Abstract
Molecular mimicry between foreign and self antigens is a mechanism of T cell receptor cross-reactivity and is thought to contribute to the development of autoimmunity. The αβ TCR A6 recognizes the foreign antigen Tax from the virus HTLV-1 when presented by the class I MHC HLA-A2. In a possible link with the autoimmune disease HAM/TSP, A6 also recognizes a self peptide from the neuronal protein HuD in the context of HLA-A2. We found here that the complexes of the HuD and Tax epitopes with HLA-A2 are close but imperfect structural mimics, and that in contrast with other recent structures of TCRs with self antigens, A6 engages the HuD antigen with the same traditional binding mode used to engage Tax. Although peptide and MHC conformational changes are needed for recognition of HuD but not Tax and the difference of a single hydroxyl triggers an altered TCR loop conformation, TCR affinity towards HuD is still within the range believed to result in negative selection. Probing further, we found that the HuD/HLA-A2 complex is only weakly stable. Overall, these findings help clarify how molecular mimicry can drive self/non-self cross-reactivity and illustrate how low peptide/MHC stability can permit the survival of T cells expressing self-reactive TCRs that nonetheless bind with a traditional binding mode.
Introduction
T cell receptor (TCR) recognition of peptide antigens presented by class I or class II major histocompatibility complex (MHC) proteins is a cornerstone of cellular immunity. Recognition of foreign antigens forms the basis for defense against viruses and other pathogens, yet recognition of self antigens can lead to autoimmunity. Although the central tolerance mechanism of negative selection plays a protective role against autoimmunity, the high level of cross-reactivity present within the T cell repertoire can promote autoimmunity if T cells are activated by foreign antigens that share key structural or chemical features with self antigens. This concept of molecular mimicry has been implicated in a number of autoimmune pathologies, including type 1 diabetes, lupus, and multiple sclerosis (1–3).
Previously determined structures of TCRs in complex with self antigens associated with autoimmunity have shown deviations from the binding mode traditionally seen in TCR recognition of foreign antigens. The traditional binding mode, typified by the structure of the αβ A6 TCR with the HTLV-1 Tax peptide presented by HLA-A2 (4), has the TCR centered diagonally over the pMHC, with the CDR3α and CDR3β loops positioned near the center of the peptide. Rather than adopting this configuration, the Ob.1A12 and 3A6 TCRs bind near the N-terminus of the peptide, such that the focus of the TCRs is on the second and third positions of the peptide rather than the center residues (5, 6). In another case, recognition of the MBP Ac1-11 self-antigen by three different TCRs proceeds with a more traditional binding mode (7, 8), but the Ac1-11 peptide only partially occupies the MHC peptide binding groove (9), resulting in atypical TCR-peptide interactions resembling those seen with the OB.1A12 and 3A6 TCRs. Although each of the examples above involves TCRs restricted to class II MHCs, there is evidence of similar behavior in class I systems (10). A hypothesis stemming from these observations is that deviations from the “traditional” binding mode used to engage foreign antigens are responsible for low affinity TCR binding, allowing T cell escape from negative selection and contributing to the development of autoimmunity (11, 12).
However, the extent to which altered TCR binding would apply within the confines of molecular mimicry is unclear. Generally speaking, differential recognition of self and non-self by the same TCR would seem inconsistent with molecular mimicry as traditionally envisioned, i.e., the conservation of key features translating into a similar mode of recognition. Examinations of molecular mimicry in other systems, including TCR recognition of foreign mimics as well as other protein-protein interactions, have routinely shown a conservation of the overall binding mode (13–15). Indeed, although the two antigens were not classified beforehand as molecular mimics, the structure of the Ob.1A12 bound to a microbial antigen recapitulated the unusual binding mode seen with the MBP self antigen (16). In another demonstration, the murine TCR 2C cross-reacts with the foreign and self SIYR and dEV8 antigens via a common, traditional binding mode (17).
Yet differential docking of a single TCR on foreign and self pMHC ligands can occur, as demonstrated by the different orientation 2C adopts on the foreign QL9/H-2Ld ligand(18). Although this latter case is not one of molecular mimicry, unanticipated complexities observed in recent structural analyses of molecular mimics, including TCR, peptide, and MHC conformational changes and poor translation of sequence into structural mimicry (13, 14), warrants caution when presuming that foreign and self mimics will be recognized similarly.
Here we examined cross-recognition between self and non-self molecular mimics by the A6 TCR. As noted above, A6 recognizes the HTLV-1 Tax antigen presented by HLA-A2 (positions 11–19 of the Tax protein; sequence LLFGYPVYV). Although it is not highly promiscuous (19), A6 cross-reacts with a variety of modified Tax variants that share previously identified key residues, as well as microbial peptides such as the yeast Tel1p peptide (13, 19–22). In a possible link with autoimmunity, A6 also recognizes a self antigen from an immunodominant region of the human neuronal protein HuD(positions 87–95; sequence LGYGFVNYI) (21, 23). T cell recognition of epitopes from HuD have been suggested to play a role in the neurological disorder HAM/TSP (HTLV-1 associated myelopathy/tropical spastic paraparesis), a demyelinating autoimmune disease affecting a minority of HTLV-1 infected individuals(24, 25). Patients with HAM/TSP that are HLA-A2+ have persistent, high levels of CD8+ T cells specific for the Tax epitope in their peripheral blood and cerebral spinal fluid (26, 27), and the A6 T cell clone was isolated from an HAM/TSP affected individual (28). The Tax and HuD peptides have related sequences (Fig. 1A), sharing a key glycine at position 4 as well as aromatics at positions 5 and 8, which play predominant roles in the crystallographic structure of Tax/HLA-A2 bound to A6 (4).
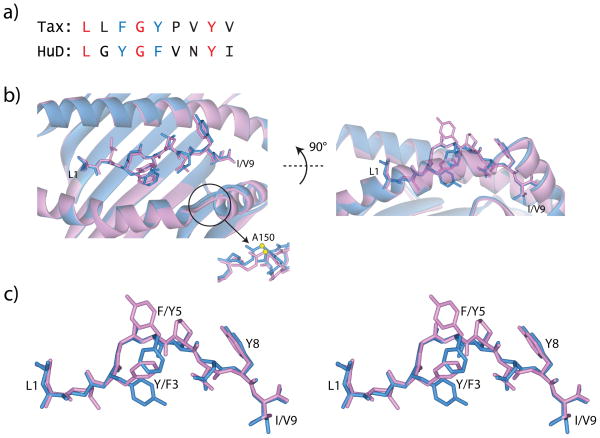
Figure 1.
The Tax and HuD complexes withHLA-A2 are close but imperfect mimics. A) Sequences of the Tax and HuD peptides. The conserved amino acids at positions 2, 4, and 8 are in red, the homologous tyrosine/phenylalanine aromatics at positions 3 and 5 are in blue. B) Overview of the peptides and peptide binding grooves in the Tax/HLA-A2 and HuD/HLA-A2 complexes. The Tax/HLA-A2 complex is pink and the HuD/HLA-A2 complex is light blue. Superimposition is through the backbones of amino acids 1-180 of the HLA-A2 heavy chains and all nine atoms of the peptides. The circle highlights a small shift in the α2 helix “linker region.” The shift is maximal at the α carbon of Ala150, as shown in the backbone trace below the ribbon diagram. C) Cross-eyed stereo view of the peptides from the superimposition in panel B. The backbones of the two peptides are identical. The only significant side chain differences occur at the side chains of positions 3 and 5 (phenylalanine and tyrosine in Tax, tyrosine and phenylalanine in HuD).
