Abstract
Histone and protein acetylation catalyzed by p300/CBP transcriptional coactivator regulates a variety of key biological pathways. This study investigates the proposed Theorell-Chance or “hit-and-run” catalytic mechanism of p300/CBP histone acetyltransferase (HAT) using bisubstrate analogs. A range of histone peptide tail peptide-CoA conjugates with different length linkers were synthesized and evaluated as inhibitors of p300 HAT. We show that longer linkers between the histone tail peptide and the CoA substrate moieties appear to allow for dual engagement of the two binding surfaces. Results with D1625R/D1628R double mutant p300 HAT further confirm the requirement for a negatively charged surface on the enzyme to interact with the histone tail.
1. Introduction
The p300/CBP histone acetyltransferase (HAT) catalyzes the lysine acetylation of more than 75 different cellular proteins [1-5] including itself [6]. Beyond histones, such acetylation events have wide-ranging effects on cell growth and gene expression, influencing cancer, the immune system, endocrine and metabolic pathways, host-pathogen interactions, and cardiovascular physiology [7-10]. Within the field of epigenetics, there has been increasing interest in developing potent and specific inhibitors of p300/CBP HAT as potential pharmacologic agents for a number of disease indications [11-15].
Compared to other HATs, p300/CBP has been reported to follow an unusual Theorell-Chance kinetic mechanism [16-18]. The well-characterized HATs including PCAF/GCN5 and Esa1 employ a sequential mechanism in which acetyl-CoA binding to enzyme precedes peptide/protein substrate binding to form a ternary complex [19, 20]. There is then direct transfer of the acetyl group from acetyl-CoA to the Lys side chain with ordered product release. The Theorell-Chance mechanism proposed for p300/CBP also involves initial binding of acetyl-CoA and direct acetyl transfer to peptide/protein substrate but does not form a detectable ternary complex. In the Theorell-Chance mechanism, often called hit and run, the peptide/protein binds in to the p300/CBP-acetyl-CoA complex in too fleeting a fashion to be measured but does allow for covalent chemistry to occur [16]. The Theorell-Chance mechanism is deduced from a characteristic pattern of product inhibition in which acetylated peptide product is competitive versus peptide substrate [18].
A number of years ago, several bisubstrate analogs including H4-CoA-20 and Lys-CoA were tested as p300 HAT inhibitors and it was found that Lys-CoA, with Ki* 20 nM, was highly potent and selective whereas the more elaborate H4-CoA-20, containing 20 residues from the histone H4 tail, was far weaker as a p300 HAT inhibitor [21]. This was unexpected because p300 HAT preferentially acetylates lysine residues in H4 tail peptides rather than in isolation and because the Lys-CoA moiety is a constituent of the H4-CoA-20 framework. An X-ray crystal structure of the p300 HAT domain in complex with Lys-CoA revealed that Lys-CoA sits in an extended conformation in a narrow tunnel formed within p300 HAT and the Lys alpha amino and carboxyl groups are near the walls of the tunnel [18] (see Figure 1). This suggested a model of steric blockade in which the additional residues of H4-CoA-20 attached to the Lys would limit its p300 binding affinity.
Figure 1.
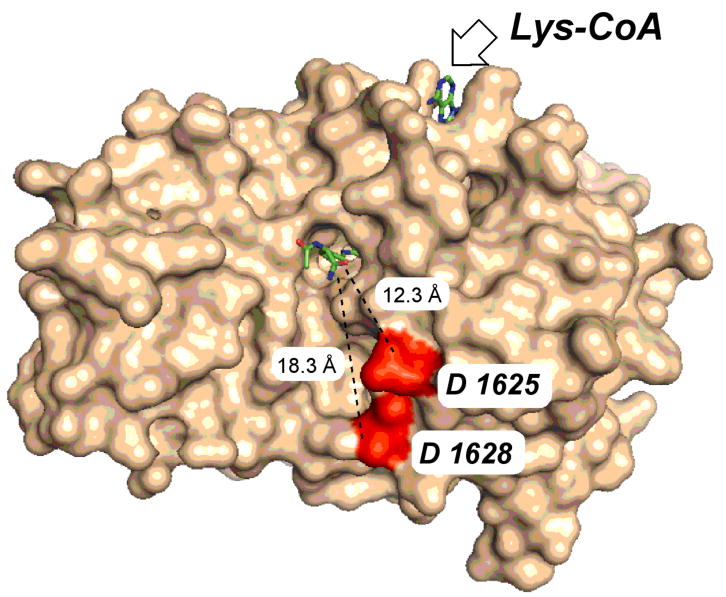
Crystal structure of p300 histone acetyltransferase complexed to Lys-CoA (PDB ID: 3BIY). The highlighted distances extend from the alpha carbon of the Lys moiety in Lys-CoA to the Asp side chains of the proposed histone tail binding residues.
