Summary
The evolution of sex chromosomes has resulted in numerous species in which females inherit two X chromosomes but males have a single X, thus requiring dosage compensation. MSL (Male-specific lethal) complex increases transcription on the single X chromosome of Drosophila males to equalize expression of X-linked genes between the sexes1. The biochemical mechanisms utilized for dosage compensation must function over a wide dynamic range of transcription levels and differential expression patterns. Lucchesi (1998)2 proposed that MSL complex regulates transcriptional elongation to control dosage compensation, a model subsequently supported by mapping of MSL complex and MSL-dependent H4K16 acetylation to the bodies of X-linked genes in males, with a bias towards 3′ ends3-7. However, experimental analysis of MSL function at the mechanistic level has been challenging due to the small magnitude of the chromosome-wide effect and the lack of an in vitro system for biochemical analysis. In this study, we use global run-on sequencing (GRO-seq)8 to examine the specific effect of MSL complex on RNA Polymerase II (RNAP II) on a genome-wide level. Results indicate that MSL complex enhances transcription by facilitating the progression of RNAP II across the bodies of active X-linked genes. Improving transcriptional output downstream of typical gene-specific control may explain how dosage compensation can be imposed on the diverse set of genes along an entire chromosome.
To investigate how MSL complex specifically elevates transcription of X-linked genes, we performed GRO-seq in SL2 cells, a male Drosophila cell line that has been extensively characterized for MSL function4,9. To display the average enrichment across genes, a 3 kb ‘metagene’ profile was plotted in which the internal regions were rescaled so that all genes appear to have the same length (Fig. 1). Analysis was restricted to expressed genes that were sufficiently large (> 2.5 kb) so that gene-body effects could be clearly assessed (822 X-linked genes, 3420 autosomal genes), and all gene profiles were normalized by their copy-number as determined by analysis of SL2 DNA content10. High correlation coefficients were observed between replicate libraries (Pearson correlation coefficient: ≥ 0.98; Fig. S1). The metagene profiles revealed a prominent 5′ peak of paused RNAP II consistent with previous ChIP and RNA-seq analysis of short 5′ RNAs11,12. In addition, a peak of RNAP II density downstream of the metagene 3′ processing site is evident, possibly due to slow release in regions of transcription termination8. The 3′ peak is present even when the influence of neighboring gene transcription is eliminated (Fig. S2).
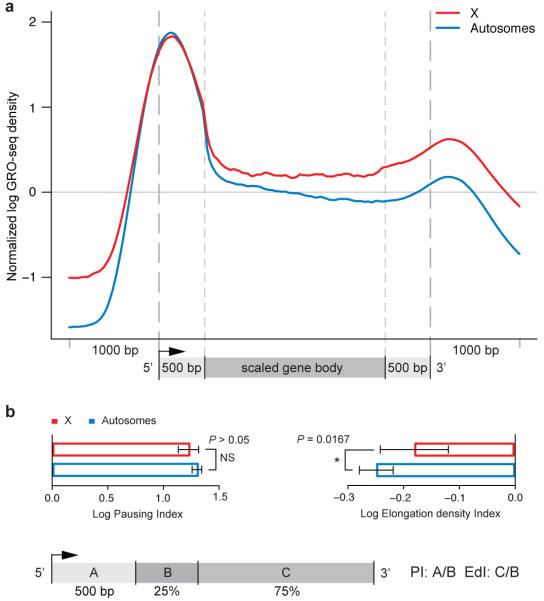
Figure 1. The male X chromosome has higher levels of engaged RNAP II over gene bodies relative to autosomes.
(a) Average GRO-seq profiles of expressed genes are shown for X (red) and autosomes (blue). Read counts on all chromosomes were normalized to genomic read coverage to control for copy number variation, mappability and other potential biases. To construct a metagene profile, genes are scaled as follows: 1) the 5′ end (1 kb upstream of the transcription start site (TSS) to 500 bp downstream) and the 3′ end (500 bp upstream of the transcript termination site (TTS) to 1 kb downstream) were unscaled; 2) The remainder of the gene is scaled to 2 kb (see Supplementary Methods). (b) Pausing indices (PI) do not differ between X (red bar) and autosomal genes (blue bar). Elongation density indices (EdI) are significantly different between X (red bar) and autosomal genes (blue bar). Error bars represent a 95% confidence interval for the mean PI or EdI (1.96*SE: n = 1344 [X-genes]; n = 6090 [A-genes]. The definitions of PI and EdI are shown in the schematic. The PI and EdI are calculated with unscaled GRO-seq tag counts.