To provide further insight into self/non-self recognition within the context of molecular mimicry and help inform the possible link between TCR recognition of Tax and HuD in HAM/TSP, here we determined the structural basis for T cell cross-reactivity between the Tax and HuD epitopes. The structure of the HuD/HLA-A2 complex revealed it is a close but imperfect structural mimic of the Tax/HLA-A2 complex. Yet, the A6 TCR engaged the HuD ligand in the same “anti-foreign” manner as how it engages the Tax ligand, with no unusual features in the TCR-pMHC interface. Interestingly though, recognition of HuD required peptide and MHC conformational adjustments not needed for recognition of Tax, and the difference of a single hydroxyl group between the peptides triggered a different conformation for theA6 CDR3β loop. The structural changes contributed to a large, 70-fold weaker affinity of A6 for the HuD ligand. However, TCR affinity towards HuD was still within the range shown to result in negative selection in other systems (29). Probing further, we found that the HuD/HLA-A2 complex is only weakly stable, explaining why HuD-reactive T cells can be found in the periphery despite the existence of T cells that engage them with the traditional, “anti-foreign” binding mode.
Materials & Methods
Proteins and peptides
Soluble HLA-A2 and A6 TCR were refolded from bacterially expressed inclusion bodies as previously described (30). Peptides were synthesized locally or purchased from Genscript; peptide purity and identify were confirmed by LC-MS. Streptavidin was purchased from Rockland, Inc. The A6 TCR used included an engineered disulfide bond across the constant domains for improved heterodimer stability (31).
Crystallization and X-ray analysis
Peptide/HLA-A2 crystals were grown from 24% PEG 3350 in 25 mM MES, pH 6.5, 0.1 M NaF or 0.1 M NaCl. A6-HuD/HLA-A2 crystals were grown from15% PEG 4000 in 0.1 M Tris, pH 8.5, 0.2 M MgCl2. Cryo-protection for all crystals consisted of 20–25% glycerol. Diffraction data were collected at Argonne National Laboratory at the indicated beamlines at 100 K using a wavelength of 0.979 Å. Data reduction, structure solution, refinement, and structure validation was performed as previously described (19). Coordinates for search models for the pMHC structures were from PDB entries 1TVB (32)for pMHC and 2GJ6 (19) for TCR-pMHC. Data collection and refinement statistics are presented in Table 1. Surface area analysis was performed with the program surface racer using a probe radius of 1.4 Å (33). Shape complementarity (34) was calculated as implemented in CCP4 (35). Coordinates and structure factors are available from the Protein Data Bank (PDB; www.rscb.org). PDB accession numbers are 3PWLfor HuD/HLA-A2, 3PWPfor A6-HuD/HLA-A2, 3PWNfor HuDG2L/HLA-A2, and 3PWJ for HuDG2L/I9V/HLA-A2.
Table 1.
X-ray data collection and refinement statistics
| HuD/HLA-A2 | A6-HuD/HLA-A2 | HuDG2L/HLA-A2 | HuDG2L/I9V/HLA-A2 | |
|---|---|---|---|---|
| Data collection | ||||
| Source | APS 19BM | APS 31ID | APS 19BM | APS 19BM |
| Space group | P21 | C2 | P21 | P1 |
| Cell dimensions | ||||
| a, b, c (Å) | 58.3, 84.2, 84.0 | 224.0, 49.1, 93.7 | 58.2, 84.4, 83.9 | 50.2, 62.9, 74.7 |
| α, β, γ (°) | 90.0, 90.1, 90.0 | 90.0, 90.1, 90.0 | 90.0, 90.0, 90.0 | 82.0, 76.2, 78.1 |
| Resolution (Å) | 20–1.65 (1.71–1.65)* | 20–2.70 (2.75–2.70) | 20–1.60 (1.63–1.60) | 20–1.70 (1.76–1.70) |
| Rsym or Rmerge | 0.077 (0.53) | 0.078 (0.37) | 0.075 (0.507) | 0.042 (0.349) |
| I/σI | 15.5 (2) | 17.0 (2.0) | 18.8 (2.1) | 18.0 (1.9) |
| Completeness (%) | 100.0 (100.0) | 94.4 (63.2) | 96.1 (85.9) | 96.2 (92.0) |
| Redundancy | 3.7 (3.6) | 3.6 (2.7) | 3.6 (2.7) | 1.9 (1.8) |
| Refinement | ||||
| Resolution (Å) | 20–1.65 | 20–2.70 | 20–1.60 | 20–1.70 |
| No. reflections | 97644 | 27035 | 102861 | 91395 |
| Rwork/Rfree | 0.158/0.195 | 0.198/0.259 | 0.180/0.214 | 0.180/0.215 |
| No. atoms | ||||
| Protein | 6442 | 6644 | 6409 | 6406 |
| Ligand/ion | 54 | 47 | 30 | 24 |
| Water | 1072 | 66 | 777 | 693 |
| B-factors | ||||
| Protein | 13.3 | 55 | 15.7 | 13.2 |
| Ligand/ion | 37.8 | 70 | 36 | 46.5 |
| Water | 25.9 | 50.8 | 25.8 | 21.3 |
| R.M.S. deviations | ||||
| Bond lengths (Å) | 0.017 | 0.013 | 0.016 | 0.017 |
| Bond angles (°) | 1.725 | 1.69 | 1.67 | 1.773 |
| Ramachandran statistics (%) | ||||
| Most favored | 93.3 | 87.1 | 93.1 | 93.5 |
| Allowed | 6.4 | 12.4 | 6.6 | 6.2 |
| Generously allowed | 0.3 | 0.3 | 0.3 | 0.3 |
Values in parentheses are for highest-resolution shell.
Surface plasmon resonance
Surface plasmon resonance measurements were performed with a Biacore 3000 instrument as previously described (30). All data were collected at 25 °C. Solution conditions were 10 mM HEPES, 3 mM EDTA, 150 mM NaCl, 0.005% surfactant P-20, pH 7.4. Peptide/MHC complexes were coupled to the sensor surface via a biotin-streptavidin linkage, with the streptavidin amine coupled to a CM5 sensor chip. Biotinylated HuD/HLA-A2, HuDG2L/HLA-A2, and HuDG2L/I9V/HLA-A2 were coupled to three of the flow cells, with the fourth serving as a reference. Soluble A6 TCR was then injected at 5 μL/min until steady-state was reached. Each injection series was repeated twice and the data globally analyzed. Data were processed in BIAevaluation 4.1 and analyzed with Origin 7.5.
Thermal stability
Circular dichroism measurements of pMHC thermal stability were performed using a Jasco J-815 spectrometer monitoring 218 nm. Solution conditions were 20 mM phosphate and 75 mM NaCl (pH 7.4). Sample concentrations were 5 μM. A temperature increment of 1 °C/min was used, maintaining the temperature within 0.1 °C for 5 seconds before sampling. As peptide/HLA-A2complexes do not show reversible unfolding, the data were fit to a nine order polynomial and the apparent Tm taken from the maximum of the first derivative of the fitted curve.
Results
The HuD/HLA-A2 self-complex is a close but imperfect mimic of the Tax/HLA-A2 non-self complex
We first determined the structure of the HuD/HLA-A2 complex at 1.65 Å resolution (Table 1). The complex displayed the usual class I pMHC architecture with two molecules per asymmetric unit (Fig. 1A). There are no substantial differences between the two molecules in the unit cell (all atoms of the two peptides superimpose with an RMSD of 0.1 Å; the backbones of the HLA-A2 peptide binding domains superimpose with an RMSD of 0.4 Å). Electron density images for the peptides in the HLA-A2 peptide binding grooves are in Supplemental Fig. 1.
