Abstract
Although the majority of metazoan mitochondrial genomes (mtDNAs) contain the same 37 genes, including 22 encoding transfer RNAs (tRNAs), the recognition of orthologs is not always straightforward. Here we demonstrate that inferring tRNA orthologs among taxa by using anticodon triplets and deduced secondary structure can be misleading: through a process of tRNA duplication and mutation in the anticodon triplet, remolded leucine (LUUR) tRNA genes have repeatedly taken over the role of isoaccepting LCUN leucine tRNAs within metazoan mtDNA. In the present work, data from within the gastropods and a broad survey of metazoan mtDNA suggest that tRNA leucine duplication and remolding events have occurred independently at least seven times within three major animal lineages. In all cases where the mechanism of gene remolding can be inferred with confidence, the direction is the same: from LUUR to LCUN. Gene remolding and its apparent asymmetry have significant implications for the use of mitochondrial tRNA gene orders as phylogenetic markers. Remolding complicates the identification of orthologs and can result in convergence in gene order. Careful sequence-based analysis of tRNAs can help to recognize this homoplasy, improving gene-order-based phylogenetic hypotheses and underscoring the importance of careful homology assessment. tRNA remolding also provides an additional mechanism by which gene order changes can occur within mtDNA: through the changing identity of tRNA genes themselves. Recognition of these remolding events can lead to new interpretations of gene order changes, as well as the discovery of phylogenetically relevant gene dynamics that are hidden at the level of gene order alone.
Mitochondrial gene order data are being used with increasing frequency as robust molecular characters in deep-level metazoan phylogenetic studies. A fundamental assumption of molecular systematics is that orthologous genes can be recognized unambiguously. The facts that most animal mtDNAs described to date are composed of 37 genes (22 tRNAs, 2 rRNAs, and 13 protein subunits), that these genes play roles essential for oxidative metabolism, and that putative homologs have been identified in the mtDNAs of other eukaryotes (1, 2) suggest that identifying orthologs among metazoan lineages should not be problematic. In practice, however, establishing homology between mtDNA genes of distantly related organisms can be difficult at the nucleotide level because of high rates of sequence evolution. This difficulty in identifying homologs is particularly acute for short (≈70–80 bp) tRNA genes. Typically, the anticodon and features of secondary structure are used to establish tRNA identity, but these can be misleading. Some tRNA genes may, through a process of duplication and point mutation(s) in the anticodon triplet, assume the identity of other tRNAs within mtDNA (3). The potential for gene remolding has been corroborated by in vitro tRNA gene knockout experiments in prokaryotic systems (4), and in studies of human mitochondrial-based diseases associated with mutations in tRNA genes (5). Such studies have demonstrated that an anticodon point mutation in one tRNA gene can enable this gene to take over the role of a second disabled tRNA, despite differences in sequence composition and structural elements. The extent to which tRNA remolding plays a role in the evolutionary dynamics of animal mitochondrial genomes, however, remains unexplored.
If remolding of mitochondrial tRNA genes has occurred during metazoan evolution, one might expect the frequency of such events to be highest between: (i) tRNA genes that share similar sequence and structural elements involved in recognition by their cognate aminoacyl-tRNA synthetase, and (ii) genes that can change identity through a single point mutation in their anticodon triplet. Two tRNA genes that share these characteristics are the isoaccepting leucine tRNA (LCUN, LUUR) genes. Although these tRNAs have different mRNA codon selectivities (LCUN with anticodon UAG recognizes four codons, CUA, CUG, CUC, and CUU, within mRNA transcripts; LUUR with anticodon UAA recognizes two codons, UUA and UUG), both genes code for tRNAs that accept the same amino acid and both are likely recognized by the same aminoacyl-tRNA synthetase, as shown for isoaccepting serine tRNAs within animal mtDNA (6). Likewise, the identity of these tRNA genes can be switched through a single point mutation in the third base of the anticodon triplet (e.g., TAA to TAG). Because these two leucine tRNA genes occur within mtDNA of nearly all metazoans (7), and homologs have been identified in other eukaryotes (e.g., ref. 1), these two genes are assumed to have diverged from an ancestral form more than 600 million years ago. Sequence differences between leucine tRNA pairs within metazoan mtDNA therefore should reflect substitutional changes that have accumulated over evolutionary time, taking into account the underlying functional constraints shared by these genes. Consequently, unexpectedly high levels of sequence resemblance between LCUN and LUUR genes may reflect recent gene duplication and identity change within the mitochondrial genome.
Here, through sequence comparisons of leucine tRNA genes within a family of aquatic gastropods, the Ampullariidae, we report the strongest evidence to date of tRNA remolding in metazoan mtDNA and highlight cases of repeated gene identity change within some lineages. By expanding our taxonomic sampling to include more distantly related gastropods, as well as published GenBank sequences of other metazoans, we have found that changes in the identity of tRNA genes through mutational remolding may be more pervasive within mtDNA than previously assumed and the effects may be widespread among disparate taxonomic groups. Our results also demonstrate that tRNA remolding is an important mechanism of gene order change and may lead to the repeated evolution of identical gene orders within portions of animal mitochondrial genomes.