The central question with regard to dosage compensation is how genes on the X chromosome differ on average from genes on autosomes. Overall, we found that RNAP II density on active X-linked genes was higher than on autosomal genes, specifically over gene bodies (Fig. 1a). The increase in tag density over the bodies of X-linked genes compared to autosomal genes was approximately 1.4-fold, consistent with previous estimates of MSL-dependent dosage compensation9,10,13. We also performed RNAP II ChIP in SL2 cells, confirming higher occupancy on X-linked genes compared to autosomes but with lower resolution and reduced sensitivity (Fig. S3). Therefore, we proceeded with GRO-seq to analyze X and autosomal differences.
To measure how X and autosomes differed on average in distribution of elongating RNAP II, we segmented genes into their 5′ 500 bp and the remainder of the coding region. We further subdivided the remainder of the coding region into 5′ and 3′ segments (25% and 75% respectively). Using this segmentation, we quantified RNAP II pausing and elongation separately based on unscaled GRO-seq signal (Fig. 1b). The pausing index (PI) was previously defined as the ratio of GRO-seq signal at the 5′ peak to the average signal over gene bodies8. Here, we calculated the PI for X and autosomal genes as the ratio of the 5′ peak (segment A) to the first 25% of the remaining gene body (segment B), and found no statistically significant difference (Fig. 1b).
To separately examine transcription elongation across gene bodies, we defined the Elongation density Index (EdI) as the ratio of tag density in the 3′ region of each gene (segment C) compared to its 5′ region after the first 500 bp (segment B). In contrast to our analysis of 5′ pausing, we found statistically significant differences in EdI (P-value < 0.0162) between X and autosomes (Fig. 1b), regardless of how 5′ and 3′ regions of genes were divided (Table S1). As defined, the average PI (log scale) is a positive number because RNAP II generally is enriched at 5′ ends compared to gene bodies; the average EdI (log scale) is a negative number, as the relative density of RNAP II typically decreases from the beginning to the end of gene bodies. We conclude that X–linked genes, on average, exhibit a significantly smaller decrease in RNAP II density along their gene bodies when compared to autosomal genes.
To measure the specific contribution of MSL complex to the increase in RNAP II within X-linked gene bodies, we used MSL2 RNAi to reduce complex levels in male SL2 cells as described previously9. Excellent correlations between replicate data sets were observed (Fig. S1). To confirm the X-specific effect of MSL2 RNAi, we computed the distributions of GRO-seq signal (averaged over the bodies of genes excluding the 5′ peak) for all genes before and after RNAi. When comparing X vs. autosomes, we found a preferential decrease on the X-chromosome, with an average control:MSL RNAi ratio of 1.4 (Fig. 2a). MSL-dependent changes in average GRO-seq density showed weak but statistically significant correlation with changes in steady-state mRNA levels assayed by expression array9 (Pearson correlation = 0.22, P-value < 1 × 10−15) or mRNA-Seq10 (Pearson correlation = 0.30, P-value < 1 × 10−15). These results confirm that MSL-dependent changes in steady-state RNA levels reflect differences in active transcription on the X chromosome.
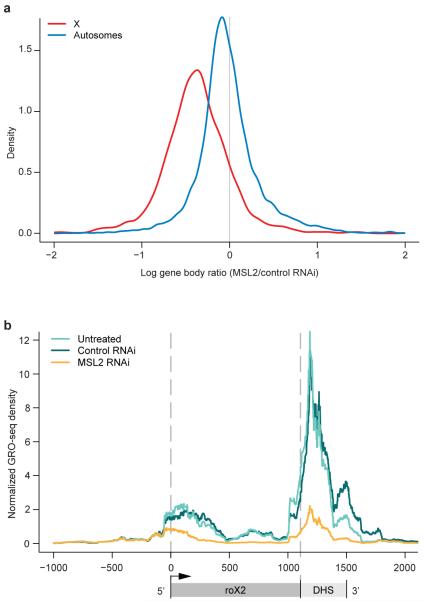
Figure 2. MSL complex increases engaged RNAP II density on the male X chromosome.
(a) The log ratio of sense-strand reads in the MSL2 RNAi sample to the control RNAi sample was computed within the body of each gene. Here, the distributions of these ratios are plotted for all genes on X and autosomes. (b) GRO-seq sense-strand read densities within the roX2 gene for the untreated, control RNAi and MSL2 RNAi samples. Schematic below GRO-seq profiles indicates the location of the DHS (DNase I Hypersensitive Site), which contains sequences that can recruit MSL complex to the X chromosome.
In addition to assessing the average decrease of X-linked RNAP II density after MSL2 RNAi, we asked whether any genes showed strong MSL-dependence, a hallmark of the roX genes that encode RNA components of the complex14,15. We found that roX2 showed a strong loss in GRO-seq density after MSL2 RNAi as predicted (9-fold) (Figs. 2b, S4). Interestingly, in the untreated or control RNAi samples, there is a prominent GRO-seq peak downstream of the major roX2 3′ end, coincident with an MSL recruitment site (see discussion below). roX1 expression is low in this isolate of SL2 cells, and no other expressed genes on X or autosomes displayed strong MSL-dependence in our assays (> 6-fold). Examples of additional individual gene profiles are shown in Figs. S5, S6.