Comparison of the HuD/HLA-A2 complex withtheTax/HLA-A2 complex (36)revealed the HuD complex to be a close but imperfect mimic of the Tax complex. Other than a small region of the α2 helix discussed below, the HLA-A2 heavy chains are in full alignment (Fig. 1B). The peptide backbones adopt the same conformation, superimposing with a0.4 Å RMSD (Fig. 1C). The majority of the peptide side chains overlay each other. However, the side chain of Tyr3 of HuD is rotated 110° compared to its analogue Phe3 in Tax, pointing the side chain towards the base of the peptide binding groove. Likewise, the side chain of Phe5 is rotated 120° compared to its analogue of Tyr5 in Tax, also pointing the side chain towards the base of the binding groove. The concerted difference in the positions 3 and 5 side chains is attributable to differences in π-stacking between the two aromatic side chains: in HuD, Tyr3 and Phe5 are arranged in a classic parallel-displaced arrangement, whereas in Tax they are arranged in a T-shaped fashion(37). The differences reflect a common structural and energetic link between non-adjacent peptide side chains in pMHC complexes(38), and alter how an incoming TCR would see the peptide, particularly at the position 5 amino acid, which in the complex of the A6 TCR with the Tax peptide is accommodated in a central pocket formed by theCDR3α and CDR3β loops (4).
The small “linker region” connecting the short and long helical elements of the α2 helix in the HuD/HLA-A2complex is slightly shifted relative to its position in Tax/HLA-A2 (highlighted in Fig. 1B; electron density images for this region are shown in Supplementary Fig. 3). Peptide-dependent shifts have been noted in this region previously (e.g., ref.39), likely reflecting an influence of the peptide on HLA-A2 flexibility as recently described with the yeast Tel1p peptide (13). The shift is small, only 1.1 Å at the α carbon of Ala150. However, as discussed below, small changes in this region impact TCR recognition of the HuD/HLA-A2 complex.
The A6-HuD/HLA-A2 ternary complex is very similar to the A6-Tax/HLA-A2 complex
We next determined the structure of the ternary complex of the A6 TCR bound to HuD/HLA-A2 at a resolution of 2.7 Å (Table 1; see Supplemental Fig. 2 for electron density images). The complex is very similar to the ternary complex of A6 with the Tax peptide, with the TCR positioning itself over HuD/HLA-A2 exactly as it does over Tax/HLA-A2, forming the same docking angle and tilt(Fig. 2A). With the exception of CDR3β, which is shifted towards the periphery of the interface as discussed below, the loops of A6 are in the same conformation in both complexes(Fig. 2B). A6 recognition of the Tax and HuD foreign and self antigens thus proceeds with the same traditional binding mode.
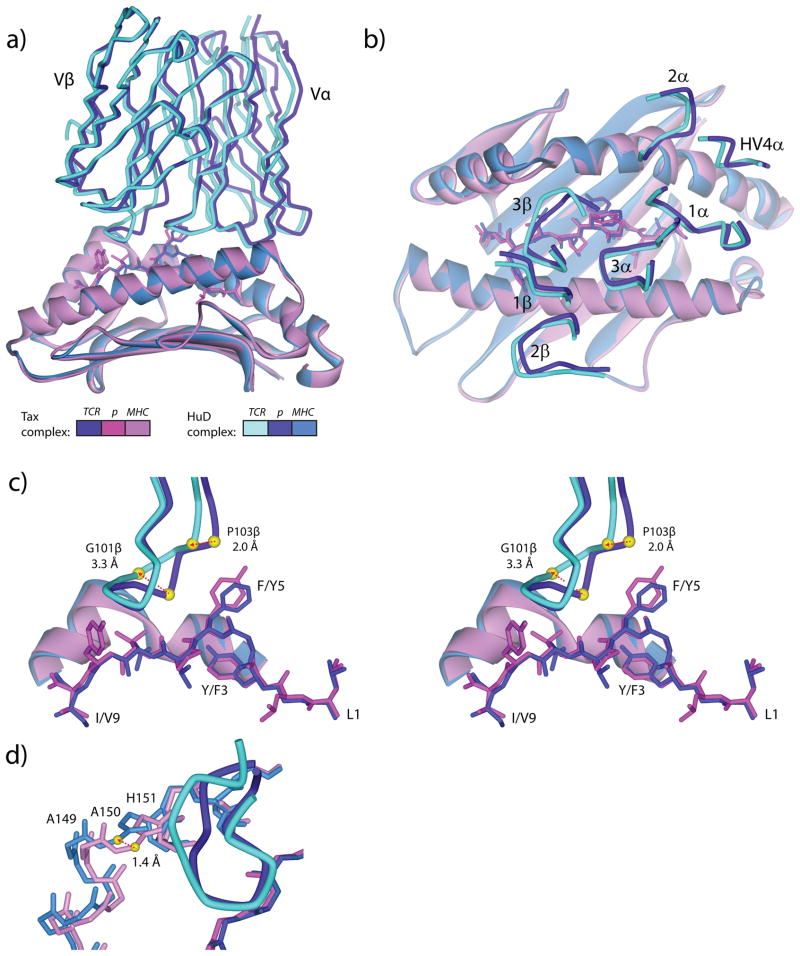
Figure 2.
The A6 TCR engages the HuD/HLA-A2 ligand identically to how it engages the Tax/HLA-A2 ligand, with a reorganization of the CDR3β loop and a small shift in the α2 helix linker region. A) Overall comparison of the A6-HuD/HLA-A2 and A6-Tax/HLA-A2 complexes, showing the TCR variable domains, peptides, and the HLA-A2 peptide binding domain. The coloring scheme is indicated below the figure and is maintained in all panels. Superimposition is through the backbones of the TCR variable domains, peptides, and HLA-A2 peptide binding domains. B) Top view of the superimposed TCR-pMHC complexes, showing the CDR loops over the pMHC molecules. Other than CDR3β, the CDR loops are in the same position in the HuD and Tax complexes. C) Cross-eyed stereo view showing the differential positioning of CDR3β in the HuD and Tax complexes. The α carbons of Gly101 and Pro103 shift by 3.3 and 2.0 Å, respectively. D) The linker region of the HLA-A2 α2 helix is displaced away from the peptide upon recognition of HuD. Measured at the α carbon of Ala150, the displacement is 1.4 Å.
The difference in the TCR CDR3β loop between the HuD and Tax complexes is maximal at the α carbon of Gly101β, which in the HuD complex is moved towards the periphery of the interface by3.3 Å (Fig. 2C). Further along the loop, Pro103β is displaced by 2.0 Å. Although the differences in the loop between the two structures is structurally significant, in context they are not unusual, as the maximal 3.3 Å shift is just below the average CDR3β movement tabulated in a recent comparison of free and bound TCRs (40). There is no obvious steric reason for the shift in CDR3β. However, CDR3β in A6 has been found to adopt a range of conformations in various structures with variants and mimics of the Tax peptide (13, 19, 20, 41). As discussed below, the loop shift here reflects a need to optimize interface electrostatics, triggered by the loss of a hydrogen bond between position 5 of the peptide and the TCR.
The shift in the α2 helix linker region noted in the structure of the free HuD/HLA-A2 complex is more pronounced in the A6-HuD/HLA-A2 complex(Fig. 2D), with the region backing away from its position in the A6-Tax/HLA-A2 complex by 1.4 Å as measured at the α carbon of Ala150. Although smaller and less of a reorganization than that seen for CDR3β, movement of the α2 helix impacts the contacts made across the interface, likely contributing to the weaker affinity A6 maintains towards HuD/HLA-A2.
Although the TCR binds with the same mode, the amino acid differences between Tax and HuD, the different CDR3β conformation, and the small shift in the α2 helix linker region lead to a number of altered contacts within the TCR-pMHC interface. The majority of the differences are between CDR3β, the center of the peptide, and the shifted region of the HLA-A2 α2 helix. There is a net loss of six hydrogen bonds or salt-bridges in the HuD structure. Despite the changes, with few exceptions the pattern of amino acids used to form the complex is similar in Tax and HuD. The “hotspot” residue of Arg65 of the HLA-A2 α2 helix remains a major participant (42, 43), as do the other amino acids comprising the proposed class I MHC “restriction triad” (Arg65, Ala69, and Gln155 in HLA-A2) (44). A detailed comparison of the structural features of the HuD and Tax TCR-pMHC interfaces is provided in Supplemental Fig. 4.