Materials and Methods
DNA Extraction, Amplification, and Sequencing. We sampled eight species within the freshwater gastropod family Ampullariidae and four representatives of a putative sister group, the Viviparidae (8). DNA was extracted and amplified by using standard protocols for molluscan tissue (ref. 9 and Supporting Materials and Methods, which is published as supporting information on the PNAS web site). To explore relationships within this family, we PCR amplified an ≈1,000-bp region spanning the 3′ end of the large subunit rRNA gene (rrnL), two leucine tRNAs, and a 5′region of the subunit 1 of the NADH dehydrogenase complex (nad1) gene. Cleaned PCR products were sequenced on an ABI 377 automated DNA sequencer.
Sequence Alignment and Phylogenetic Analyses. Sequences were aligned by using clustal x (10). This alignment was modified by eye in macclade 4.05 (11) by using secondary structure features of the rrnL gene, tRNAs, and gene boundaries. Regions of poor alignment, typically unpaired loops and bulges of various sizes bounded by stem regions or highly conserved sites, were excluded from phylogenetic analyses. Identification of tRNAs was based on their predicted secondary structure by using trnascan-se (12) and the triplet sequence in their putative anticodon loops. The nad1 sequence was recognized by the presence of a start codon (ATG or GTG) and an uninterrupted ORF, and by sequence similarity to published gastropod nad1 sequences. We excluded tRNAs and any intervening sequence from phylogenetic analyses (see below).
Maximum parsimony (MP; equal weighting) and maximum likelihood (ML) analyses were performed in paup* 4.0b10 (13), and Bayesian analyses (MB) were performed by using mrbayes Ver. 3.0B4 (14). Support for clades was determined by using nonparametric bootstrap replicates (MP and ML) and posterior probabilities (MB). Hierarchical likelihood ratio tests implemented in modeltest (15) were used to evaluate the best model of evolution to be used in ML and MB. For MB, we used equally probable or “flat” prior probabilities and ran four chains of the Markov chain Monte Carlo, starting with a random tree and sampling one tree every 100 generations for 1,000,000 generations. Convergence on a stationary distribution was determined by examination of plots of log likelihood values, and phylogenetic inferences were based only on those trees sampled after a “burn-in” of at least 10,000 generations.
Sequence Similarity Between Leucine tRNA Pairs. To explore the potential for tRNA duplication and remolding between tRNA genes, we compared the overall sequence similarity between LCUN and LUUR tRNAs within our study group. We later broadened our survey to include other distantly related gastropods for which we had sequence data, as well as GenBank records for all nonvertebrate metazoans and a subsample of vertebrates (see Table 1, which is published as supporting information on the PNAS web site). For a given taxon, annotated LCUN and LUUR sequence pairs were aligned by eye in macclade by using secondary structure diagrams obtained by using trnascan-se (12). Percentage similarity between the two aligned leucines was calculated by the number of base matches between leucines divided by the total alignment length. The third base in the anticodon triplet was excluded from pairwise comparisons; hence, a score of 100% would indicate a perfect match across all bases except the third base of the anticodon triplet.
Simulations of Leucine tRNA Divergence. To address the possibility that very similar LCUN and LUUR tRNAs might have arisen by chance rather than by gene duplication and remolding, we simulated the evolution of leucine tRNAs along our ampullariid and viviparid phylogeny (see Results and Discussion) by using LCUN and LUUR from the viviparid Campeloma decisum to represent a hypothetical ancestral sequence. Simulations were undertaken in seq-gen (16) by using the model selected by the hierarchical likelihood ratio test. Our model assumed instantaneous restoration of complementarity in base-paired regions and did not allow for indels, potentially biasing the results in favor of more similar sequences. Anticodon triplets were considered invariant. Topology and branch lengths used were those of our ML tree of rrnL and nad1 datasets. Simulations were run 100 times, resulting in 1,200 within-species LCUN–LUUR comparisons.
Evolution of LCUN and LUUR. The evolution of LCUN and LUUR genes was also examined directly by exploring phylogenetic relationships between these two gene regions sequenced for the 12 ampullariid and viviparid taxa, as well as for other groups in which there was evidence of high sequence resemblance within leucine pairs. For each taxonomic group, sequence alignments were based on secondar y structure predictions from trnascan-se (12). The variable DHU and TψC loop sequences were excluded from phylogenetic analyses in cases where alignment was difficult across species. The third base of the anticodon triplet was also excluded so that relationships could be investigated without the bias of leucine identity. Phylogenetic analyses were performed by using MB and paup*. A Shimodaira–Hasegawa test of alternative phylogenetic hypotheses was implemented in paup* on ML trees, with full likelihood optimization, 1,000 bootstrap replicates, and a one-tailed test of significance. Trees were midpoint rooted, based on the expectation that a phylogenetic analysis of two tRNAs that had diverged in the Precambrian would reveal two groups separated by a long internal branch.