A separate, negatively charged shallow pocket lined by the side chains Asp-1625 and Asp-1628 of p300 spaced about 10 Å from the Lys-CoA tunnel appears to be important for peptide substrate interaction. Mutation of Asp-1625 and Asp-1628 to Arg reduces p300's acetyltransferase efficiency toward positively charged peptide substrates but has much less of an effect on negatively charged peptide substrates [18]. Taken together with the proposed Theorell-Chance mechanism and Lys-CoA binding selectivity, it has been suggested that the p300-Lys-CoA X-ray structure captures a late stage of the reaction coordinate which represents further destabilization of H4 tail peptide interactions after a predicted weak encounter complex between p300 and a peptide substrate.
Our hypothesis emerging from these earlier studies is that it may be possible to generate more potent versions of H4-CoA-20 in which the linker between the CoA and the peptide backbone is lengthened. Such stretched analogs of H4-CoA-20 could accommodate dual interactions on p300 engaging both the Lys-CoA channel and the potential peptide substrate binding groove. Below, we describe the synthesis and experimental analysis of this novel class of analogs.
2. Materials and Methods
2.1. General
All materials for Fmoc solid phase synthesis were purchased from Novabiochem (Darmstadt, Germany) and used without additional purification. All amino acids used here were of L-configuration. Peptides were synthesized in Protein Technologies (Rainin) PS3 peptide synthesizer and purified using a Varian ProStar HPLC equipped with C18 reverse phase column. Amino acid analysis was performed in the Harvard microchemistry facility or the W. M. Keck facility at Yale University. All buffering and bacterial media reagents were purchased from Fisher Scientific, Inc. (Hampton, NH). Chitin beads were purchased from New England BioLabs. Purification of proteins were carried out using a Pharmacia Biotech (GE Healthcare) FPLC equipped with a monoS ion-exchange column (GE Healthcare). 14C acetyl-CoA was purchased from Amersham. Unlabeled acetyl-CoA was purchased from Calbiochem. All additional chemicals were purchased from Sigma-Aldrich Chemical Co. (St. Louis, MO) and used without further purification. Phosphorimage analysis was perfomed on a Typhoon 9410 variable mode imager (GE Healthcare) and quantified using Image Quant TL software.
2.2. Synthesis of Bisubstrate Analogs
2.2.1. Synthesis of bromoacyl peptides
Bisubstrate analogs used in this study are conjugates of histone H4 N–terminal (H4-20mer) peptide (NH2-SGRGKGGKGLGKGGAKRNRA-COOH) and coenzyme A. To be specific, the compounds described here vary in the linker that conjoins the peptide backbone and coenzyme A. Synthesis of compounds 1, 2, 3 and 4: using standard Fmoc SPPS, H4-20mer peptide containing lysine protected on its N(ε) with Mtt (4-methyltrityl) as the eighth residue (K8) was synthesized on a 0.1 mmol scale using wang resin as the solid phase support. At the end of the synthesis, the N-terminus was capped with acetic anhydride. The orthogonal protecting group Mtt on K8 was cleaved under mildly acidic conditions (methylene chloride: hexafluoroisopropanol: trifluoroethanol: triethylsilane = 6.5: 2: 1: 0.5 v/v) while the peptide was still attached to the solid phase (Scheme 2). The free amine was coupled to the respective bromoacid (20 equiv.) using DIC (diisopropylcarbodiimide) for 16 h. This was followed by global deprotection (trifluoroacetic acid (TFA): water: triisopropylsilane 10: 0.33: 0.33 v/v) and purification of the bromoacyl peptides using C18 reverse phase HPLC with water and acetonitrile (0.05% TFA) as the mobile phase.