Are there features in the A6-HuD/HLA-A2 interface easily identifiable as “sub-optimal?” As noted above, the shift in CDR3β is within the range of those seen previously. The amount of buried solvent accessible surface area in the HuD and Tax interfaces is nearly identical (2084 Å2 with Tax vs. 2128 Å2 with HuD), as is the proportion of buried hydrophobic/hydrophilic surface (54% hydrophobic with Tax, 55% hydrophobic with HuD). The total number of interatomic contacts is also nearly identical (130 with Tax vs. 126 with HuD). Although the number of hydrogen bonds and salt-bridges is reduced in the HuD complex compared to the Tax complex (15 with Tax, 9 with HuD), the number present is still within the range seen in other TCR-pMHC complexes with foreign antigens (45). Although the contacts are altered, the general pattern of amino acids contacted on both sides of the is similar with both the HuD and Tax peptides(see Supplemental Fig. 4). Lastly, the shape complementarity in the A6-HuD/HLA-A2 interface is actually improved compared to that in the A6-Tax/HLA-A2 interface (0.66 with HuD vs. 0.63 with Tax). Thus, a priori, the complex of A6 with HuD/HLA-A2 does not show any easily identifiable “sub-optimal” features.
The shift in CDR3β is attributable to imperfect chemical mimicry between HuD and Tax
Although the CDR3β loop is in a different conformation in the HuD structure than in the Tax structure, we noticed that the conformation of the loop is very similar to an alternate conformation observed previously with a variant of the Tax peptide. This peptide, referred to as TaxY5F(3,4FF), replaced Tyr5 of Tax with a doubly-fluorinated phenylalanine derivative, with one of the two fluorines substituting for the tyrosine hydroxyl (41)(Fig. 3A). The fact that both structures showed nearly the same altered loop conformation suggested to us a mechanism in which the conformation of the CDR3β loop is dependent upon electrostatic interactions in the interface.
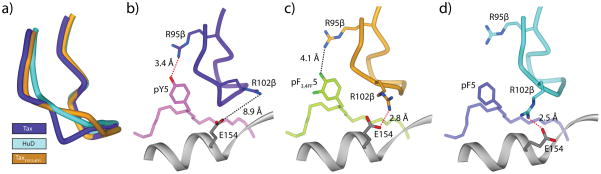
Figure 3.
The altered conformation of the CDR3β loop in the A6-HuD/HLA-A2 complex results from the loss of a single hydrogen bond and the subsequent need tore-optimize interface electrostatics. A) Comparison of CDR3β from the Tax, HuD, and TaxY5F(3,4FF) complexes, generated through superimposition of the backbones of the TCR Vα/Vβ domains. B) In the Tax complex, Tyr5 of the peptide hydrogen bonds to Arg95 of CDR3β. There is no interaction between Arg102β and Glu154 of the HLA-A2 heavy chain. C) In the HuD complex, there is no hydrogen bond to Arg95 of CDR3β. To compensate, the reorganized loop forms a salt-bridge between Arg102β and Glu154 of the HLA-A2 heavy chain. D) In the TaxYF5(3,4FF) complex, the hydrogen bond to Arg95β is also lost, and the reorganized loop forms a similar salt-bridge with Glu154 of the heavy chain.
Notably, the HuD and the TaxY5F(3,4FF) peptides alter interface electrostatics similarly. In the structure with the native Tax peptide, the side chain of Arg95of CDR3β, at the apex of the pocket formed by the CDR3α and CDR3β loops, hydrogen bonds to the Tax Tyr5 hydroxyl (Fig. 3B). Neither the HuD peptide nor the TaxY5F(3,4FF) peptide can form this hydrogen bond(fluorine is a poor hydrogen bond acceptor(46)), and in both cases Arg95β moves away from the side chain (Fig. 3C–D). The loss of this hydrogen bond in both structures is countered by the formation of a new salt bridge between Arg102β seven residues down the loop and Glu154of the HLA-A2 heavy chain. The difference in the position of the CDR3β loop in the HuD and TaxY5F(3,4FF) complexes thus results from the need to optimize electrostatics between the TCR and the pMHC, stemming from the loss of the hydrogen bond to the position five side chain. In other words, the difference of a single hydroxyl appears to trigger the altered positioning of the CDR3β loop.
Peptide conformational changes in HuD but not Tax upon A6 binding contribute to a weaker TCR affinity
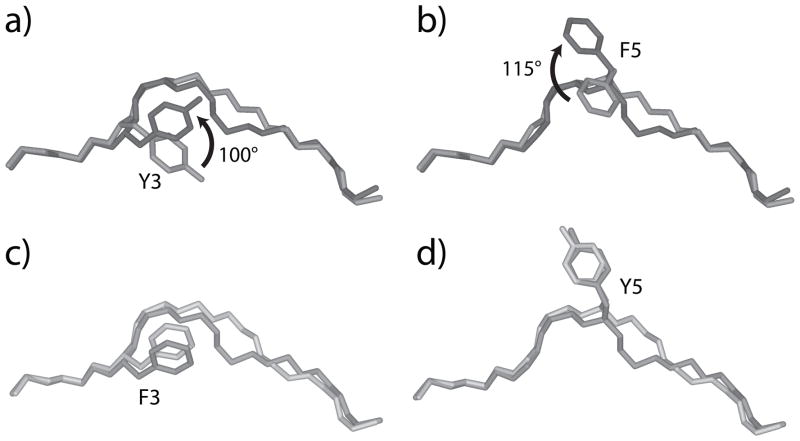
In the complex with the TCR, the HuD peptide adopts the same conformation as the Tax peptide, as is clear in Fig. 2. As the side chains of Tyr3 and Phe5 of the HuD peptide are presented by HLA-A2 in conformations different than their counterparts in the Tax peptide, they must therefore rotate in order for the ternary complex to form (Fig. 4A,B). Similar conformational changes are not needed for A6 recognition of the Tax peptide, as the side chains of Phe3 and Tyr5 are presented in conformations compatible with TCR recognition (Fig. 4C,D).
Figure 4.
Side chain conformational changes occur in A6 recognition of HuD but not Tax. A) Superimposed HuD peptides from the HuD/HLA-A2 and A6-HuD/HLA-A2 complexes, showing the 100° rotation of Tyr3 that occurs upon TCR binding. The peptide from the pMHC complex is light grey, the peptide from the TCR-pMHC complex is dark grey, with the arrow indicating the direction of rotation. B) Same as in panel A, but showing the 115° rotation in Phe5 that occurs upon TCR binding. C-D) Superimposed Tax peptides from the Tax/HLA-A2 and A6-Tax/HLA-A2 complexes, showing that unlike HuD, rotations in the positions of Phe3 (C) and Tyr5 (D) are not needed for TCR recognition. Peptides from the pMHC complex are light grey, peptides from the TCR-pMHC complex are dark grey.