Next, we compared the average RNAP II density along X and autosomal metagene profiles after control and MSL2 RNAi. Unlike our initial analysis of X and autosomes, where different gene populations were compared (Fig. 1), here we could examine the same genes in the presence and absence of MSL complex (Fig. 3). We found that after MSL2 RNAi, the density of elongating RNAP II over the bodies of X-linked genes decreased, approaching the level on autosomes (Figs. 3, S7). The presence of MSL complex affected RNAP II density starting just downstream of the 5′ peak and continuing through the bodies of X-linked genes (Figs. 3, S7). Thus, GRO-seq functional data correlate with physical association of MSL complex which is biased towards 3′ ends of active genes on the male X chromosome4,5.
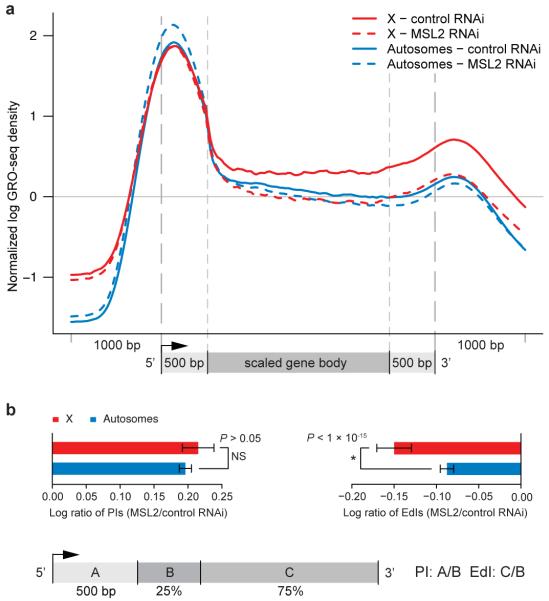
Figure 3. MSL complex facilitates the progression of engaged RNAP II across transcription units.
(a) Metagene profiles of expressed X chromosome genes and autosomal genes in control RNAi and MSL2 RNAi samples. Higher RNAP II density can be seen within the bodies of genes on the X (solid-red) compared to those on autosomes (solid-blue) in the control RNAi sample. After MSL2 RNAi, average RNAP II density on X decreases over gene bodies (dashed-red) becoming similar to autosomal gene bodies (dashed-blue). (b) Ratios of pausing indices (PI) between control and MSL2 RNAi treated cells are not significantly different for genes on the X (red bar) compared to those on autosomes (blue bar). In contrast, ratios of elongation density indices (EdI) between the control and MSL2 RNAi sample decreased significantly for genes on the X (red bar) compared to those on the autosomes (blue bar). Pausing indices (PI) and elongation density indices (EdI) were calculated as described for Figure 1. Error bars represent a 95% confidence interval for the mean PI or EdI (1.96*SE: n = 1358 [X-genes]; n = 6135 [A-genes].
To quantify the differences in density of engaged RNAP II over X-linked genes in the presence and absence of MSL complex, we calculated the pausing (PI) and elongation density indices (EdI), expressing them as ratios comparing MSL2 and control RNAi treatment. We found that both X and autosomes increased PI and decreased EdI after MSL2 RNAi treatment (Fig. S8). However, in each case the change was larger on X than on autosomes, and the most profound difference was an MSL-dependent change in EdI on the X compared with autosomes (P < 1 × 10−15; Fig. 3b). EdI was computed, as before, by defining the 5′/3′ regions as 25%/75% of the gene body after removing the 5′ peak, but the difference was statistically significant for all other values until the 3′ end was reached (Table S1). When these analyses were performed separately for two independently prepared sets of GRO-seq libraries (Fig. S9), the results were also statistically significant (P-value < 7.6 × 10−14, P-value < 1.1 × 10−4 for each of two replicates). We conclude that MSL complex causes the transcriptional elongation profiles of X-linked genes to differ from those of autosomal genes.