Results and Discussion
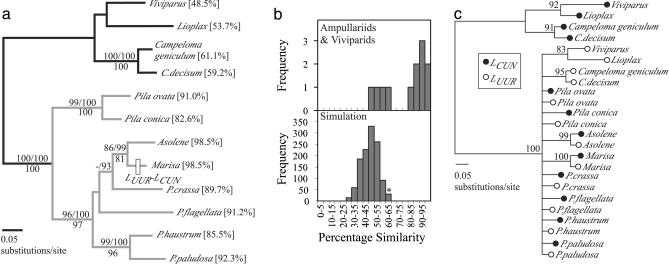
Phylogenetic Relations and Gene Order Change Within the Ampullariidae. The region of mtDNA sequenced in ampullariids and viviparids varied in size from 915 to 1,120 bp. Much of this length variation resulted from the presence of a putative noncoding region between the two leucine genes (see Table 2, which is published as supporting information on the PNAS web site). This region was particularly large, reaching from 145 to 184 bp in Marisa and Asolene, respectively; in other species this varied from 0 to 69 bp. We were unable to identify tRNA pseudogenes within the extensive noncoding region of Marisa and Asolene. The final aligned data set, excluding tRNA leucines, intervening noncoding sequence, and regions of ambiguous alignment, consisted of 678 bp. Phylogenetic analyses revealed strong support for a monophyletic Ampullariidae, with Pila as the sister group to a clade including Pomacea, Asolene, and Marisa (Fig. 1a). There was inconsistent support among analyses for the nesting of Marisa and Asolene within a paraphyletic Pomacea, although there was strong support for a sister-group relationship between Marisa and Asolene.
Fig. 1.
(a) ML phylogeny of eight ampullariid gastropods (genera Pila, Pomacea, Marisa, and Asolene) and four viviparid outgroups (genera Viviparus, Lioplax, and Campeloma) based on 678 bp of mtDNA. Support (>70%) for each clade is shown above branches for ML (100 replicates) and MB (posterior probabilities) and below for MP (1,000 replicates). ML, MB, and MP consensus trees were topologically congruent except for one clade: Parsimony analyses weakly supported a clade of Pomacea crassa and Pomacea flagellata (support: 60%) that was not recovered under ML and MB. A derived gene order change was discovered in Marisa only. Percent similarity values for LCUN vs. LUUR comparisons are given next to taxon labels; a clade with similarity values >75% is shown in gray. (b) The distribution of leucine tRNA similarity values sampled for the 12 ampullariids and viviparids (Upper) versus the simulated evolution of leucine tRNAs along the ML phylogeny (Lower). The asterisk refers to the starting value in our simulation model (61%). (c) A midpoint rooted 50% majority rule consensus phylogeny (MB) of 12 LCUN and 12 LUUR sequences from eight ampullariid taxa and four viviparid outgroups. Posterior probabilities (>70%) are shown above each node.
For all taxa, excluding Marisa cornuarietis, gene order corresponded to the ancestral pattern for gastropods (rrnL–LCUN–LUUR–nad1), as inferred from comparisons with other known molluscan sequences (7). In Marisa, the order of the two leucine tRNA genes was reversed (Fig. 1a), although both genes were transcribed in the same direction as in other taxa. This derived gene order has also evolved independently in other distantly related caenogastropods (17, 18).
The sequences of LCUN and LUUR were considerably more similar within ampullariid species than within viviparid outgroups (Fig. 1a). Aligned LCUN and LUUR sequences within species differed at 1–12 sites in the ingroup versus 28–35 sites in the outgroup (Fig. 1a). Similarities in sequence composition were most striking within the clade consisting of Marisa and Asolene, where sequences of LCUN and LUUR differed by one base in addition to the third base of the anticodon triplet. Simulations revealed that LCUN and LUUR were unlikely to converge to such high levels of similarity by chance (Fig. 1b).
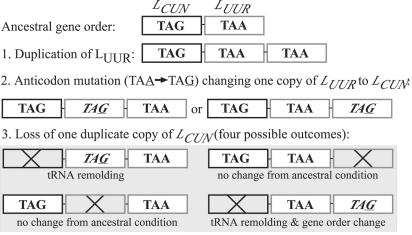
Remolding of LUUR Genes Within Ampullariid Gastropods. Phylogenetic analyses of ampullariid and viviparid leucine tRNAs did not reveal the expected pattern of monophyletic LCUN and LUUR clades. Instead, 4 viviparid LCUN sequences formed a monophyletic group distantly related to all other leucine tRNA sequences, whereas the remaining 8 ampullariid LCUN sequences were nested within a clade of 12 viviparid and ampullariid LUUR sequences (Fig. 1c). There is no rooting that would result in monophyletic LCUN and LUUR clades for these taxa, suggesting that the 8 ampullariid LCUN genes are the likely product of the duplication and remolding of LUUR (Fig. 1c). Furthermore, a Shimodaira–Hasegawa test of the difference in log likelihood scores for the seven trees with optimal log likelihood scores and the optimal log likelihood score of a tree constrained to monophyletic LCUN and LUUR clades was significant (48.39439; P = 0.003). Sister group relations and high similarities between LCUN and LUUR of Marisa (98.5%) and of Asolene (98.5%), also strongly suggested that subsequent gene remolding events had occurred recently and independently within each lineage, resulting in a gene order change in Marisa but not in Asolene. LCUN versus LUUR sequence comparisons between Marisa and Asolene revealed lower similarity values (83.8% for both comparisons; 11 differences over 69 bases), further supporting the conclusion that gene remolding events occurred independently within each lineage and not in the common ancestor of these taxa. A hypothetical mechanism for gene remolding (Fig. 2) illustrates how this process may or may not be associated with gene order change.