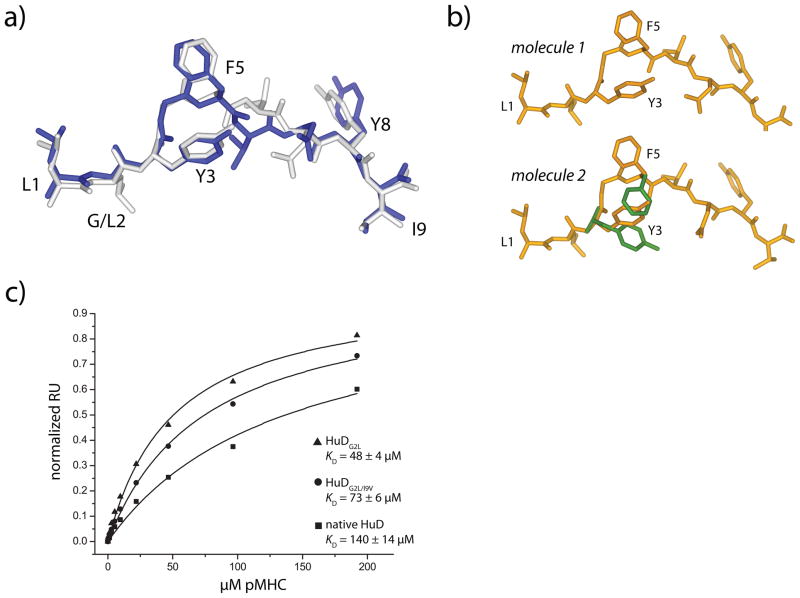
With glycine at position 2 and isoleucine at position 9, the HuD peptide has poor anchor residues for HLA-A2 (47). We were initially concerned that reduced stability of the pMHC complex would impede crystallization and thus studied anchor-modified variants of the HuD peptide. In the first variant, glycine 2 was substituted with leucine (HuDG2L). The second variant included the Gly2→Leu modification but also substituted isoleucine 9 with valine (HuDG2L/I9V). Although as demonstrated above reduced pMHC stability did not impact our studies with the native peptide, we did crystallize and determine the structures of the complexes of the two anchor-modified peptides with HLA-A2(Table 1; see Supplemental Fig. 3 for electron density images). Both complexes were identical to the complex with the native peptide except in the positioning of the side chains of Tyr3 and Phe5. Unlike the native peptide, in the HuDG2L/HLA-A2 complex, the peptide was presented with both side chains mimicking their conformations in the A6-HuD/HLA-A2 complex (this was true for both molecules in the asymmetric unit, for which all atoms of the peptide superimposed with an RMSD of 0.2 Å)(Fig. 5A). The positioning of the side chains in the HuDG2L/I9V/HLA-A2 structure was more complicated: the Tyr3 and Phe5 side chains in the first molecule in the asymmetric adopted their TCR-bound conformations, but the electron density in the second molecule in the asymmetric unit allowed the side chains to be modeled in both the TCR-bound and TCR-free conformations of the native peptide (Fig. 5B). Anchor modification thus perturbed the side chain conformational equilibrium towards the TCR-bound state, with a greater perturbation in the singly modified HuDG2L peptide. The mechanism behind the perturbations is not clear, as there are no backbone or side chain changes propagated from the anchor residues, and there are no crystallographic contacts in any of the structures that could influence side chain position. However, anchor modification of class I MHC-presented peptides has previously been shown to affect the conformation of amino acids distant from the site of substitution (48–50).
Figure 5.
Anchor modification in the HuD peptide reveals side chain conformational differences as a component of weaker TCR affinity. A) The conformation of the peptide in the HuDG2L/HLA-A2 complex is identical to the conformation of the peptide in the ternary complex with the native, unmodified HuD peptide. The anchor-modified peptide is white, and the native peptide from the ternary complex is dark blue. B) The peptide in the first molecule in the asymmetric unit of the doubly-modified G2L/I9V HuD/HLA-A2 complex adopts the conformation seen in the complex of the native peptide with the A6 TCR (top), whereas the peptide in the second molecule in the asymmetric unit could be refined in both the conformation seen when the native peptide is TCR-bound and when the native peptide is TCR-free (alternate positions in green). C) Surface plasmon resonance indicates that the affinity of the A6 TCR is higher for the anchor-modified peptide/HLA-A2 complexes than it is for the native HuD/HLA-A2 complex, consistent with the structural results showing anchor-modification biases the side chain conformational equilibrium towards a binding-competent state.
A prediction from these structural results is that the affinity of A6 should be stronger for the anchor-modified HuD ligands than it is for the native ligand. To confirm this, we examined the affinity of the A6 TCR towards the native and the two anchor-modified HuD ligands in a Biacore binding assay. TCR affinity for the native ligand was weak at 140 μM, in good agreement with a previously reported measurement (23). Fully consistent with the structural data, the affinity for the doubly modified HuDG2L/I9V variant was two-fold stronger at 70μM, whereas affinity for the singly modified HuDG2L variant was stronger still at 48 μM (Fig. 5C). For comparison, the affinity for the Tax ligand is a much stronger 2 μM(30).
The results with the anchor-modified peptides can be extrapolated to help explain the difference in the A6 TCR’s affinity towards the native HuD and Tax ligands. As A6 presents its central side chains in their TCR-bound conformation whereas HuD does not, we can conclude that one reason why A6 recognizes HuD more weakly than Tax is that, unlike Tax, the central aromatic side chains of the HuD peptide must shift into a binding-competent conformation in order for recognition to proceed.
The HuD/HLA-A2 complex is of low stability relative to the Tax/HLA-A2 complex
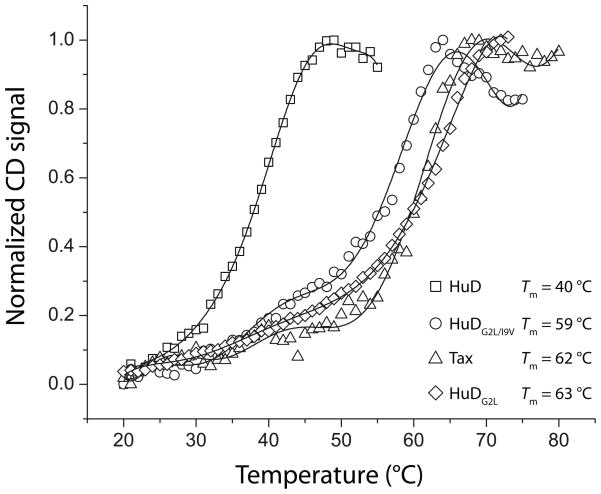
Although A6 TCR recognized the HuD/HLA-A2 complex with a weaker affinity than the Tax/HLA-A2 complex, the affinity of 140 μM is still within a range believed to result in negative selection(29). As poor antigen presentation levels can also lead to escape from negative selection (51) and the HuD peptide has suboptimal anchors for HLA-A2, we examined the thermal stability of the HuD/HLA-A2 complex using circular dichroism (CD) spectroscopy. CD has long been used to examine peptide/MHC stability, and measurements of thermal stability correlate with peptide-MHC binding affinity(52). The Tm of the native HuD/HLA-A2 complex was measured as 40°C(Fig. 6). As expected, anchor modification of the HuD peptide enhanced stability, with the Tm of the doubly-modified HuDG2L/I9V complex measured as 59 °C and the Tm of the singly modified HuDG2L complex measured as 63 °C. For comparison, the Tm of the Tax/HLA-A2 complex was measured as 62 °C, in close agreement with previous studies (13, 36). Thus, the native HuD/HLA-A2 complex has exceedingly low stability relative to the Tax/HLA-A2 complex.
Figure 6.
The HuD/HLA-A2 complex is substantially less stable than the Tax complex as well as the anchor-modified HuDG2L and HuDG2L/I9V complexes as demonstrated by measurements of thermal stability monitored by CD spectroscopy. The solid lines represent polynomial fits to the data, and the apparent Tm values, taken from the first derivatives of the fitted curves, are indicated in the legend.
Discussion
T cell cross-reactivity between foreign and self antigens has been implicated in a number of autoimmune pathologies, and molecular mimicry between self and non-self is frequently discussed as an underlying component. Yet structures of TCRs bound to self antigens implicated in autoimmunity have shown unusual features that distinguish them from structures of TCRs bound to foreign antigens (5–8). These observations have led to hypotheses that relate unusual TCR binding to reduced TCR affinity and T cell escape from negative selection, permitting the development of autoimmunity(11, 12). The extent to which unusual binding would apply in the context of molecular mimicry however is unclear: unusual recognition of a self antigen together with more traditional recognition of a foreign antigen by the same TCR is inconsistent with molecular mimicry as traditionally envisioned, i.e., the conservation of key structural or chemical features translating into a similar mode of recognition. Although it is possible that conservation of “hot spots” could still permit differential TCR binding, resembling to some extent how the 2C TCR cross-reacts with the QL9/H-2Ld and dEV8/H-2Kb ligands (18), this still stretches the expectations of a molecular mimicry mechanism.