Compound 5 was synthesized by first coupling the epsilon amino functionality of K8 with L-N(α)-Fmoc-N-Mtt lysine (4 equiv.) using HCTU ((2-(6-Chloro-1H-benzotriazole-1-yl)-1,1,3,3-tetramethylaminium hexafluorophosphate, 4 equiv.). This was followed by deprotection of Fmoc (20% piperidine in DMF (dimethylformamide), 2-3 times) and reprotection with acetic anhydride (20 equiv.) in the presence of catalytic amounts of DMAP (dimethylaminopyridine) and excess Hunigs base in DMF. Following this, the N(ε)-Mtt protecting group was cleaved under mildly acidic conditions.
Bromoacetylation of the deprotected amine was performed with 20 equiv. of bromoacetic anhydride in the presence of excess Hunigs base in DMF. This was followed by global deprotection and purification using C18 reverse phase HPLC. The purified bromoacyl peptides (Table 1a) were subjected to conjugation with CoA.
Table 1.
| Compound | Bromopeptide | ||
|---|---|---|---|
| Calculated (Da) | Found (Da) | ||
| 1 | [M+H]+ | 2076.04 | 2076.11 |
| 2 | [M+H]+ | 2090.17 | 2089.9 |
| 3 | [M+H]+ | 2131.89 | 2132.1 |
| 4 | [M+H]+ | 2160.13 | 2160.02 |
| 5 | [M+H]+ | 2246.14 | 2245.88 |
| 6 | [M+H]+ | 2204.27 | 2203.61 |
| 7 | [M+H]+ | 2330.2 | 2330.30 |
| Table 1a. MALDI-TOF data for the bromo peptides | |||
| Compound | CoA Conjugate | ||
|---|---|---|---|
| Calculated (Da) | Found (Da) | ||
| 1 | [M+H]+ | 2762.23 | 2761.52 |
| 2 | [M+Na]+ | 2798.23 | 2798.12 |
| 3 | [M+Na]+ | 2839.85 | 2839.62 |
| 4 | [M+H]+ | 2846.92 | 2845.82 |
| 5 | [M+H]+ | 2932.97 | 2932.02 |
| 6 | [M+H]+ | 2890.89 | 2889.87 |
| 7 | [M+Na]+ | 3039.06 | 3038.64 |
| Table 1b. MALDI-TOF data for the bisubstrate analogs | |||
For the synthesis of bisubstrate analog 6, L-Nα-Fmoc-N(β-Alloc diaminopropionic acid was incorporated as the 8th residue (instead of lysine) in H4-20mer peptide (0.1 mmol scale). The alloc protecting group was then removed using Pd(PPh3)4 (excess, dissolved in a cocktail of chloroform : N-methyl-morpholine : acetic acid 37: 1: 2 v/v using sonication) over 16 h. The rest of the synthesis was identical to that of bisubstrate analog 5. Similarly, using HCTU, Compound 7 was made by coupling Fmoc-4-aminobutyric acid to K8 of the H4-20mer peptide. After Fmoc removal, the peptide was coupled to L-N(α)-Fmoc-N(ε)-Mtt lysine. The rest of the synthesis was identical to that of compound 5. Note: After every chemical step, the resin was thoroughly washed with DMF and methylene chloride, alternatively.
2.2.2. Conjugation of coenzyme A (CoA) to bromoacyl peptides
As the final step, conjugation of CoA was performed by reacting CoA (20 equiv.) with the purified bromoacyl peptides in triethylammonium-bicarbonate buffer (pH 8.0) for 2 days at 35°C. The crude product was acidified to ∼ pH 3 and purified using C18 reverse phase HPLC with water and acetonitrile (0.05% TFA) as the mobile phase. The fractions were analyzed using MALDI-TOF (THAP (trihydroxyacetophenone) matrix), pooled, concentrated under reduced pressure and lyophilized. The lyophilized sample was applied to P4 Biogel gel filtration column and eluted with 20 mM ammonium acetate (pH 7.5) buffer at a rate of 0.3 mL/min. The fractions (2 mL) were analyzed by UV absorption at 259 nm, concentrated and lyophilized. The purity and concentration of the conjugates were further confirmed by amino acid analysis. MALDI-TOF data for the conjugates are provided in Table 1b.