Fig. 2.
Hypothetical model of leucine tRNA gene duplication and remolding within the Ampullariidae based on previous models of tandem duplication and deletion (e.g., see ref. 43). Where remolding occurs, it can result in gene order conservation or gene order change.
Gene duplication and remolding events thus appear to have occurred at least three times within the Ampullariidae: once within a stem lineage and twice within the derived genera Marisa and Asolene. Multiple cases of LUUR duplication and identity change within this family may have arisen from the ancestral remolding event within the stem ampullariid lineage which resulted in adjacent tRNAs with very similar nucleotide sequence; the close proximity of identical sequence elements likely predisposed this region to further slipped-strand mispairing during mtDNA replication (19), thus providing additional opportunities for gene duplications and remolding events. Because the viviparid/ampullariid split may be quite ancient, however, gene remolding events common to ampullariids may be shared by other basal caenogastropods, such as the Campanilidae and Cyclophoridae. Interestingly, the LCUN of Campanile symbolicum (family Campanilidae) appears to be a remolded LUUR gene (80.9% similarity), whereas the cyclophorid Aperostoma has only LCUN between rrnL and nad1, suggesting a different outcome of gene dynamics within this region of mtDNA.
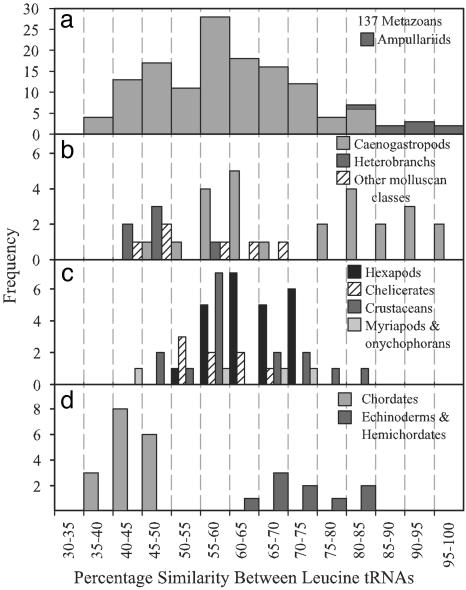
Leucine tRNA Remolding Within the Metazoa. Comparisons of percent similarity between leucine tRNA pairs within the mtDNA of 137 metazoans revealed a broad range of values from 35.1% (Alligator mississippiensis) to 98.5% (Marisa cornuarietis and Asolene spixii) over aligned lengths from 54 to 77 bases (Fig. 3a and see Table 1). Because the distribution of values was not obviously bimodal, there was little evidence for a distinct separation between leucine tRNA pairs that represented orthologous versus paralogous genes, and thus no definitive boundary above which one leucine tRNA could be classed as a “remolded duplicate” of the other. Such a boundary would not be expected, however, if gene duplication and remolding events have occurred at different time depths during the evolutionary history of metazoans. The nonrandom sampling of taxonomic groups invalidated statistical analyses to determine mean similarity values across the Metazoa and to identify outliers and possible remolded leucine tRNAs. Nevertheless, very similar LCUN and LUUR genes within taxonomic groups such as the Mollusca (98.5%; Fig. 3b), Arthropoda (82.3%; Fig. 3c), and the Deuterostomia (84.5%; Fig. 3d) suggest that recent gene remolding events have likely occurred among some representatives of these groups.
Fig. 3.
Frequency distribution of percent sequence similarity values from pairwise comparisons of leucine tRNA genes (LCUN,LUUR) across 137 metazoan taxa (including ampullariids) (a); 37 molluscan taxa, grouped into caenogastropods, heterobranch gastropods, and other molluscan classes (four bivalves, one chiton, one cephalopod) (b); 51 arthropods and 1 onychophoran, grouped by arthropod subphyla (c); and 26 deuterostome taxa, including 8 echinoderms, 1 hemichordate, and 17 chordates (d).
Leucine tRNA Remolding Within the Mollusca. High similarity values (>75%) were confined to the Caenogastropoda (range: 48.5–98.5%, n = 25), whereas members of the Heterobranchia, a sister clade to the Caenogastropoda, had consistently low values (42.6–55.0%; n = 6), as did other molluscan classes sampled (44.4–66.7%; n = 6). Within the Caenogastropoda, high sequence resemblances were evident only in members of the family Ampullariidae, three of seven species sampled within the family Vermetidae, and representatives of two other families (Littorinidae: Littorina saxatilis; and Campanilidae: Campanile symbolicum). High leucine tRNA similarities were sometimes associated with switches in position relative to the ancestral caenogastropod condition (e.g., Marisa, Littorina, and Serpulorbis), but not always (e.g., Pomacea and Asolene). Current classifications of gastropods (20) place ampullariids and vermetids in different orders, suggesting that the remolding of leucine tRNAs has occurred independently within these caenogastropod lineages.