Cross-reactivity by the A6 TCR between the foreign antigen Tax and the self antigen HuD provided us with an opportunity to examine the structural basis for self/non-self cross-reactivity within the context of molecular mimicry. Consistent with a molecular mimicry mechanism, HLA-A2 presents the Tax and HuD peptides in very similar conformations, with differences in the positions of only two side chains. Also consistent with molecular mimicry, the A6 TCR engagesHuD/HLA-A2 with the same center-focused, diagonal binding mode it uses to engageTax/HLA-A2, with no differences in position or orientation. A traditional molecular mimicry mechanism in terms of both antigen presentation as well as receptor recognition can therefore account for TCR cross-reactivity between the Tax and HuD antigens.
Although the traditional “anti-foreign” TCR binding mode is conserved, there are differences in the interaction betweenA6 and the Tax and HuD complexes with HLA-A2. Unlike Tax, the central aromatic side chains of the HuD peptide must undergo conformational changes in order for the TCR to bind. There is a shift in the α2 helix that occurs upon recognition of HuD but not Tax. The inability of the central phenylalanine side chain of the HuD peptide to form a hydrogen bond within the CDR3α/CDR3β pocket results in a repositioning of the CDR3β loop. It is thus only due to the structural plasticity available to both the TCR and the pMHC that A6 is able to cross-react between the two ligands. This conformational “melding” of both receptor and ligand is not unique to this case: A6 cross-reacts between Tax and its foreign mimic from the yeast Tel1p protein via similar, albeit larger, conformational changes in the TCR, peptide, and the MHC (13). The LC13 TCR cross-reacts between foreign peptides with related sequences but divergent structures by forcing them to adopt similar conformations in the TCR-bound state(14), and the CDR3α and CDR3β loops of LC13 undergo significant changes from their unbound forms (53, 54). Recognition of the pBM8/H-2kbm8 ligand by the BM3.3 TCR occurs with changes in peptide and MHC, and although the structure of the free BM3.3 TCR is not available, the CDR3α and CDR3β loops adopt different conformations upon recognition of other ligands(55). To a lesser extent, conformational changes occur in foreign and self peptide/DR2b complexes upon recognition by the Ob.1A12, and again, there are differences in CDR loop positions in the foreign and self ternary complexes, and binding proceeds with kinetic and thermodynamic indicators of conformational changes (5, 16, 56). Thus, although previous work highlighted the need to focus on structural as opposed to just sequence similarities when considering molecular mimicry (2, 3, 57), it is becoming increasingly clear that both ligand and receptor adaptability or flexibility and the resultant conformational melding often needed for binding add an additional layer of complexity.
Although CD8+ T cell recognition of HuD antigens has been implicated in HAM/TSP, and although the A6 TCR was cloned from a HAM/TSP patient, whether T cell recognition of cells presenting the HuD peptide is a component of disease is controversial. Nonetheless, HuD-specific T cells such as those expressing A6 must still escape the process of negative selection. If low affinity stemming from an altered TCR binding mode cannot explain this, what mechanisms can? The structural melding needed for A6 engagement of HuD weakens affinity 70-foldcompared to the Tax ligand. However, the affinity of approximately 140 μM may not be sufficiently weak to allow escape, as TCR-pMHC interactions with affinities in this range can still result in negative selection (29). Amore likely explanation is found in the low stability of the HuD/HLA-A2 complex: in addition to weak TCR binding affinities, low levels of antigen presentation can also permit T cell escape from negative selection (51), and poorly stable pMHC complexes require correspondingly higher affinity TCR affinities for productive signaling (58). The HuD self antigen thus provides an example of how weak peptide binding to MHC can permit the survival of T cells with receptors capable of recognizing self antigens with a traditional anti-foreign binding mode. Similar observations have been made with the 2C TCR (17), suggesting such behavior may be common. Thus, cross-recognition of weakly stable self mimics of foreign antigens via conserved, traditional binding modes likely contribute to the development and progression of autoimmunity.
Supplementary Material
Acknowledgments
Supported by GM067079 from NIGMS, NIH. KHP was supported by the Notre Dame CBBI training program, funded by GM075762 from NIGMS, NIH. OYB was supported by a fellowship from the Walther Cancer Foundation. Results shown in this report are derived from work performed at the Structural Biology Center, LRL-CAT, and LS-CAT at the Advanced Photon Source, Argonne National Laboratory. Argonne is operated by UChicago Argonne, LLC, for the U.S. Department of Energy under contract DE-AC02-06CH11357. Use of the LRL-CAT facilities at Sector 31 of the Advanced Photon Source was provided by Eli Lilly & Company who operates the facility.
We thank Cynthia Piepenbrink for outstanding technical assistance.
References
- 1.Oldstone MBA. Molecular mimicry and immune-mediated diseases. FASEB J. 1998;12:1255–1265. doi: 10.1096/fasebj.12.13.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kohm AP, Fuller KG, Miller SD. Mimicking the way to autoimmunity: an evolving theory of sequence and structural homology. Trends Microbiol. 2003;11:101–105. doi: 10.1016/s0966-842x(03)00006-4. [DOI] [PubMed] [Google Scholar]
- 3.Wucherpfennig KW. Structural Basis of Molecular Mimicry. Journal of Autoimmunity. 2001;16:293–302. doi: 10.1006/jaut.2000.0499. [DOI] [PubMed] [Google Scholar]
- 4.Garboczi DN, Ghosh P, Utz U, Fan QR, Biddison WE, Wiley DC. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996;384:134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- 5.Hahn M, Nicholson MJ, Pyrdol J, Wucherpfennig KW. Unconventional topology of self peptide-major histocompatibility complex binding by a human autoimmune T cell receptor. Nat Immunol. 2005;6:490–496. doi: 10.1038/ni1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y, Huang Y, Lue J, Quandt JA, Martin R, Mariuzza RA. Structure of a human autoimmune TCR bound to a myelin basic protein self-peptide and a multiple sclerosis-associated MHC class II molecule. EMBO J. 2005;24:2968–2979. doi: 10.1038/sj.emboj.7600771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maynard J, Petersson K, Wilson DH, Adams EJ, Blondelle SE, Boulanger MJ, Wilson DB, Garcia KC. Structure of an autoimmune T cell receptor complexed with class II peptide-MHC: insights into MHC bias and antigen specificity. Immunity. 2005;22:81–92. doi: 10.1016/j.immuni.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 8.Feng D, Bond CJ, Ely LK, Maynard J, Garcia KC. Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction ‘codon’. Nat Immunol. 2007;8:975–983. doi: 10.1038/ni1502. [DOI] [PubMed] [Google Scholar]