2.3. Histone acetyltransferase (HAT) assays
These reactions were performed analogous to previously described procedures [17]. Assay buffer contained in a final volume of 30 μL: 50 mM HEPES pH 7.9, 0.1mM EDTA, 50 μg/mL BSA, 1 mM DTT, 100 μM H4-15mer (substrate peptide), 10 nM p300 HAT, 0 to various concentrations of bisubstrate analogs and 20 μM of 14C-acetyl-CoA. The mixture except 14C-acetyl-CoA was incubated at 30°C for 7 min. The reaction was initiated by the addition of 6 μL (∼2.8 μCi/mL) of 14C-acetyl-CoA at 30°C. After 7-14 min, reactions were quenched by vortexing with 6 μL of gel-loading dye. The quenched reaction mixtures (13 or 15 μL) were loaded onto a 16% Tris-Tricine gel and the acetylated peptide was electrophoretically separated from the protein. The gel was thoroughly washed with water to remove excess 14C-acetyl-CoA and dried in a gel drier. The dried gel was then exposed to a phosphorimager plate for ∼ 36 h and quantified relative to a 14C-BSA standard. Product formation was shown to be linear with time and enzyme concentration in the range investigated and less than 10% of the limiting substrate was converted to product. Duplicate measurements were generally within 20%. The observed rates were plotted as 1/V0 vs [I] to obtain IC50 values (Table 2). Also, for selected bisubstrate analogs, the relative HAT activity at various concentrations is presented in Fig. 3.
Table 2.
IC50 values of bisubstrate analogs (1-7).
| Compound | IC50 (μM) |
|---|---|
| Lys-CoA | 0.060 ± 0.008 |
| 1 | 2.6 ± 0.24 |
| 2 | 3.2 ± 0.4 |
| 3 | 2.4 ± 0.3 |
| 4 | 1.6 ± 0.2 |
| 5 | 1.2 ± 0.1 |
| 6 | 0.97 ± 0.1 |
| 7 | 0.4 ± 0.05 |
| Compound 7 | IC50 (μM) |
| wt p300 HAT | 0.4 ± 0.05 |
| D1625R; D1628R | 6.7 ± 0.8 |
| Lys-CoA | IC50 (μM) |
| wt p300 HAT | 0.060 ± 0.008 |
| D1625R; D1628R | 0.209 ± 0.015 |
Figure 3.
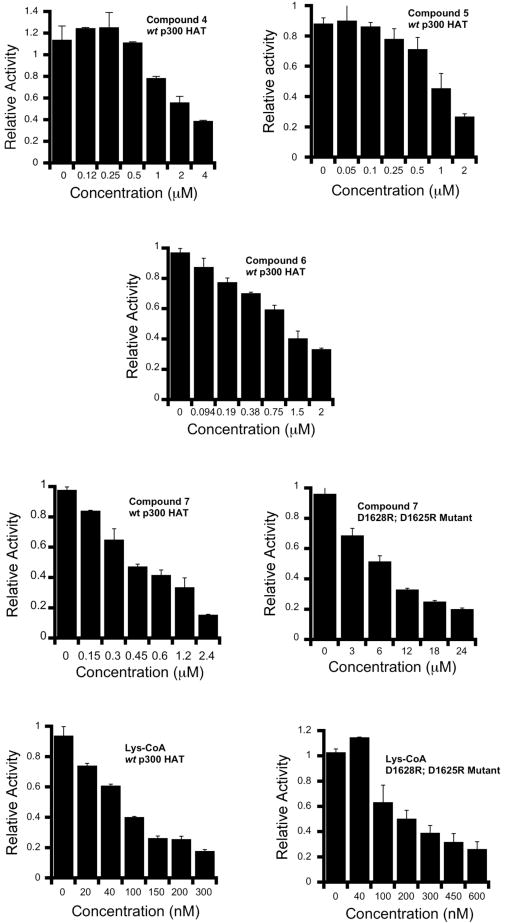
Inhibition of p300 HAT with selected bisubstrate analogs; bars represent the relative HAT activity at various inhibitor concentrations employed. Error bars represent the span of duplicate measurements.
