × 105, as estimated from four neutral loci (K. M. Olsen, North Carolina State University, Raleigh, NC, personal communication), and thus the mean time of species-wide fixation of a duplicate locus by neutral-drift processes would be ≈1.6 million years. This is larger than the age of all of the duplicate loci in our study, and thus it might be possible to ascertain what evolutionary forces may be associated with the recent fixation of these duplicate gene copies.
The estimated levels of molecular variation are low for all duplicated gene pairs (Table 1). Levels of silent site nucleotide diversity, π (20), are 0.001 for GD1-B and GD3-B, and π = 0.002 for GD2-B. For GD1 and GD3, levels of silent site nucleotide diversity are higher for the progenitor A copies of these loci, with π = 0.003 and 0.004, respectively. In contrast, π for the progenitor GD2-A is lower (π = 0.001) than for the duplicate B copy (Table 1). In all cases, the levels of silent site nucleotide diversity for these duplicate gene pairs are lower than the mean of π = 0.007 estimated for A. thaliana nuclear genes (25).
The reduced levels of variation for several of these duplicate gene copies suggest that they may have undergone a recent adaptive sweep (31). Whether these reduced levels of nucleotide polymorphisms do indeed deviate significantly from expectations based on the neutral-equilibrium model of molecular evolution can be tested with the HKA test (23). This test is based on the prediction that levels of intraspecific polymorphisms and interspecific divergence are correlated under a neutral-drift process, and it relies on comparisons of test genes with neutral reference loci. Six previously studied A. thaliana nuclear genes were used as the neutral reference loci for HKA tests (24–27); these six genes were chosen based on evidence that their levels and patterns of variation are consistent with the neutral model.
Based on a multilocus HKA test using silent sites, none of these six reference genes has levels of polymorphisms that differ significantly from each other (X2 = 1.261, P < 0.94). In contrast, a multilocus HKA test that includes the six gene duplicate pairs as well as the six reference loci is significant (X2 = 23.193, P < 0.05), indicating that several of these duplicate loci have levels of variation that differ significantly from those of the neutral reference loci or from each other (Fig. 6, which is published as supporting information on the PNAS web site). The duplicate copy genes, GD1-B and GD3-B, provide the largest contributions to the multilocus HKA test statistic, collectively accounting for >52% of the test statistic X2. For these two genes, the deviations of nucleotide polymorphisms from neutral expectations arise from a deficit of observed intraspecific polymorphisms within A. thaliana.
Because the multilocus HKA test result is significant, pairwise HKA tests were conducted for each of the duplicated loci against the six reference loci to determine which specific loci are responsible for the significant deviation from the neutral drift model (Table 2). The results from these separate tests support the multilocus HKA analysis. HKA tests for GD1-B are significant at the nominal level (P < 0.05) for all of the six reference genes. For GD3-B, HKA tests are significant at the nominal level (P < 0.05) against the AP3 locus and marginally significant (P < 0.1) for three other genes. We corrected the P values for these multiple nonindependent tests across all six tests for the gene duplicates by using Sime's method (28). The Sime's corrected probabilities are significant at the nominal and Bonferroni-corrected levels for GD1-B (P < 0.004) and at the nominal level for GD3-B (P < 0.018; Table 2).
Table 2. Probability values from pairwise HKA tests.
| Reference locus
|
Sime's corrected probability
|
||||||
|---|---|---|---|---|---|---|---|
| Gene | AP3 | F3H | PI | API | CAL | FAH | |
| GD1 | |||||||
| A | 0.043† | 0.063* | 0.062* | 0.081* | 0.110 | 0.190 | 0.02† |
| B | 0.008‡ | 0.009‡ | 0.011† | 0.014† | 0.016† | 0.028† | 0.003‡ |
| GD2 | |||||||
| A | 0.072* | 0.110 | 0.110 | 0.150 | 0.180 | 0.270 | 0.04† |
| A (minus Ita-0) | 0.008‡ | 0.012† | 0.015† | 0.018† | 0.020† | 0.026† | 0.004‡ |
| B | 0.100 | 0.150 | 0.150 | 0.190 | 0.320 | 0.400 | 0.046† |
| GD3 | |||||||
| A | 0.120 | 0.170 | 0.160 | 0.210 | 0.270 | 0.410 | 0.053* |
| B | 0.042† | 0.053* | 0.054* | 0.075* | 0.110 | 0.130 | 0.018† |
Nominal significance levels: *, P < 0.10; †, P < 0.05; ‡, P < 0.01; §, P < 0.001.