To visualize the location along gene bodies at which MSL complex functions, we calculated control:MSL2 RNAi GRO-seq ratios and generated a metagene profile (Fig. 4a). Here, values above zero represent higher relative amounts of engaged RNAP II in the presence of MSL complex compared to after RNAi treatment. In contrast, values below zero represent a relative increase in engaged RNAP II after MSL2 RNAi. In the absence of MSL complex, there is a relative increase in the amount of RNAP II localized to 5′ ends of both autosomal (blue) and X-linked genes (red), perhaps due to relocalization of RNAP II from the bodies of X-linked genes (Fig. 4a). A limitation of the GRO-seq assay is that we cannot currently distinguish between initiating and 5′ paused polymerase, so we cannot assign a definitive role for this 5′ increase in RNAP II after MSL2 RNAi treatment. However, relative RNAP II levels over autosomal gene bodies do not increase, suggesting that any relocalized enzyme in this experiment is likely to remain paused rather than progressing across transcription units. This is consistent with a model in which the functional outcome of MSL2 RNAi is to shift RNAP II density away from productive transcription through X-linked gene bodies.
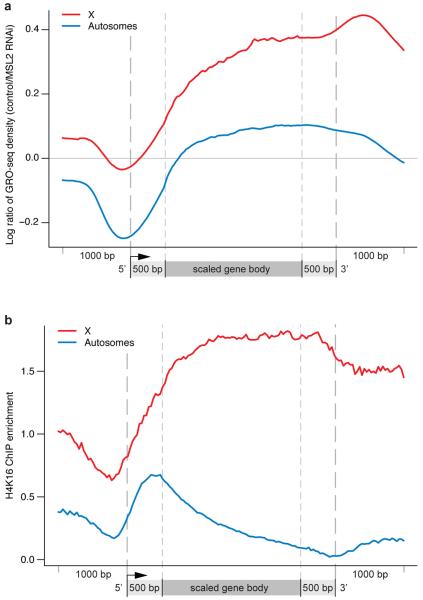
Figure 4. MSL function correlates with the presence of H4K16 acetylation.
(a) The MSL2-dependent effect on RNAP II density as shown by metagene profiles of control: MSL2 RNAi GRO-seq sense-strand reads shown on log scale (base 2). The black line (y = 0) indicates no change after MSL2 RNAi treatment. The cumulative effect of MSL2 RNAi treatment peaks toward the 3′ ends of X-linked genes (red) while having less effect on autosomal genes (blue). (b) Similar to the effect of MSL complex on engaged RNAP II, H4K16 acetylation on the male X chromosome localizes to the bodies of active genes with at 3′ bias (red). On autosomes, H4K16 acetylation is present at 5′ ends (blue) as described previously7.
We plotted the local effect of MSL complex in Fig. 4a to compare it to the status of H4K16 acetylation (Fig. 4b), catalyzed by the MOF component of MSL complex3,16. H4K16 acetylation typically is enriched at 5′ ends of most active genes in mammals and flies6,17; in contrast, a 3′ bias of this mark is a distinctive characteristic of the dosage compensated male X chromosome in Drosophila3,6,7. Interestingly, there is an overall coincidence across gene bodies between MSL complex-dependent GRO-seq signal and the presence of H4K16 acetylation (Fig. 4a;7). How might H4K16 acetylation biased toward the 3′ end of genes generate the improved transcriptional elongation indicated by our GRO-seq results? During transcription elongation, nucleosomes are thought to comprise a barrier to the progress of RNAP II18-20 and several well studied elongation factors, including Spt6 and the FACT complex, are proposed to function by removing nucleosomes that block RNAP II progression and replacing them in the wake of transcription18,21. Interestingly, H4K16 acetylation of nucleosomes has been observed to act in opposition to the formation of higher order chromatin structure in vitro22,23. Thus, H4K16 acetylation is likely to further reduce the steric hindrance to RNAP II progression through chromatin. Improving the entry of RNAP II into the bodies of genes may allow 5′, gene-specific events to proceed at an increased but still regulated rate. Furthermore, reduction in the repressive effect of nucleosomes could increase mRNA output by improving the processivity of RNAP II on each template. Available methodologies cannot distinguish between these mechanisms in vivo, and therefore future approaches will be required to assess their relative contributions to dosage compensation.
In addition to increasing the transcription of X-linked genes for dosage compensation, MSL complex also positively regulates the roX noncoding RNA components of the complex, to promote their male-specificity14,15. roX1 expression is low in our SL2 cell line, but our GRO-seq data indicate that active transcription of roX2 is highly dependent on MSL2 as predicted (Fig. 2b; Fig. S4). Interestingly, there is a strong GRO-seq peak at the 3′ roX2 DHS (DNase I Hypersensitive Site) which contains sequences important for targeting MSL complex to the X chromosome. Sites of roX gene transcription are thought to be critical for MSL complex assembly24,25. Therefore, it is possible that paused RNAP II at the roX2 DHS could promote an open chromatin structure that facilitates MSL complex targeting or incorporation of noncoding roX2 RNA into the complex.
In summary, we hypothesize that MSL complex functions on the male X chromosome to promote progression and processivity of RNAP II through the nucleosomal template. Improving transcriptional output downstream of typical gene-specific regulation makes biological sense when compensating the diverse set of genes found along an entire chromosome.