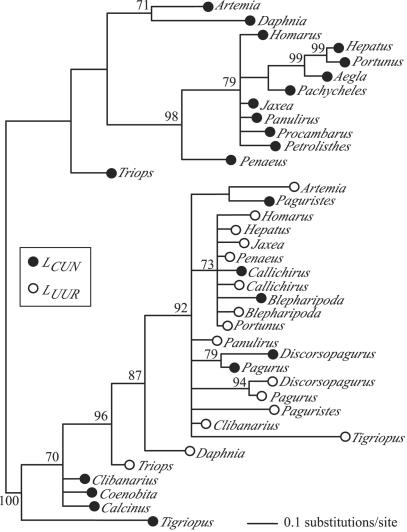
Leucine tRNA Remolding Within the Crustacea. Within the Arthropoda, the highest leucine tRNA similarities were evident within the Crustacea. A phylogeny of LCUN and LUUR sequences from 22 crustacean taxa (Fig. 4) suggested that LCUN tRNAs within 5 decapod species are remolded LUUR genes; there is no rooting that would result in monophyletic LCUN and LUUR clades. In four of these taxa, Blepharipoda, Paguristes, Discorsopagurus, and Pagurus, these tRNAs are located between cox1 and cox2 in the order LCUN–LUUR (Fig. 4). This arrangement has been inferred to be the result of a translocation of LCUN from its ancestral position between rrnL and nad1 and is taken to be a synapomorphy of a major clade of anomuran crustaceans including the Hippoidea, Paguroidea, and Coenobitoidea (21). Our tRNA phylogeny suggests a different interpretation, in which the adjacent positions of LCUN and LUUR result from a duplication of LUUR, followed by a remolding of the duplicate to take on the function of the LCUN (Fig. 4). Based on the complete mtDNA of Pagurus longicarpus (22), the original LCUN appears to have been lost. Within Clibanarius, Calcinus, and Coenobita, believed to form the crown of the anomuran clade, LCUN and LUUR are no longer adjacent (21). If the phylogeny proposed (21) is correct, the LCUN tRNAs of these derived taxa should be descendants of the LUUR duplication and remolding event. Our results are ambiguous on this point. Midpoint rooting places the LCUN tRNAs of these hermit crabs with the LUUR clade, consistent with the inference of gene duplication and the proposed phylogeny, although there is an alternative rooting in which the LCUN tRNAs of these taxa are part of a monophyletic clade containing only LCUN tRNAs. In the fifth decapod species, Callichirus, the two leucine tRNAs are adjacent, again between cox1 and cox2, but in this case in the order LUUR–LCUN. The high sequence similarity of the Callichirus leucine tRNAs (82.3%) and the inferred phylogenetic position relative to the previously discussed taxa (21) suggest that this may be an independent duplication and remolding event.
Fig. 4.
Midpoint rooted 50% majority rule consensus phylogeny (MB) of LCUN and LUUR sequences from 16 crustacean taxa for which both sequences were available in GenBank, as well as 6 additional taxa for which only the LCUN sequence was available. Support indices shown are posterior probabilities (>70%).
Leucine tRNA similarity comparisons within the Crustacea exemplify the weakness of identifying remolding events by sequence similarity alone. Of the five decapod species with putative remolded leucine tRNAs, four had similarity values under 75% (Paguristes 59.4%, Clibanarius 68.8%, Pagurus 71.6%, and Discorsopagurus 74.6%). Consequently, high similarity values are likely to identify only relatively recent remolding events. Phylogenetic analyses of leucine tRNA sequences provide a more powerful approach to identifying gene duplication and remolding events, but they may also be limited by saturation of phylogenetic signal over time. Given that substitutions in stem regions of tRNA molecules may not become saturated for nearly 100 million years for transitions and 350 million years for transversions (23), however, there is hope that careful phylogenetic analyses of tRNA gene pairs will allow further documentation of tRNA gene remolding over deep evolutionary time scales. Other points of similarity among tRNAs, such as secondary structural features (24) and shared sequence motifs, may also strengthen the inference of homology.
Leucine tRNA Remolding Within the Deuterostomia. High sequence similarities between leucine tRNAs within the mtDNAs of echinoderms and a hemichordate (Fig. 3d), and the results of a phylogenetic analysis of deuterostome leucine tRNA sequences (see Fig. 5, which is published as supporting information on the PNAS web site), suggest that a gene duplication and remolding event took place within a common ancestor to these two sister phyla, as suggested by Castresana et al. (25). Evidence for this history is further supported by apparent vestiges of the original LCUN gene concatenated to the 5′ end of nad5 in echinoderms and hemichordates (3, 25).