The multilocus HKA results, and, to some extent, the pairwise HKA tests thus reveal that significantly reduced variations characteristic of adaptive sweeps are associated with the early evolution, and likely the fixation, of these two gene copies in the A. thaliana genome and that positive selection may act soon after gene duplication occurs. In contrast, the level of silent site nucleotide diversity for the duplicate copy GD2-B is not significantly lower than predicted from the neutral-drift model (Table 2). None of the HKA tests is significant for this locus (P < 0.10–0.40), although the Sime's corrected probability is significant at the nominal level (P < 0.046).
Evidence for Positive Selection at Progenitor Loci. Positive selection also appears to govern the evolutionary dynamics of at least two progenitor loci early in the establishment of gene duplicate pairs in the A. thaliana genome. Both the GD1-A and GD2-A progenitor gene copies have molecular signatures associated with the action of adaptive selection. The GD1-A gene has significantly reduced levels of nucleotide variation. The HKA tests for this gene were significant at the nominal level (P < 0.05), compared to the AP3 locus, and marginally significant at the nominal level (P < 0.1) for three other loci (Table 2). The Sime's corrected probability of P < 0.02 is significant only at the nominal level.
The pattern of polymorphism for GD2-A is characteristic of a partial selective sweep. All six polymorphisms in this gene are attributable to the presence of a single allele from the Moroccan Ita-0 accession; all of the other 15 alleles have no nucleotide polymorphisms. If we exclude the Ita-0 allele from the analysis, then π = 0.000 for GD2-A, all HKA tests compared with the six reference loci are significant at the nominal level (P < 0.008–0.026), and the Sime's corrected probability is significant at both the nominal and Bonferroni-corrected levels (P < 0.003; Table 2).
In contrast, the results of the tests for selection for the GD3-A progenitor gene suggest that it is evolving neutrally. HKA tests do not show any significant deviations in nucleotide diversity levels between this gene and any of the six neutral reference loci (P < 0.13–0.22; Table 2), with a Sime's corrected probability that is only marginally significant at the nominal level (P < 0.053). Sequence analysis reveals that two single base insertion events in the GD3-A coding region in 5 of 16 A. thaliana ecotypes result in frame-shift mutations and premature stop codons. Thus, it appears that GD3-A is in the process of becoming a pseudogene.
Evolutionary Forces Associated with Recent Gene Duplication. Determining the evolutionary forces that surround the fixation and early evolution of duplicate loci provides crucial insights into the dynamics of genome diversification by means of gene duplication events. Our study indicates that the signature of recent positive selection is observed for the duplicate gene copies in two of three gene pairs examined, suggesting that selective sweeps can play a pivotal role in the early evolution of these duplicated copies. Given the recent duplication events (<0.5 mya) associated with the origin of these two gene pairs, it is possible that these selective sweeps are associated with the fixation of the duplicate copies. We can estimate the selection coefficients for these sweeps based on the time of fixation of these duplicate loci. From the neutral theory, the mean time to fixation of a selected allele is equal to (2/s) ln(2Ne), where s is the selection coefficient and Ne is the effective population size (30). Assuming Ne of 4 × 105 (see above) and that the fixation of the duplicate locus required the entire period from the duplication event to the present to complete, we estimate the selection coefficients for the possible adaptive fixation of these duplicate gene copies as s = 10–4 to 10–5. This is a conservative estimate, given that the fixation time for the duplicate gene copy is likely shorter.