Methods Summary
To measure the density of engaged RNAP II, GRO-Seq experiments were conducted on DRSC SL2 cells grown in Schneider’s medium with 10% FBS8. To determine how MSL complex contributes to dosage compensation, MSL2 and control (GFP) RNAi treatments were conducted using a bathing protocol9. Nuclei were subjected to GRO-seq analysis after RNAi treatment. Two biological replicates were performed for the untreated, control RNAi, and MSL2 RNAi experiments.
Supplementary Material
Acknowledgments
We thank Fred M. Winston, Steve Buratowski, Artyom Alekseyenko, Marnie Gelbart, Charlotte Wang, and Andrey Gortchakov for helpful comments on the manuscript, and are very grateful to Nils Gehlenborg for graphic design expertise. This work was supported by the following NIH grants: GM45744 (M.I.K.), GM082798 (P.J.P.) and HG4845 (J.T.L.). E.L. was supported by a Charles A. King Trust fellowship from the Medical Foundation.
Appendix
METHODS
RNAi and cell culture methods
Control and MSL2 RNAi were performed in SL2-DRSC cells as described in Gelbart et al. 20097. The control RNAi construct targeted the eGFP gene that is not present in SL2 cells, and the experimental RNAi construct targeted the MSL2 gene (www.flyrnai.org: DRSC 00829). Primer sequences for generation of the eGFP dsRNA template by PCR from pEGFP-N1(Clontech) were: forward, 5′–TAATACGACTCACTATAGGGAGAGGTGAGCAAGGGCGA- GGAGCT-3′, and reverse, 5′–TAATACGACTCACTATAGGGAGATCTTGAAGTTCACCTTGATGC-CG-3′. The primers used for amplifying the MSL2 gene from Drosophila genomic DNA were: 5′-TAATACGACTCACTATAGGGAGAGTTGGCTGTGCTGGCTG-3′ and reverse, 5′-TAATACGACTCACTATAGGGAGATGTTGGCTCGTCACTGTC-3.
dsRNA was synthesized from PCR products containing T7 promoters using the Ambion MEGAscript kit, and 225 ug of dsRNA was added to 2 × 107 cells in a T225 flask. RNAi treatment was performed for 6 days after which mRNA was prepared and transcriptionally active nuclear extracts were generated as described below. mRNA preparation, cDNA synthesis and qPCR analysis of roX2 and msl2 RNA compared with the PKA normalization control were performed as described in Gelbart et. al. 20097. A 12.3-fold average decrease of msl2 mRNA was observed after MSL2 RNAi treatment when compared with the control treatment.
Preparation of GRO-seq libraries for next-generation sequencing
Preparation of transcriptionally active nuclei from Drosophila SL2-DRSC cells after RNAi treatment was conducted as follows: SL2 cells grown in a T225 tissue culture flask were scraped and 1 × 108 cells were pelleted at 500 g for 3 minutes at 4 °C. Then, cells were washed in 10 ml of cold PBS and spun at 500 g for 3 minutes at 4 °C. Cells were swelled by resuspending gently in 10 mls ice cold swelling buffer (10 mM Tris (pH = 7.5); 2 mM MgCl2; 3 mM CaCl2) and placed on ice for 5 minutes. Next, cells were pelleted at 600 g for 10 minutes at 4 °C. Pelleted cells were resuspended in 1 ml lysis buffer (10 mM Tris (pH = 7.5), 2 mM MgCl2, 3 mM CaCl2, 10% Glycerol, 0.5% NP40, 2 U/ml SUPERaseIN (Invitrogen)) and pipetted 20 times with a P1000 tip with the end cut off. 9 ml lysis buffer was added and nuclei were pelleted at 600 g for 5 minutes. Nuclei were washed in 1 ml lysis buffer and then 9 mls was added followed by pelleting for 5 min at 600 g at 4 °C. A small aliquot was taken for Trypan blue staining to check that lysis occurred and nuclei were still intact. Next, nuclei were resuspended in 1 ml freezing buffer (50 mM Tris-Cl (pH = 8.3); 40% glycerol; 5 mM MgCl2; 0.1 mM EDTA) using a P1000 tip with the end cut off. Nuclei were pelleted for 1 minute and resuspended in 500 ul of freezing buffer and aliquoted into 100 ul aliquots and frozen in liquid nitrogen. All solutions were prepared with DEPC-treated water.