- 9.He X-l, Radu C, Sidney J, Sette A, Ward ES, Garcia KC. Structural Snapshot of Aberrant Antigen Presentation Linked to Autoimmunity: The Immunodominant Epitope of MBP Complexed with I-Au. Immunity. 2002;17:83–94. doi: 10.1016/s1074-7613(02)00340-0. [DOI] [PubMed] [Google Scholar]
- 10.Gras S, Saulquin X, Reiser JB, Debeaupuis E, Echasserieau K, Kissenpfennig A, Legoux F, Chouquet A, Le Gorrec M, Machillot P, Neveu B, Thielens N, Malissen B, Bonneville M, Housset D. Structural Bases for the Affinity-Driven Selection of a Public TCR against a Dominant Human Cytomegalovirus Epitope. J Immunol. 2009;183:430–437. doi: 10.4049/jimmunol.0900556. [DOI] [PubMed] [Google Scholar]
- 11.Wucherpfennig KW, Call MJ, Deng L, Mariuzza R. Structural alterations in peptide-MHC recognition by self-reactive T cell receptors. Curr Opin Immunol. 2009;21:590–595. doi: 10.1016/j.coi.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng L, Mariuzza RA. Recognition of self-peptide-MHC complexes by autoimmune T-cell receptors. Trends in Biochemical Sciences. 2007;32:500–508. doi: 10.1016/j.tibs.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borbulevych OY, Piepenbrink KH, Gloor BE, Scott DR, Sommese RF, Cole DK, Sewell AK, Baker BM. T cell receptor cross-reactivity directed by antigen-dependent tuning of peptide-MHC molecular flexibility. Immunity. 2009;31:885–896. doi: 10.1016/j.immuni.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macdonald WA, Chen Z, Gras S, Archbold JK, Tynan FE, Clements CS, Bharadwaj M, Kjer-Nielsen L, Saunders PM, Wilce MCJ, Crawford F, Stadinsky B, Jackson D, Brooks AG, Purcell AW, Kappler JW, Burrows SR, Rossjohn J, McCluskey J. T Cell Allorecognition via Molecular Mimicry. 2009;31:897–908. doi: 10.1016/j.immuni.2009.09.025. [DOI] [PubMed] [Google Scholar]
- 15.Beckett D. Functional Switches in Transcription Regulation; Molecular Mimicry and Plasticity in Protein-Protein Interactions. Biochemistry. 2004;43:7983–7991. doi: 10.1021/bi049890b. [DOI] [PubMed] [Google Scholar]
- 16.Harkiolaki M, Holmes SL, Svendsen P, Gregersen JW, Jensen LT, McMahon R, Friese MA, van Boxel G, Etzensperger R, Tzartos JS, Kranc K, Sainsbury S, Harlos K, Mellins ED, Palace J, Esiri MM, van der Merwe PA, Jones EY, Fugger L. T cell-mediated autoimmune disease due to low-affinity crossreactivity to common microbial peptides. Immunity. 2009;30:348–357. doi: 10.1016/j.immuni.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 17.Degano M, Garcia KC, Apostolopoulos V, Rudolph MG, Teyton L, Wilson IA. A functional hot spot for antigen recognition in a superagonist TCR/MHC complex. Immunity. 2000;12:251–261. doi: 10.1016/s1074-7613(00)80178-8. [DOI] [PubMed] [Google Scholar]
- 18.Colf LA, Bankovich AJ, Hanick NA, Bowerman NA, Jones LL, Kranz DM, Garcia KC. How a Single T Cell Receptor Recognizes Both Self and Foreign MHC. Cell. 2007;129:135–146. doi: 10.1016/j.cell.2007.01.048. [DOI] [PubMed] [Google Scholar]
- 19.Gagnon SJ, Borbulevych OY, Davis-Harrison RL, Turner RV, Damirjian M, Wojnarowicz A, Biddison WE, Baker BM. T Cell Receptor Recognition via Cooperative Conformational Plasticity. Journal of Molecular Biology. 2006;363:228–243. doi: 10.1016/j.jmb.2006.08.045. [DOI] [PubMed] [Google Scholar]
- 20.Ding YH, Baker BM, Garboczi DN, Biddison WE, Wiley DC. Four A6-TCR/peptide/HLA-A2 structures that generate very different T cell signals are nearly identical. Immunity. 1999;11:45–56. doi: 10.1016/s1074-7613(00)80080-1. [DOI] [PubMed] [Google Scholar]
- 21.Hausmann S, Biddison WE, Smith KJ, Ding YH, Garboczi DN, Utz U, Wiley DC, Wucherpfennig KW. Peptide recognition by two HLA-A2/Tax11-19-specific T cell clones in relationship to their MHC/peptide/TCR crystal structures. J Immunol. 1999;162:5389–5397. [PubMed] [Google Scholar]
- 22.Gagnon SJ, Turner RV, Shiue MG, Damirjian M, Biddison WE. Extensive T cell receptor cross-reactivity on structurally diverse haptenated peptides presented by HLA-A2. Molecular Immunology. 2006;43:346–356. doi: 10.1016/j.molimm.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 23.Laugel B, Boulter JM, Lissin N, Vuidepot A, Li Y, Gostick E, Crotty LE, Douek DC, Hemelaar J, Price DA, Jakobsen BK, Sewell AK. Design of Soluble Recombinant T Cell Receptors for Antigen Targeting and T Cell Inhibition. J Biol Chem. 2005;280:1882–1892. doi: 10.1074/jbc.M409427200. [DOI] [PubMed] [Google Scholar]
- 24.Levin MC, Jacobson S. HTLV-I associated myelopathy/tropical spastic paraparesis (HAM/TSP): a chronic progressive neurologic disease associated with immunologically mediated damage to the central nervous system. J Neurovirol. 1997;3:126–140. doi: 10.3109/13550289709015802. [DOI] [PubMed] [Google Scholar]
- 25.Plumelle Y. HAM/TSP pathogenesis hypothesis. Medical Hypotheses. 1999;52:595–604. doi: 10.1054/mehy.1998.0714. [DOI] [PubMed] [Google Scholar]
- 26.Greten TF, Slansky JE, Kubota R, Soldan SS, Jaffee EM, Leist TP, Pardoll DM, Jacobson S, Schneck JP. Direct visualization of antigen-specific T cells: HTLV-1 Tax11-19-specific CD8+ T cells are activated in peripheral blood and accumulate in cerebrospinal fluid from HAM/TSP patients. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:7568–7573. doi: 10.1073/pnas.95.13.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bieganowska K, Hollsberg P, Buckle GJ, Lim DG, Greten TF, Schneck J, Altman JD, Jacobson S, Ledis SL, Hanchard B, Chin J, Morgan O, Roth PA, Hafler DA. Direct Analysis of Viral-Specific CD8+ T Cells with Soluble HLA-A2/Tax11-19 Tetramer Complexes in Patients with Human T Cell Lymphotropic Virus-Associated Myelopathy. J Immunol. 1999;162:1765–1771. [PubMed] [Google Scholar]
- 28.Utz U, Banks D, Jacobson S, Biddison WE. Analysis of the T-cell receptor repertoire of human T-cell leukemia virus type 1 (HTLV-1) Tax-specific CD8+ cytotoxic T lymphocytes from patients with HTLV-1-associated disease: evidence for oligoclonal expansion. J Virol. 1996;70:843–851. doi: 10.1128/jvi.70.2.843-851.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Motyka B, Teh HS. Naturally Occurring Low Affinity Peptide/MHC Class I Ligands Can Mediate Negative Selection and T Cell Activation. J Immunol. 1998;160:77–86. [PubMed] [Google Scholar]
- 30.Davis-Harrison RL, Armstrong KM, Baker BM. Two Different T Cell Receptors use Different Thermodynamic Strategies to Recognize the Same Peptide/MHC Ligand. Journal of Molecular Biology. 2005;346:533–550. doi: 10.1016/j.jmb.2004.11.063. [DOI] [PubMed] [Google Scholar]
- 31.Boulter JM, Glick M, Todorov PT, Baston E, Sami M, Rizkallah P, Jakobsen BK. Stable, soluble T-cell receptor molecules for crystallization and therapeutics. Protein Eng. 2003;16:707–711. doi: 10.1093/protein/gzg087. [DOI] [PubMed] [Google Scholar]
- 32.Borbulevych OY, Baxter TK, Yu Z, Restifo NP, Baker BM. Increased Immunogenicity of an Anchor-Modified Tumor-Associated Antigen Is Due to the Enhanced Stability of the Peptide/MHC Complex: Implications for Vaccine Design. J Immunol. 2005;174:4812–4820. doi: 10.4049/jimmunol.174.8.4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsodikov OV, Record MT, Jr, Sergeev YV. Novel computer program for fast exact calculation of accessible and molecular surface areas and average surface curvature. J Comput Chem. 2002;23:600–609. doi: 10.1002/jcc.10061. [DOI] [PubMed] [Google Scholar]