The finding that the fixation and preservation of at least some duplicate genes in the Arabidopsis genome is driven by positive selection allows us to differentiate between alternative models of duplicate gene preservation (10, 13). Neofunctionalization, or the evolution of functional divergence between duplicate genes, is driven by positive selection on the duplicate copy (11, 13). In contrast, subfunctionalization as a mechanism for duplicate gene preservation is thought to occur primarily through neutral-drift processes at unlinked loci (10, 13), a pattern observed in only one of the three duplicate gene copies in our study. The association of adaptive sweeps with at least some of the duplicated loci may not be too surprising, given that theoretical work suggests that the probability of fixation of dispersed, unlinked duplicate loci tends to be higher for neofunctional duplicate gene pairs than for subfunctional loci (13). It would be interesting to determine the full extent of differentiation in expression and phenotypic patterns between these duplicate loci.
The progenitor gene GD2-A and, to some extent, GD1-A also show evidence for positive selection. The role of selective forces in the early evolutionary divergence of progenitor genes after a duplication event should depend on the specific functional features associated with the presence of the duplicate loci. Concurrent selection on progenitor and duplicate copies, as observed in the GD1 gene pair, is predicted by early studies that indicate that positive selection on both copies can occur simultaneously if the ancestral gene is segregating for selectively maintained alleles (32, 33). Furthermore, the incipient pseudo-genization of the neutrally evolving GD3-A locus suggests that gene loss may occur early in the process of duplicate gene evolution.
It should be noted that, because the duplicate loci were identified by using data from only one individual, the identification of duplicate gene pairs may potentially be biased toward those that are at high frequency in the species and away from those present at low frequency that could potentially be fixed by drift. Nevertheless, the pervasive action of positive selection associated with the early evolution of the duplicate gene pairs in this study generally agrees with genome-wide surveys, which demonstrate that duplicate loci persist for long periods of evolutionary time in A. thaliana (5, 6). The long mean half-life of Arabidopsis duplicate genes is ≈22 million years (5, 6), which may reflect the selective advantages of duplicate loci in this plant genome that can counteract and/or delay the loss of gene duplicates by pseudogene formation and subsequent neutral genetic drift. In contrast, half-lives for Drosophila duplicate genes are <3 million years (5, 6), and it would be instructive to determine whether there are also differences in the role of selection in the fixation of duplicate genes in these two species correlated with the differences in the rates of duplicate gene retention.
This is the first systematic study of the molecular evolutionary forces associated with the establishment of recently duplicated genes in eukaryotic genomes. Most studies of duplicate gene evolution focus on the role of selection on long-established gene pairs, providing insights into the macroevolutionary dynamics of these loci. Our investigation, which supports recent theoretical work (10, 13, 32), indicates that fates of duplicate gene pairs may be determined in the initial phases of duplicate gene evolution, and that positive selection plays a prominent role in the very early evolutionary histories of duplicated nuclear genes.
Supplementary Material
Acknowledgments
We thank members of the Purugganan Laboratory, Amy Lawton-Rauh, and Bruce Walsh for stimulating discussions and Jennifer Shoemaker and Bruce Weir for discussions on statistical methodologies. This work was supported in part by a National Institutes of Health Post-doctoral Fellowship to R.C.M. and a National Science Foundation grant to M.D.P.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CAPS, cleaved amplified polymorphic sequence; HKA, Hudson–Kreitman–Aguade; mya, million years ago; GD, gene duplicate.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY469987 and AY470073)
References
- 1.Ohno, S. (1970) Evolution by Gene Duplication (Springer, New York).