GRO-seq libraries were prepared as described in Core et al. 2008 with the following changes: 1) Glycoblue (3 ul: 15mg/ml) (Ambion) was used in all of the ethanol precipitations to assure the release of the nascent RNAs from the interior surface of Eppendorf tubes; 2) Wash buffers for BrU immunoprecipitation differ from those described in Core et al. 8 as follows: 1) High salt wash buffer for anti-BrdU (0.25x SSPE, 1 mM EDTA, 0.05% Tween, 137.5 mM NaCl; 2) Binding buffer for anti-BrdU (0.25x SSPE, 1 mM EDTA, 0.05% Tween, 37.5 mM NaCl); 3) Elution buffer (20 mM DTT, 300 mM NaCl, 50 mM Tris-Cl pH 7.5, 1 mM EDTA, 0.1% SDS). 3) All immunoprecipitation wash buffers contain superRNAsin (1 ul/5 ml) (Invitrogen) to block degradation that can occur during the immunoprecipitation process.
Computational analysis of GRO-seq data
Data generation & quality assessment
Sequencing was performed on an Illumina Genome Analyzer IIx. Two independent biological replicates were generated for each of the three experiments (untreated, control RNAi, and MSL2 RNAi). Data are available from Gene Expression Omnibus (GEO) with accession numbers GSE25321 and GSE25887. Reads were aligned to the D. melanogaster genome (dm3) using the Bowtie alignment software26. Only uniquely mapping reads with no more than one mismatch were retained. We obtained 10.6 million aligned reads from the untreated samples (7.1 M from replicate I; 3.5 M from replicate II), 25.2 million aligned reads from the control RNAi samples (20.5 M from replicate I, 4.7 M from replicate II), and 28.4 million from the MSL2 RNAi samples (22.4 M from replicate I, 6.0 M from replicate II). To assess the agreement between replicates, a correlation coefficient was computed between sense-strand read densities across genes in the two replicates for each of the three treatments. The agreement between replicates is excellent, with the following correlation coefficients: 1) Untreated: Spearman: 0.97; Pearson: 0.98; 2) control RNAi: Spearman: 0.99; Pearson: 0.98; 3) MSL2 RNAi: Spearman: 0.99; Pearson: 0.98 (Fig. S1). For most of the analysis, the two replicates were combined and processed together to increase statistical power. Key results were also confirmed in each replicate separately.
Generating average profiles
To examine the difference between RNAi and control as well as between the X and autosomes, it was important to derive accurate ‘metagene’ profiles. To improve existing TSS annotations, previously published small (< 100 bp), capped nuclear RNA-seq data27 was used. This dataset contains RNA isolated from 5′ ends of transcripts. Starting with Flybase build 5.23, start sites for each annotated transcript were adjusted by up to 150 bp from the original location. The position within the 301 bp window centered on the existing TSS annotation with the highest number of reads from this capped nuclear RNA-seq dataset was annotated as the new TSS for that transcript. In the event that two positions within the search space had the same number of reads, the most 5′ position was designated the TSS. Finally, transcripts with identical start sites were filtered out, ensuring each annotation is unique.
To derive the metagene profile, we first computed the profile for each gene before computing the average. For each gene, the GRO-seq read profile on each strand was normalized to total sequencing depth and was smoothed using Gaussian smoothing with a bandwidth of 200 bp. To adjust for copy number variations, alignability and sequencing biases, the GRO-seq read density was further normalized by the analogous density of genomic sequencing reads 10. Specifically, each gene was divided into 200 bins and the log ratio (base 2) between GRO-seq and genomic sequencing read densities were computed for each bin. To avoid ratios becoming infinity when the denominator is zero, we applied the common technique of adding a pseudocount (1 in this case) to both numerator and denominator. To average the log ratios across genes for the metagene profile, the 5′ end (1 kb upstream of the TSS to 500 bp downstream) and the 3′ end (500 bp upstream of the transcript termination site (TTS) to 1 kb downstream) were unscaled. The region within the gene body extending from 500 bp downstream of the TSS and 500 bp upstream of the TTS was scaled to 2 kb (see Figure 1a).
Only genes longer than 2.5 kb were considered to avoid short genes in which the 5′ peak is difficult to distinguish from the body of the gene. In addition, genes with less than one RPKM/gene copy in the untreated GRO-seq sample were considered unexpressed and thus excluded. In a number of genes, the read distribution downstream of the 5′ peak contained high peaks, possibly due to unannotated internal TSS that distorted the average profiles. To mitigate the effect of these outliers, we removed 5% of the genes in which the highest density peak was downstream of the first 500 bp. These genes were not removed when computing P-values or for other analyses.
The ChIP-chip metagene profiles (Fig. S3) were computed from array data by the same scaling method used for the GRO-seq metagene profiles. There was no need for further normalization in these profiles because we also normalized to array input, thereby controlling for copy number. Individual gene profiles (Fig. 2b, S4-6) were computed in a similar manner to the metagene profiles, only no scaling was performed and a 100 bp sliding window was used to smooth the reads instead of Gaussian smoothing. As before, read density was normalized to total sequencing depth and for copy number using genomic sequencing reads as in the GRO-seq metagene profile calculations.