The Unidirectional Pattern of Leucine tRNA Remolding. In all cases of leucine tRNA identity change where the direction of remolding could be inferred with confidence, remolding was unidirectional, from LUUR to LCUN, and never vice versa. This asymmetry is impressive, given that this pattern of gene remolding appears the same within caenogastropods, decapod crustaceans, and a lineage of deuterostomes. The only reported instance of gene remolding from LCUN to LUUR is within human lung carcinoma cybrid cells, where point mutations in LUUR leading to a severe pathological condition are compensated in some cells by a point mutation within LCUN, converting this tRNA to LUUR and restoring the wild-type phenotype (5). Why remolding should be predominantly unidirectional within leucine tRNAs across metazoan groups is unclear. Base compositional bias is unlikely to provide an explanation for this asymmetry, because the direction of change, from A to G, is opposite the pattern of base compositional bias typically seen in mtDNA of invertebrate groups such as gastropods. Nevertheless, the implications of this are important. A biased outcome of duplication and remolding events can lead to a greater probability of convergence in gene orders, as seen within gastropod molluscs. Models underlying gene-order-based phylogenetic methods will need to incorporate the possibility of this transformational asymmetry.
Prevalence of Gene Remolding in Animal mtDNA. The remolding of duplicated tRNA genes has been commonplace within the evolution of animal mitochondrial genomes. In fact, the lower number of identity elements involved in the recognition of animal mitochondrial tRNAs by their cognate aminoacyl-tRNA synthetases compared with their nuclear counterparts (23, 26, 27) may predispose the mitochondrial genome to such changes in tRNA identity through time. Evolutionary changes in the genetic code within animal mtDNA also attest to the malleability of tRNA genes and flexibility of aminoacyl-tRNA synthetases in the characteristics of tRNAs that they recognize (28). While here we have documented the repeated duplication and remolding of the leucine tRNA gene LUUR within one family of gastropods, a survey of 137 metazoan leucine tRNA pairs suggests that similar events have occurred within other gastropod clades, twice within a clade of decapod crustaceans, and independently in a lineage of deuterostomes leading to hemichordates and echinoderms. We suspect that additional instances of such events will become evident as the complete sequences of more mtDNAs are published.
Are gene remolding events characteristic only of isoaccepting leucine tRNAs or do such events occur between other tRNAs? Of the 22 tRNA genes within typical animal mtDNA, two isoaccepting tRNA genes code for the amino acids leucine and serine, respectively. These tRNAs may be more susceptible to remolding events than other tRNA genes because, like isoaccepting leucine and serine prokaryotic tRNAs (29, 30), they appear to be recognized by a single aminoacyl-tRNA synthetase, which does not use the anticodon triplet as a recognition site for tRNA identity (5, 6, 31, 32). Consequently, recognition of leucine tRNAs by their cognate synthetase should be based on structural features common to both LUUR and LCUN and not hindered by mutational changes in the anticodon triplet to or from TAG to TAA. By this same reasoning, gene remolding might also be expected to occur between isoaccepting serine tRNAs. Indeed, an apparent switch in position of serine tRNAs (anticodons TCT and TGA) in the mtDNA of the nematode Trichinella spiralis relative to an inferred ancestral condition has the signature of gene identity change (33). Although high levels of sequence similarity have also been noted between specific structural regions of serine tRNA pairs within Katharina, Mytilus, Drosophila, and Lumbricus, these similarities have been attributed to selective constraints associated with interactions with a common aminoacyl-tRNA synthetase (34). Although such constraints might account for shared identity elements between isoaccepting tRNAs, the wide range of similarities between LCUN and LUUR tRNAs within animal groups (Fig. 3), and the few recognition sites actually involved in tRNA recognition by aminoacyl-tRNA synthetases (29) argue against selection as an explanation for nearly identical isoaccepting tRNAs.
The extent to which gene reassignments may have occurred between nonisoaccepting tRNAs during metazoan evolution remains to be determined through comparative genomic approaches. The propensity for such events may, however, depend both on the frequency of duplication of tRNAs, providing “spare” tRNA templates for remolding, and on the fidelity of aminoacyl-tRNA synthetases for their cognate tRNAs, based on the specificity and position of recognition sites (29). Given that studies of bacterial, yeast, and human cytosolic tRNAs have demonstrated that gene reassignments can occur among nonisoaccepting pairs of tRNAs (4, 29, 35), and that aminoacyl-tRNA synthetase recognition of some tRNA molecules may be dependent on only a few identity elements (4, 29, 31, 35), gene remolding may also be applicable to some nonisoaccepting tRNA gene pairs within animal mtDNA.
tRNA Gene Remolding and Mitochondrial Gene Rearrangements. Given the enormous number of possible mitochondrial tRNA gene arrangements, and the resulting improbability of chance convergence, confidence has been expressed in the robustness of tRNA gene order characters (e.g., refs. 36 and 37). This confidence is based on the assumption that homologous genes are being compared, and that we understand the mechanisms by which gene orders change, allowing us to develop realistic models and methods of phylogenetic inference for these data. These models help us to determine the likelihood of homoplasious arrangements, and consequently, the reliability of specific markers. While, in theory, the probability that two mitochondrial genomes share the same derived genome organization by chance alone is low (38), functional constraints may make some tRNA arrangements more likely than others (39), resulting in convergence in gene order within portions of the mitochondrial genome (40, 41). Whereas four mechanisms have been invoked to explain mtDNA rearrangements [duplication/random loss, illicit priming by tRNA genes, recombination, and duplication/nonrandom loss (39)], a fifth involving duplication/anticodon mutation (3) has not been considered important, given the paucity of evidence for this within particularly well-studied taxonomic groups (41).