3. Results
We designed and synthesized a set of three analogs of H4-CoA-20 (1) in which the acetyl bridge between Lys and CoA is replaced by a propionyl, a hexanoyl, and an octanoyl lipid linker (Fig. 2, 2-4). Two additional analogs incorporate a protected lysyl linker between either the natural Lys at position 8 of the H4-tail (5) or an amino-alanine at position 8 of the H4-tail (6) followed by CoA attachment. One final analog (7) incorporates two consecutive lysyl linkers attached to the natural Lys-8. In general, the extended linker length between the H4 tail backbone increases from compound 2 through 7 (Fig. 2). Compound 7 provides the overall longest spacing between the H4 tail backbone and the CoA. Each of these bisubstrate analogs was prepared by solid phase synthesis as shown in Scheme 1 and purity and chemical structures determined by HPLC and mass spectrometry. Concentrations of all bisubstrate analogs 1-7 were ascertained using amino acid analysis.
Figure 2.
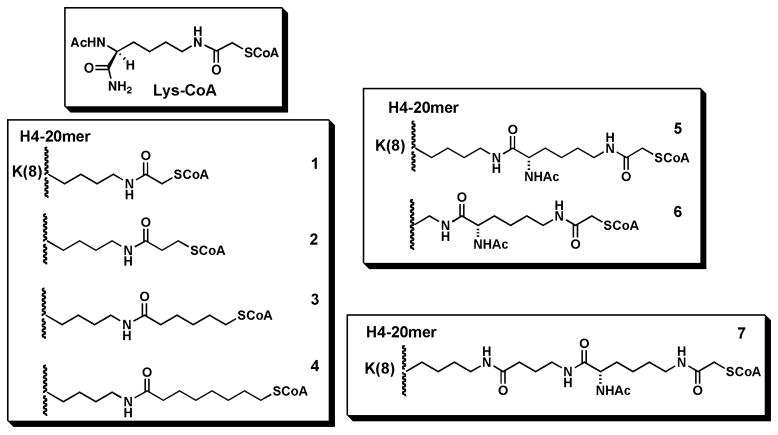
Bisubstrate analogs studied in this article.
Scheme 1.
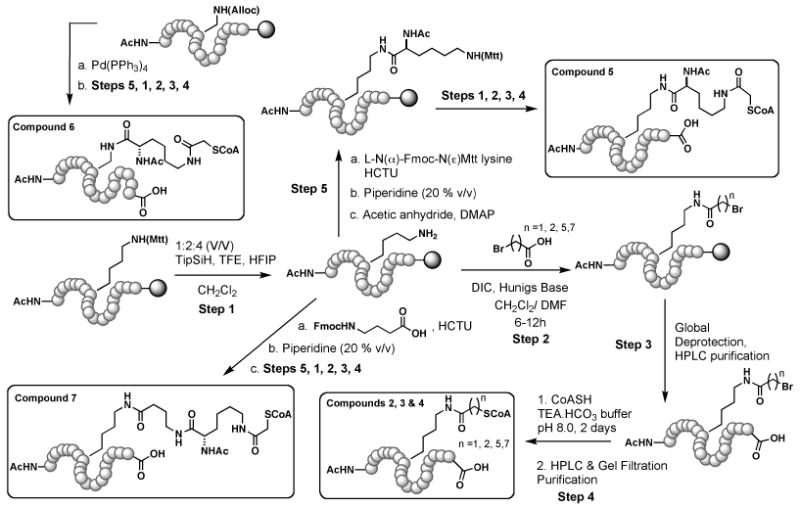
Synthesis of bisubstrate analogs (1-7). The shaded circle represents wang resin (solid phase support). The H4-20mer peptide sequence is as follows. NH2-SGRGKGGKGLGKGGAKRNRA-COOH
Compounds 1-7 along with Lys-CoA were tested as inhibitors of semisynthetic p300 HAT acetyltransferase using a direct radioactive assay in which 14C-acetyl-CoA is separated fron 14C-labeled product peptide (acetylated H4-15) using Tris Tricine gel electrophoresis. IC50 values were determined in the presence of 20 μM acetyl-CoA and 100 μM peptide and referenced to Lys-CoA which showed an IC50 of 0.06 μM under these conditions. We believe that the 7 min pre-incubation time with inhibitor along with the 7 min assay time is sufficient for equilibrium inhibition to be achieved based on prior studies [17]. As can be seen in Table 2, each of the compounds 1-7 was inferior in potency to Lys-CoA with IC50 values ranging from 0.4 to 3.2 μM. Notably, there was a clear correlation between the linker length and potency with the compounds with the shorter linker displaying the weaker IC50. In fact, compound 7 with the longest linker had an IC50 of 0.4 μM, only 7-fold weaker than Lys-CoA.