The control/MSL2 RNAi log ratio metagene plot (Fig. 4a) was produced by taking the log ratio of the Gaussian-smoothed read densities in MSL2 RNAi and control samples across the body of each gene. The log ratios (base 2) were computed for each gene before scaling (with pseudocount of 1) and then averaged across genes (thus this ratio is not simply the ratio of the profiles in Fig. 3a). Overall, higher values in Fig. 4a represent a greater drop in GRO-seq signal after MSL2 RNAi treatment.
Computing the Pausing Index (PI) and Elongation density Index (EdI)
Pausing Index (PI)
To compare the level of RNAP II at the 5′ ends of genes compared with that progressing into gene bodies, we defined a ‘pausing index’ (PI) as the ratio of 5′ GRO-seq read density within the first 500 bp downstream of the transcriptional start site to the read density within the next 25% of the gene body. The 5′ read density is calculated as the number of sense strand reads in the 5′ region divided by the number of uniquely mappable positions (as determined using PeakSeq28) in this same region. A position is “mappable” if, given only the 36 bp sequence at that position, the position in the genome can be uniquely identified. Correcting for mappability in this manner prevents regions that have no reads because they are unmappable from biasing the analysis. A similar calculation is performed to determine the density in the next 25% of the gene. A high PI indicates that RNAP II is biased toward the 5′ end.
Elongation density Index (EdI)
To analyze the distribution of active RNAP II within a given gene, we calculated an ‘elongation density index’ (EdI) by taking the ratio of the 3′ read density to the 5′ read density. The first 500 bp of the gene is excluded from this calculation to eliminate the effect of the large 5′ peak frequently associated with paused polymerase. The remainder of the gene is then split into two portions, the 5′ region and the 3′ region. We state the main results with the 5′ region containing the first 25% of the gene (after the first 500 bp) and the 3′ region the remaining 75%, but multiple points of division were tested (Table S1). The 3′ density is calculated as it was done above. A low EdI indicates that RNAP II is biased towards the 5′ end, while a larger value indicates greater RNAP II towards the 3′ end.
The gene set considered in the analysis of EdIs is similar to that used to produce the profile plots, except that no outliers were removed and only short genes less than 500 bp (instead of 2.5 kb) were excluded. These criteria were relaxed to make our analysis more conservative. To avoid outlier ratios that can result from a small number of reads, genes with fewer than 3 reads in the first 500 bp of the gene, the 5′ region or in the 3′ region were removed. A one-sided Wilcoxon test was used to test whether EdIs on the X chromosome are significantly greater than on autosomes in the untreated sample. To compare the elongation density indices for the MSL2 RNAi with the control RNAi, the same procedure was followed, except that only genes with an EdI defined in both samples were considered.
To determine whether removing outliers (as defined previously for the metagene profiles) alters our results, we compared EdI ratios (MSL2/control RNAi) with and without outlier removal. When outliers were removed, the shift in the distribution of EdI ratios on X relative to autosomes remained significant (P-value < 1 × 10−15). Likewise, the difference between the EdI distribution on X relative to autosomes in the untreated sample remains significant after outlier removal (P-value < 0.017 before removal, P-value < 0.020 after outlier removal). Overall, outlier removal has little effect on the statistical significance of our EdI comparisons.
Comparing GRO-seq data with mRNA-seq
To compare our data with previous experiments9 that measured the effect of MSL2 RNAi on expression levels, we examined GRO-seq read densities before and after treatment with MSL2 RNAi. Ratios of gene expression levels before and after MSL2 RNAi obtained by RNA-Seq experiments10 were compared to analogous GRO-seq ratios. GRO-seq ratios were computed only from reads mapping to the gene bodies. The region extending from the TSS to 500 bp downstream was excluded from these calculations so that the 5′ peak around the TSS would not bias the results. Read densities for each gene with at least 10 reads in both the MSL2 RNAi dataset and the control RNAi dataset were normalized to dataset size, and then a ratio was computed. The Pearson correlation coefficient between GRO-seq ratios and those derived from RNA-seq is highly significant (P-value < 1 × 10−15), but with relatively low absolute magnitude (R = 0.30). If only X-linked genes are considered, the Pearson correlation remains unchanged (R = 0.30) and is still highly significant (P-value < 1 × 10−15). When a similar comparison was performed between GRO-seq ratios and expression array data9, a significant Pearson correlation of R = 0.22 was observed (P-value < 1 × 10−15).