- 2.Gu, Z. L., Steinmetz, L. M., Gu, X., Scharfe, C., Davis, R. W. & Li, W. H. (2003) Nature 421, 63–66. [DOI] [PubMed] [Google Scholar]
- 3.Long, M. Y. & Langley, C. H. (1993) Science 260, 91–95. [DOI] [PubMed] [Google Scholar]
- 4.Gilbert, W., deSouza, S. J. & Long, M. Y. (1997) Proc. Natl. Acad. Sci. USA 94, 7698–7703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lynch, M. & Conery, J. S. (2000) Science 290, 1151–1155. [DOI] [PubMed] [Google Scholar]
- 6.Lynch, M. & Conery, J. C. (2001) Science 293, U2–U3. [Google Scholar]
- 7.Zhang, J. Z., Zhang, Y. P. & Rosenberg, H. F. (2002) Nat. Genet. 30, 411–415. [DOI] [PubMed] [Google Scholar]
- 8.Dermitzakis, E. T. & Clark, A. G. (2001) Mol. Biol. Evol. 18, 557–562. [DOI] [PubMed] [Google Scholar]
- 9.Lawton-Rauh, A., Buckler, E. S. & Purugganan, M. D. (1999) Mol. Biol. Evol. 16, 1037–1045. [DOI] [PubMed] [Google Scholar]
- 10.Lynch, M. & Force, A. (2000) Genetics 154, 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walsh, J. B. (1995) Genetics 139, 421–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nowak, M. A., Boerlijst, M., Cooke, J. & Smith, J. M. (1997) Nature 388, 167–171. [DOI] [PubMed] [Google Scholar]
- 13.Lynch, M., O'Hely, M., Walsh, B. & Force, A. (2001) Genetics 159, 1789–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu, Y. G. & Whittier, R. F. (1995) Genomics 25, 674–681. [DOI] [PubMed] [Google Scholar]
- 15.Konieczny, A. & Ausubel, F. M. (1993) Plant J. 4, 403–410. [DOI] [PubMed] [Google Scholar]
- 16.Neff, M. M., Neff, J. D., Chory J. & Pepper A. E. (1998) Plant J. 14, 387–392. [DOI] [PubMed] [Google Scholar]
- 17.Arabidopsis Genome Initiative (2000) Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- 18.Kumar, S., Tamura, K., Jakobsen, I. & Nei, M. (2001) Bioinformatics 17, 1244–1245. [DOI] [PubMed] [Google Scholar]
- 19.Rozas, J. & Rozas, R. (1999) Bioinformatics 15, 174–175. [DOI] [PubMed] [Google Scholar]
- 20.Nei, M. (1987) Molecular Evolutionary Genetics (Columbia Univ. Press, New York).
- 21.Watterson, G. A. (1975) Theor. Popul. Biol. 7, 256–276. [DOI] [PubMed] [Google Scholar]
- 22.Swofford, D. L. (2002) PAUP* 4.0: Phylogenetic Analysis Using Parsimony (*and Other Methods) (Sinauer, Sunderland, MA).
- 23.Hudson, R., Kreitman, M. & Aguade, M. (1987) Genetics 116, 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olsen, K. M., Womack, A., Garrett, A. R., Suddith, J. I. & Purugganan, M. D. (2002) Genetics 160, 1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purugganan, M. D. & Suddith, J. I. (1999) Genetics 151, 839–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purugganan, M. D. & Suddith, J. I. (1998) Proc. Natl. Acad. Sci. USA 95, 8130–8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aguade, M. (2001) Mol. Biol. Evol. 18, 1–9. [DOI] [PubMed] [Google Scholar]
- 28.Simes, R. J. (1986). Biometrika 73, 751–754. [Google Scholar]
- 29.Koch, M. A., Haubold, B. & Mitchell-Olds, T. (2000) Mol. Biol. Evol. 17, 1483–1498. [DOI] [PubMed] [Google Scholar]
- 30.Kimura, M. (1983) The Neutral Theory of Molecular Evolution (Cambridge Univ. Press, Cambridge, U.K.).
- 31.Barton, N. H. (1998) Genet. Res. 72, 123–133. [Google Scholar]
- 32.Walsh, B. (2003) Genetica (The Hague) 118, 279–294. [PubMed] [Google Scholar]
- 33.Spofford, J. B. (1969) Am. Nat. 103, 407–432. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.