To further probe the inhibitory properties of 7, we investigated its potency against the D1625R/D1628R double mutant of p300 HAT. Compared to its inhibition of wt p300 HAT, compound 7 showed a 16-fold weakening of its IC50 value (Table 2, 6.7 μM) with the double mutant enzyme. A related analysis of Lys-CoA showed that its potency was only 3.5-fold weaker with the D1625R/D1628R p300 HAT compared with wt p300 HAT. Taken together, these data suggest that the negatively charged, putative peptide binding pocket of p300 HAT is more important for the inhibition by compound 7 than for Lys-CoA. Thus it is likely that the long linker of compound 7 allows for dual engagement of the Lys-CoA tunnel and peptide binding pocket which could account for the enhanced potency of 7 versus the parent compound Lys-CoA.
4. Discussion
Bisubstrate analog inhibitors for enzymes that catalyze reactions involving two substrates can be useful structural and functional tools [22-26]. Among the major challenges in bisubstrate analog inhibitor design is constructing the appropriate linker that can allow for optimal engagement of the two substrate binding sites while limiting entropic loss associated with forming a noncovalent tripartite complex. Here, we have addressed the paradoxical observation that a “partial” bisubstrate analog Lys-CoA is a much more potent p300/CBP HAT inhibitor as opposed to the more complete H4-CoA-20 bisubstrate analog. This unusual behavior may reflect the atypical kinetic mechanism of p300/CBP which appears to involve a fleeting interaction of the histone peptide substrate. Nevertheless, inspection of the X-ray structure of p300 HAT complexed to Lys-CoA suggested it could be possible to find a bisubstrate analog with a longer linker that stretched to cover the presumed peptide binding pocket.
Compound 7, an H4-CoA-20 analog with two consecutive Lys residues incorporated into the linker, proved to be about 7-fold more potent than the parent compound H4-CoA-20. We deduce that this linker allows for interactions of compound 7 to both the Lys-CoA tunnel as well as the peptide binding pocket, based on the enhanced affinity as well as the mutagenesis data, although it is formally possible that other binding interactions are involved. It is interesting that the predicted extended length of the long linker of 7 plus the three histone peptide residues to the next basic residue (a total of at least 25 angstroms) far exceeds the distance between the two binding surfaces apparent in the X-ray structure of p300 HAT. Several of the analogs of 2-6 would have been expected to have linkers sufficient to traverse this distance yet they showed reduced affinity compared to 7. It is thus likely that the distance to ensure correct orientation of the two ligand moieties of the peptide-CoA conjugates needs to be somewhat larger than the minimum distance. Yet, there is a penalty associated with excessively long linkers because of the diminishing returns on the entropic benefit of the tether.
While an improvement over H4-CoA-20 (1), the affinity of compound 7 as well as the other analogs (2-6) prepared in this study were far less than the simple and early Lys-CoA analog. This suggests that the presumed benefit from engaging the peptide binding pocket is outweighed by the potential clashing of the linker in some unfavorable fashion with the enzyme. Our findings may underscore the specialized challenge of designing bivalent inhibitors for enzymes that use Theorell-Chance catalytic mechanisms. The substrate promiscuity engendered by such a mechanism may allow p300/CBP HAT to play such a broad variety of roles in biology.