References
- 1.Gelbart ME, Kuroda MI. Drosophila dosage compensation: a complex voyage to the X chromosome. Development. 2009;136:1399–1410. doi: 10.1242/dev.029645. doi:136/9/1399 [pii] 10.1242/dev.029645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lucchesi JC. Dosage compensation in flies and worms: the ups and downs of X-chromosome regulation. Curr Opin Genet Dev. 1998;8:179–184. doi: 10.1016/s0959-437x(98)80139-1. [DOI] [PubMed] [Google Scholar]
- 3.Smith ER, Allis CD, Lucchesi JC. Linking global histone acetylation to the transcription enhancement of X-chromosomal genes in Drosophila males. J Biol Chem. 2001;276:31483–31486. doi: 10.1074/jbc.C100351200. [DOI] [PubMed] [Google Scholar]
- 4.Alekseyenko AA, Larschan E, Lai WR, Park PJ, Kuroda MI. High-resolution ChIP-chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome. Genes Dev. 2006;20:848–857. doi: 10.1101/gad.1400206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilfillan GD, et al. Chromosome-wide gene-specific targeting of the Drosophila dosage compensation complex. Genes Dev. 2006;20:858–870. doi: 10.1101/gad.1399406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kind J, et al. Genome-wide analysis reveals MOF as a key regulator of dosage compensation and gene expression in Drosophila. Cell. 2008;133:813–828. doi: 10.1016/j.cell.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 7.Gelbart ME, Larschan E, Peng S, Park PJ, Kuroda MI. Drosophila MSL complex globally acetylates H4K16 on the male X chromosome for dosage compensation. Nature structural & molecular biology. 2009;16:825–832. doi: 10.1038/nsmb.1644. doi:nsmb.1644 [pii] 10.1038/nsmb.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. doi:1162228 [pii]10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamada FN, Park PJ, Gordadze PR, Kuroda MI. Global regulation of X chromosomal genes by the MSL complex in Drosophila melanogaster. Genes Dev. 2005;19:2289–2294. doi: 10.1101/gad.1343705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, et al. Expression in aneuploid Drosophila S2 cells. PLoS Biol. 2010;8:e1000320. doi: 10.1371/journal.pbio.1000320. doi:10.1371/journal.pbio.1000320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muse GW, et al. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeitlinger J, et al. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belote JM, Lucchesi JC. Male-specific lethal mutations of Drosophila melanogaster. Genetics. 1980;96:165–186. doi: 10.1093/genetics/96.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bai X, Alekseyenko AA, Kuroda MI. Sequence-specific targeting of MSL complex regulates transcription of the roX RNA genes. Embo J. 2004;23:2853–2861. doi: 10.1038/sj.emboj.7600299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meller VH, Rattner BP. The roX genes encode redundant male-specific lethal transcripts required for targeting of the MSL complex. Embo J. 2002;21:1084–1091. doi: 10.1093/emboj/21.5.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hilfiker A, Hilfiker-Kleiner D, Pannuti A, Lucchesi JC. mof, a putative acetyl transferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. Embo J. 1997;16:2054–2060. doi: 10.1093/emboj/16.8.2054. doi:10.1093/emboj/16.8.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaplan CD, Laprade L, Winston F. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003;301:1096–1099. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- 19.Mavrich TN, et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–362. doi: 10.1038/nature06929. doi:nature06929 [pii]10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mavrich TN, et al. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome research. 2008;18:1073–1083. doi: 10.1101/gr.078261.108. doi:gr.078261.108 [pii]10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belotserkovskaya R, et al. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. doi:10.1126/science.1085703301/5636/1090 [pii] [DOI] [PubMed] [Google Scholar]
- 22.Robinson PJ, et al. 30 nm chromatin fibre decompaction requires both H4-K16 acetylation and linker histone eviction. J Mol Biol. 2008;381:816–825. doi: 10.1016/j.jmb.2008.04.050. doi:S00222836(08)00508-1 [pii]10.1016/j.jmb.2008.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shogren-Knaak M, et al. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 24.Park Y, Kelley RL, Oh H, Kuroda MI, Meller VH. Extent of chromatin spreading determined by roX RNA recruitment of MSL proteins. Science. 2002;298:1620–1623. doi: 10.1126/science.1076686. [DOI] [PubMed] [Google Scholar]
- 25.Oh H, Park Y, Kuroda MI. Local spreading of MSL complexes from roX genes on the Drosophila X chromosome. Genes Dev. 2003;17:1334–1339. doi: 10.1101/gad.1082003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. doi:gb2009-10-3-r25 [pii]10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nechaev S, et al. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science. 2010;327:335–338. doi: 10.1126/science.1181421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rozowsky J, et al. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat Biotechnol. 2009;27:66–75. doi: 10.1038/nbt.1518. doi:nbt.1518 [pii]10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.