Here, we argue that gene remolding events should be considered to be an important mechanism of gene rearrangement: remolding of tRNA genes does occur among disparate taxonomic groups and can also result in changes in gene order. Likewise, because of the propensity of LUUR to assume the function of the LCUN gene within animal mtDNA, this mechanism of rearrangement can lead to incorrect homology assessment and gene order homoplasy. For instance, duplication and remolding may help to explain why nearest-neighbor tRNA gene orders have been considered to be less reliable phylogenetic characters (42). Failure to recognize gene remolding as an important mechanism at work within mtDNA can also lead to a misinterpretation of the mechanics of some gene order changes: previous interpretations of translocations of leucine tRNA genes within decapod crustaceans (21) are likely due instead to gene duplication and identity change. Interpretation of gene remolding events also holds the potential for uncovering gene dynamics that are hidden at the level of gene order alone, as in the Ampullariidae. Hence, the addition of tRNA gene remolding to the toolkit of the mtDNA rearrangement mechanisms can help us to understand more completely the tempo and mode of mitochondrial gene dynamics.
Implications of Gene Remolding for Metazoan Phylogeny. Given the apparent prevalence of tRNA gene duplication and remolding events within the evolution of metazoan mitochondria, it is surprising that these have received so little attention (but see refs. 39 and 41). Yet, if tRNA duplication and remolding are commonplace within animal mtDNAs, this has important implications. First, the remolding of tRNA genes can blur the distinction between orthologous and paralogous genes, between perceived gene order changes and gene identity changes, and between homologous and analogous characters in phylogenetic analyses. Changes of tRNA identity, of course, do not invalidate the use of mitochondrial tRNA gene orders in phylogenetic analysis, but do suggest that the assessment of homology of tRNAs at higher taxonomic levels may be more difficult than is typically assumed, and may require careful study at lower levels. In the worst case, identification of tRNA homologs may be problematic. Second, if the original tRNA whose function is coopted by a duplicate is lost, homologous comparisons for the original tRNA will no longer be possible. This situation will create difficulties for methods that require a full complement of genes for all taxa in an analysis. Third, methods that use gene order data in phylogenetic analysis will have to incorporate gene duplication and remolding and its apparent asymmetry to develop realistic models. Further research is necessary to explore the frequency of gene remolding events within metazoans and to determine the extent to which this pattern of gene duplication/identity change can be generalized to other tRNAs within the mitochondrial genome.
Supplementary Material
Acknowledgments
We thank Cliff Cunningham, Mark Dowton, Dennis Lavrov, Mónica Medina, and Gavin Naylor for comments. This work was supported by National Science Foundation Grant DEB-9509324 (to R.B. and T.M.C.) and is contribution no. 68 of the program in Tropical Biology at Florida International University.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: LCUN and LUUR, isoaccepting leucine tRNAs; MP, maximum parsimony; ML, maximum likelihood; MB, Bayesian analysis.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY449491–AY449518).
References
- 1.Gray, M. W., Lang, B. F., Cedergren, R., Golding, G. B., Lemieux, C., Sankoff, D., Turmel, M., Brossard, N., Delage, E., Littlejohn, T. G., et al. (1998) Nucleic Acids Res. 26, 865–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paquin, B., Laforest, M.-J., Forget, L., Roewer, I., Wang, Z., Longcore, J. & Lang, B. F. (1997) Curr. Genet. 31, 380–395. [DOI] [PubMed] [Google Scholar]
- 3.Cantatore, P., Gadaleta, M. N., Roberti, M., Saccone, C. & Wilson, A. C. (1987) Nature 329, 853–855. [DOI] [PubMed] [Google Scholar]
- 4.Saks, M. E., Sampson, J. R. & Abelson, J. (1998) Science 279, 1665–1670. [DOI] [PubMed] [Google Scholar]
- 5.El Meziane, A., Lehtinen, S. K., Hance, N., Nijtmans, L. G. J., Dunbar, D., Holt, I. J. & Jacobs, H. T. (1998) Nat. Genet. 18, 350–353. [DOI] [PubMed] [Google Scholar]
- 6.Yokogawa, T., Shimada, N., Takeuchi, N., Benkowski, L., Suzuki, T., Omori, A., Ueda, T., Nishikawa, K., Spremulli, L. L. & Watanabe, K. (2000) J. Biol. Chem. 275, 19913–19920. [DOI] [PubMed] [Google Scholar]
- 7.Boore, J. L. (1999) Nucleic Acids Res. 27, 1767–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berthold, T. (1991) Abhandlungen des Naturwissenschaftlichen Vereins in Hamburg 29, 1–256. [Google Scholar]
- 9.Collins, T. M., Frazer, K., Palmer, A. R., Vermeij, G. J. & Brown, W. M. (1996) Evolution 50, 2287–2304. [DOI] [PubMed] [Google Scholar]
- 10.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maddison, D. R. & Maddison, W. P. (2002) MACCLADE, Analysis of Phylogeny and Character evolution (Sinauer, Sunderland, MA), Version 4.05.