Acknowledgments
We thank NIH for funding this study. Also, we thank Dr. Ling Wang for her assistance with the purification of p300 HAT and HAT assays.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 2.Bannister AJ, Kouzarides T. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 3.Mellert HS, McMahon SB. Trends Biochem Sci. 2009;34:571–578. doi: 10.1016/j.tibs.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang L, Tang Y, Cole PA, Marmorstein R. Curr Op Struct Biol. 2008;18:741–747. doi: 10.1016/j.sbi.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gu W, Roeder RG. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 6.Thompson PR, Wang D, Wang L, Fulco M, Pediconi N, Zhang D, An W, Ge Q, Roeder RG, Wong J, Levrero M, Sartorelli V, Cotter RJ, Cole PA. Nat Struct Mol Biol. 2004;11:308–315. doi: 10.1038/nsmb740. [DOI] [PubMed] [Google Scholar]
- 7.Goodman RH, S S. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 8.Iyer NG, Oezdag H, Caldas C. Oncogene. 2004;23:4225–4231. doi: 10.1038/sj.onc.1207118. [DOI] [PubMed] [Google Scholar]
- 9.B A, Martinez-Balbás MA, Martin K, Haus-Seuffert P, Meisterernst M, Kouzarides T. EMBO J. 1998;17:2886–2893. doi: 10.1093/emboj/17.10.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kishimoto KT, Okudela K, Otsuka A, Sasaki H, Tanabe C, Sakiyama T, Hirama C, Kitabayashi I, Minna JD, Takenoshita S, Yokota J. Clin Cancer Res. 2005;11:512–519. [PubMed] [Google Scholar]
- 11.Cebrat M, Kim CM, Thompson PR, Daugherty M, Cole PA. Bioorg Med Chem. 2003;11:3307–3313. doi: 10.1016/s0968-0896(03)00265-7. [DOI] [PubMed] [Google Scholar]
- 12.Arif M, Pradhan SK, T GR, Vedamurthy BM, Agrawal S, Dasgupta D, Kundu TK. J Med Chem. 2009;52:267–277. doi: 10.1021/jm800657z. [DOI] [PubMed] [Google Scholar]
- 13.Bowers EM, Yan G, Mukherjee C, Orry A, Wang L, Holbert MA, Crump NT, Hazzalin CA, Liszczak G, Yuan H, Larocca C, Saldanha SA, Abagyan R, Sun Y, Meyers DJ, Marmorstein R, Mahadevan LC, Alani RM, Cole PA. Chem Biol. 2010;17:471–482. doi: 10.1016/j.chembiol.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cole PA. Nature Chem Biol. 2008;4:590–597. doi: 10.1038/nchembio.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng Y, Balasubramanyam K, Cebrat M, Buck D, Guidez F, Zelent A, Alani RM, Cole PA. J Am Chem Soc. 2005;127:17182–17183. doi: 10.1021/ja0558544. [DOI] [PubMed] [Google Scholar]
- 16.McKinley-McKee JS. Acta Chem Scand. 1963;17:S339–S342. [Google Scholar]
- 17.Thompson PR, Kurooka H, Nakatani Y, Cole PA. J Biol Chem. 2001;276:33721–33729. doi: 10.1074/jbc.M104736200. [DOI] [PubMed] [Google Scholar]
- 18.Liu X, Wang L, Zhao K, Thompson PR, Hwang Y, Marmorstein R, Cole PA. Nature. 2008;451:846–850. doi: 10.1038/nature06546. [DOI] [PubMed] [Google Scholar]
- 19.Tanner KG, Langer MR, Kim Y, Denu JM. J Biol Chem. 2000;275:22048–22055. doi: 10.1074/jbc.M002893200. [DOI] [PubMed] [Google Scholar]
- 20.Berndsen CE, Selleck W, McBryant SJ, Hansen JC, Tan S, Denu JM. Biochemistry. 2007;46:2091–2099. doi: 10.1021/bi602366n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lau OD, Kundu TK, Soccio RE, Ait-Si-Ali S, Khalil EM, Vassilev A, Wolffe AP, Nakatani Y, Roeder RG, Cole PA. Mol Cell. 2000;5:589–595. doi: 10.1016/s1097-2765(00)80452-9. [DOI] [PubMed] [Google Scholar]
- 22.Chung S, Parker JB, Bianchet M, Amzel LM, Stivers JT. Nat Chem Biol. 2009;5:407–413. doi: 10.1038/nchembio.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poux AN, Cebrat M, Kim CM, Cole PA, Marmorstein R. Proc Natl Acad Sci U S A. 2002;99:14065–14070. doi: 10.1073/pnas.222373899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu M, Magalhaes ML, Cook PF, Blanchard JS. Biochemistry. 2006;45:14788–14794. doi: 10.1021/bi061621t. [DOI] [PubMed] [Google Scholar]
- 25.Wu J, Xie N, Wu Z, Zhang Y, Zheng YG. Bioorg Med Chem. 2009;17:1381–6. doi: 10.1016/j.bmc.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 26.Osborne T, Roska RL, Rajski SR, Thompson PR. J Am Chem Soc. 2008;130:4574–4575. doi: 10.1021/ja077104v. [DOI] [PubMed] [Google Scholar]