- 34.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 35.4, C. C. P. N. . The CCP4 Suite: Programs for Protein Crystallography. Acta Cryst. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 36.Khan AR, Baker BM, Ghosh P, Biddison WE, Wiley DC. The structure and stability of an HLA-A*0201/octameric tax peptide complex with an empty conserved peptide-N-terminal binding site. J Immunol. 2000;164:6398–6405. doi: 10.4049/jimmunol.164.12.6398. [DOI] [PubMed] [Google Scholar]
- 37.McGaughey GB, Gagne M, Rappe AK. Pi-Stacking Interactions: alive and well in proteins. Journal of Biological Chemistry. 1998;273:15458–15463. doi: 10.1074/jbc.273.25.15458. [DOI] [PubMed] [Google Scholar]
- 38.Theodossis A, Guillonneau C, Welland A, Ely LK, Clements CS, Williamson NA, Webb AI, Wilce JA, Mulder RJ, Dunstone MA, Doherty PC, McCluskey J, Purcell AW, Turner SJ, Rossjohn J. Constraints within major histocompatibility complex class I restricted peptides: Presentation and consequences for T-cell recognition. Proceedings of the National Academy of Sciences. 107:5534–5539. doi: 10.1073/pnas.1000032107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith KJ, Reid SW, Stuart DI, McMichael AJ, Jones EY, Bell JI. An Altered Position of the [alpha]2 Helix of MHC Class I Is Revealed by the Crystal Structure of HLA-B*3501. Immunity. 1996;4:203–213. doi: 10.1016/s1074-7613(00)80429-x. [DOI] [PubMed] [Google Scholar]
- 40.Armstrong KM, Piepenbrink KH, Baker BM. Conformational changes and flexibility in T-cell receptor recognition of peptide-MHC complexes. Biochem J. 2008;415:183–196. doi: 10.1042/BJ20080850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piepenbrink KH, Borbulevych OY, Sommese RF, Clemens J, Armstrong KM, Desmond C, Do P, Baker BM. Fluorine substitutions in an antigenic peptide selectively modulate T-cell receptor binding in a minimally perturbing manner. Biochemical Journal. 2009;423:353–361. doi: 10.1042/BJ20090732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baxter TK, Gagnon SJ, Davis-Harrison RL, Beck JC, Binz AK, Turner RV, Biddison WE, Baker BM. Strategic mutations in the class I MHC HLA-A2 independently affect both peptide binding and T cell receptor recognition. J Biol Chem. 2004;279:29175–29184. doi: 10.1074/jbc.M403372200. [DOI] [PubMed] [Google Scholar]
- 43.Baker BM, Turner RV, Gagnon SJ, Wiley DC, Biddison WE. Identification of a Crucial Energetic Footprint on the α1 Helix of Human Histocompatibility Leukocyte Antigen (HLA)-A2 That Provides Functional Interactions for Recognition by Tax Peptide/HLA-A2-specific T Cell Receptors. J Exp Med. 2001;193:551–562. doi: 10.1084/jem.193.5.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clements CS, Dunstone MA, Macdonald WA, McCluskey J, Rossjohn J. Specificity on a knife-edge: the [alpha][beta] T cell receptor. Current Opinion in Structural Biology. 2006;16:787–795. doi: 10.1016/j.sbi.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 45.Rudolph MG, Stanfield RL, Wilson IA. How TCRs Bind MHCs, Peptides, and Coreceptors. Annu Rev Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- 46.Dunitz JD. Organic Fluorine: Odd Man Out. Chem Bio Chem. 2004;5:614–621. doi: 10.1002/cbic.200300801. [DOI] [PubMed] [Google Scholar]
- 47.Madden DR. The Three-Dimensional Structure of Peptide-MHC Complexes. Annual Review of Immunology. 1995;13:587–622. doi: 10.1146/annurev.iy.13.040195.003103. [DOI] [PubMed] [Google Scholar]
- 48.Kersh GJ, Miley MJ, Nelson CA, Grakoui A, Horvath S, Donermeyer DL, Kappler J, Allen PM, Fremont DH. Structural and Functional Consequences of Altering a Peptide MHC Anchor Residue. J Immunol. 2001;166:3345–3354. doi: 10.4049/jimmunol.166.5.3345. [DOI] [PubMed] [Google Scholar]
- 49.Borbulevych OY, Insaidoo FK, Baxter TK, Powell DJ, Jr, Johnson LA, Restifo NP, Baker BM. Structures of MART-1(26/27-35) Peptide/HLA-A2 Complexes Reveal a Remarkable Disconnect between Antigen Structural Homology and T Cell Recognition. J Mol Biol. 2007;372:1123–1136. doi: 10.1016/j.jmb.2007.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sharma AK, Kuhns JJ, Yan S, Friedline RH, Long B, Tisch R, Collins EJ. Class I Major Histocompatibility Complex Anchor Substitutions Alter the Conformation of T Cell Receptor Contacts. J Biol Chem. 2001;276:21443–21449. doi: 10.1074/jbc.M010791200. [DOI] [PubMed] [Google Scholar]
- 51.Liu GY, Fairchild PJ, Smith RM, Prowle JR, Kioussis D, Wraith DC. Low avidity recognition of self-antigen by T cells permits escape from central tolerance. Immunity. 1995;3:407–415. doi: 10.1016/1074-7613(95)90170-1. [DOI] [PubMed] [Google Scholar]
- 52.Morgan CS, Holton JM, Olafson BD, Bjorkman PJ, Mayo SL. Circular dichroism determination of class I MHC-peptide equilibrium dissociation constants. Protein Sci. 1997;6:1771–1773. doi: 10.1002/pro.5560060819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kjer-Nielsen L, Clements CS, Brooks AG, Purcell AW, Fontes MR, McCluskey J, Rossjohn J. The Structure of HLA-B8 Complexed to an Immunodominant Viral Determinant: Peptide-Induced Conformational Changes and a Mode of MHC Class I Dimerization. J Immunol. 2002;169:5153–5160. doi: 10.4049/jimmunol.169.9.5153. [DOI] [PubMed] [Google Scholar]
- 54.Kjer-Nielsen L, Clements CS, Brooks AG, Purcell AW, McCluskey J, Rossjohn J. The 1.5 A crystal structure of a highly selected antiviral T cell receptor provides evidence for a structural basis of immunodominance. Structure. 2002;10:1521–1532. doi: 10.1016/s0969-2126(02)00878-x. [DOI] [PubMed] [Google Scholar]
- 55.Mazza C, Auphan-Anezin N, Gregoire C, Guimezanes A, Kellenberger C, Roussel A, Kearney A, van der Merwe PA, Schmitt-Verhulst AM, Malissen B. How much can a T-cell antigen receptor adapt to structurally distinct antigenic peptides? EMBO J. 2007;26:1972–1983. doi: 10.1038/sj.emboj.7601605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith KJ, Pyrdol J, Gauthier L, Wiley DC, Wucherpfennig KW. Crystal Structure of HLA-DR2 (DRA*0101, DRB1*1501) Complexed with a Peptide from Human Myelin Basic Protein. The Journal of Experimental Medicine. 1998;188:1511–1520. doi: 10.1084/jem.188.8.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lang HL, Jacobsen H, Ikemizu S, Andersson C, Harlos K, Madsen L, Hjorth P, Sondergaard L, Svejgaard A, Wucherpfennig K, Stuart DI, Bell JI, Jones EY, Fugger L. A functional and structural basis for TCR cross-reactivity in multiple sclerosis. Nat Immunol. 2002;3:940–943. doi: 10.1038/ni835. [DOI] [PubMed] [Google Scholar]
- 58.Holler PD, Kranz DM. Quantitative Analysis of the Contribution of TCR/pepMHC Affinity and CD8 to T Cell Activation. Immunity. 2003;18:255–264. doi: 10.1016/s1074-7613(03)00019-0. [DOI] [PubMed] [Google Scholar]
- 59.Cole DK, Yuan F, Rizkallah PJ, Miles JJ, Gostick E, Price DA, Gao GF, Jakobsen BK, Sewell AK. Germline-governed recognition of a cancer epitope by an immunodominant human T-cell receptor. Journal of Biological Chemistry. 2009;284:27281–27289. doi: 10.1074/jbc.M109.022509. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.