- 12.Lowe, T. M. & Eddy, S. R. (1997) Nucleic Acids Res. 25, 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swofford, D. L. (2002) paup*, Phylogenetic Analysis Using Parsimony (*and Other Methods) (Sinauer, Sunderland, MA), Version 40b10.
- 14.Huelsenbeck, J. P. & Ronquist, F. (2001) Bioinformatics 17, 754–755. [DOI] [PubMed] [Google Scholar]
- 15.Posada, D. & Crandall, K. A. (1998) Bioinformatics 14, 817–818. [DOI] [PubMed] [Google Scholar]
- 16.Rambaut, A. & Grassly, N. C. (1997) Comput. Appl. Biosci. 13, 235–238. [DOI] [PubMed] [Google Scholar]
- 17.Wilding, C. S., Mill, P. J. & Grahame, J. (1999) J. Mol. Evol. 48, 348–359. [DOI] [PubMed] [Google Scholar]
- 18.Rawlings, T. A., Collins, T. M. & Bieler, R. (2001) Mol. Biol. Evol. 18, 1604–1609. [DOI] [PubMed] [Google Scholar]
- 19.Levinson, G. & Gutman, G. A. (1987) Mol. Biol. Evol. 4, 203–221. [DOI] [PubMed] [Google Scholar]
- 20.Ponder, W. F. & Warén, A. (1988) Malacological Rev., Suppl. 4, 288–317.
- 21.Morrison, C. L., Harvey, A. W., Lavery, S., Tieu, K., Huang, Y. & Cunningham, C. W. (2001) Proc. R. Soc. London Ser. B 269, 345–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hickerson, M. J. & Cunningham, C. W. (2000) Mol. Biol. Evol. 17, 639–644. [DOI] [PubMed] [Google Scholar]
- 23.Kumazawa, Y. & Nishida, M. (1993) J. Mol. Evol. 37, 380–398. [DOI] [PubMed] [Google Scholar]
- 24.Murrell, A., Campbell, N. J. & Barker, S. C. (2003) Syst. Biol. 52, 296–310. [DOI] [PubMed] [Google Scholar]
- 25.Castresana, J., Feldmaier-Fuchs, G., Yokobori, S., Satoh, N. & Pääbo, S. (1998) Genetics 150, 1115–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumazawa, Y., Himeno, H., Miura, K. & Watanabe, K. (1991) J. Biochem. (Tokyo) 109, 421–427. [DOI] [PubMed] [Google Scholar]
- 27.De Georgi, C., Martiradonna, A. & Saccone, C. (1996) Curr. Genet. 30, 191–199. [DOI] [PubMed] [Google Scholar]
- 28.Yokobori, S., Suzuki, T. & Watanabe, K. (2001) J. Mol. Evol. 53, 314–326. [DOI] [PubMed] [Google Scholar]
- 29.Giege, R., Sissler, M. & Florentz, C. (1998) Nucleic Acids Res. 26, 5017–5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larkin, D. C., Williams, A. M., Martinis, S. A. & Fox, G. E. (2002) Nucleic Acids Res. 30, 2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimada, N., Suzuki, T. & Watanabe, K. (2001) J. Biol. Chem. 276, 46770–46778. [DOI] [PubMed] [Google Scholar]
- 32.Sohm, B., Frugier, M., Brulé, H., Olszak, K., Przykorska, A. & Florentz, C. (2003) J. Mol. Biol. 328, 995–1010. [DOI] [PubMed] [Google Scholar]
- 33.Lavrov, D. V. & Brown, W. M. (2001) Genetics 157, 621–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boore, J. L. & Brown, W. M. (1995) Genetics 141, 305–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tocchini-Valentini, G., Saks, M. E. & Abelson, J. (2000) J. Mol. Biol. 298, 779–793. [DOI] [PubMed] [Google Scholar]
- 36.Boore, J. L., Collins, T. M., Stanton, D., Daehler, L. L. & Brown, W. M. (1995) Nature 376, 163–165. [DOI] [PubMed] [Google Scholar]
- 37.Boore, J. L., Lavrov, D. V. & Brown, W. M. (1998) Nature 392, 667–668. [DOI] [PubMed] [Google Scholar]
- 38.Dowton, M., Castro, L. R. & Austin, A. D. (2002) Invertebr. Syst. 16, 345–356. [Google Scholar]
- 39.Dowton, M., Castro, L. R., Campbell, S. L., Bargon, S. D. & Austin, A. D. (2003) J. Mol. Evol. 56, 517–526. [DOI] [PubMed] [Google Scholar]
- 40.Flook, P., Rowell, H. & Gellissen, G. (1995) Naturwissenschaften 82, 336–337. [Google Scholar]
- 41.Dowton, M. & Austin, A. D. (1999) Mol. Biol. Evol. 16, 298–309. [DOI] [PubMed] [Google Scholar]
- 42.Boore, J. L. & Brown, W. M. (1998) Curr. Opin. Genet. Dev. 8, 668–674. [DOI] [PubMed] [Google Scholar]
- 43.Moritz, C., Dowling, T. E. & Brown, W. M. (1987) Ann. Rev. Ecol. Syst. 18, 269–292. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.