Abstract
The mouse meiotic mutant Mei1 was isolated in a screen for infertile mice descended from chemically mutagenized embryonic stem cells. Homozygotes of both sexes are sterile due to meiotic arrest caused by defects in chromosome synapsis. Notably, RAD51 protein does not load onto Mei1 mutant meiotic chromosomes, suggesting that there is a defect in either recombinational repair or the production of double-strand breaks (DSBs) that require such repair. Here, we show that treatment of mutant males with cisplatin restores RAD51 loading, suggesting that mutant spermatocytes have intact recombinational repair mechanisms. Levels of histone H2AX phosphorylation (γH2AX) at leptonema are significantly reduced compared with wild-type controls but comparable to that seen in animals deficient for SPO11, the molecule required for catalyzing DSB formation during meiosis. These observations provide evidence that genetically programmed DSB induction is defective in Mei1 leptotene spermatocytes. We also report the positional cloning of Mei1, which encodes a product without significant homology to any known protein. Expressed almost exclusively in gonads, Mei1 has no apparent homologs in yeast, worms, or flies. However, Mei1 orthologs are present in the genomes of mammals, chickens, and zebrafish. Thus, Mei1 is required for vertebrate meiosis. To our knowledge, Mei1 is the first meiosis-specific mutation identified by forward genetic approaches in mammals.
The genetic regulation of meiosis in mammals is poorly understood relative to that in lower eukaryotes such as yeast. Although the basic events of meiosis, homologous chromosome pairing, synapsis, recombination, and the reduction to haploidy, are conserved from fungi to humans, mammalian gametogenesis involves unique and complex developmental events compared with sporulation of yeast. These include specification of the germ lineage, sex determination, gonad formation, and creation of highly differentiated gametes. The identification of genes that play roles in these processes is hindered by the fact that the phenotypic consequence of mutations in such genes, infertility, does not lend itself to simple genetic analyses in humans (unless sex-linked) or in mice, where the identification of spontaneous infertility mutations has been essentially nonexistent.
Substantial progress toward understanding mammalian meiosis has been made by exploiting the conservation that exists between Saccharomyces cerevisiae and mammals. Mutations affecting all stages of the meiotic process in yeast have been identified. Mammalian orthologs of key meiotic genes such as DMC1, SPO11, MSH4, and MSH5 have been identified and disrupted by homologous recombination in embryonic stem (ES) cells (1–6). For the most part, the phenotypes of mutant mice confirmed the substantial mechanistic conservation with yeast. However, such a reverse-genetics strategy cannot be used to identify genes that are important to mammalian meiosis but have no yeast ortholog. On occasion, genes such as the breast cancer suppressor gene, BRCA1, are identified by virtue of their involvement in somatic processes. In mice, BRCA1 is detectable as foci along synaptonemal complexes during pachynema. Spermatocytes deficient in BRCA1 (from mice in which embryonic lethality was rescued by p53 nullizygosity) exhibit severe meiotic disruptions and infertility (7).
To identify additional genes required for meiosis, we initiated phenotype-based screens for mutations causing infertility as a consequence of meiotic defects (8). One mutant that was isolated in these experiments, Mei1 (the allele of which is now designated Mei1m1Jcs), was particularly interesting. Mei1m1Jcs mutants of both sexes were sterile and displayed defective chromosome synapsis. However, there were distinct differences in the degree of meiotic progression between spermatocytes and oocytes; whereas male meiosis invariably arrested at entry into pachynema, oocytes often progressed to an abnormal metaphase I (9). Another striking characteristic of mutant meiocytes was the absence of RAD51 binding to meiotic chromosomes. This observation, coupled with the detection of some H2AX phosphorylation (γH2AX) in the nuclei of mutant zygotene spermatocytes, was suggestive of an inability to repair double-strand breaks (DSBs) via a RAD51-mediated repair pathway. Alternatively, MEI1 might be required for production of genetically programmed meiotic DSBs to a level or a state that triggers detectable RAD51 binding to meiotic chromosomes. Mei1m1Jcs was mapped to a 2-Mb interval on central chromosome 15 that has conserved synteny with human 22q13 (9). Here, we report the positional cloning of Mei1, which encodes a protein present only in vertebrates. Additional experiments, including a quantitative study of γH2AX binding to leptotene chromosomes in both Mei1 and Spo11 mutant mice, suggest that formation of genetically programmed DSBs in MEI1-deficient mice is disrupted or abolished.
Methods
Genotyping. Approximately 2-mm tail tips were lysed by incubation at 55°C in 100 μl of buffer PBND [50 mM KCl/10 mM Tris, pH 8.3/2.5 mM MgCl2/0.1 mg/ml gelatin/0.45% (vol/vol) Nonidet P-40/0.45% (vol/vol) Tween 20]. One microliter was used directly for PCR, and products were electrophoresed on 4.0% MetaPhor (FMC) gels. Primers sequences for D15Jcs50 are 5′-ATACAGCCCAGACTGGCCTCAAATG-3′ and 5′-GCTCAGTTGGTAAATGCCTGCCG-3′, and primer sequences for D15Jcs153 are 5′-TACCTTGCATTCAACACAGATCACG-3′ and 5′-AGATGTTTGCAGACATGGAGCTAGC.
RT-PCR. Wild-type and mutant cDNA was generated from 0.5 μg of total RNA by using the SuperScript Preamplification kit (GIBCO/BRL). Targets were amplified from 1 μl of cDNA product in standard PCR reactions (35 cycles of 94°C for 30 sec; 55°C for 30 sec; 72°C for 45 sec). Primer sequences for generating RT-PCR products or Northern blot hybridization probes are as follows: M1C-3F, 5′-GAGCATCTTTCGTCCTCCAGCGAAA-3′; M1C-6R, 5′-AGGGACCTCCTGTTCAGAGGGGTC-3′; M1C-5BF, 5′-CGACTGACCCTGGAGCTGAAGGCC-3′; M1C-6BR, 5′-CCCCACGCAGGTCTGGGATGCCC-3′.
Sequence Analysis. PCR and RT-PCR amplicons were purified with the QIAquick PCR Purification kit (Qiagen, Chatsworth, CA) before sequencing. The Mei1m1Jcs A-to-T transversion was detected in 342 bp of PCR amplicons produced with the primers: MOMS-A2F (5′-TTGTCAGGCTGTGGGCATTCAGGGG-3′) and MOMS-CR (5′-TGTCTCAGGCAGTTGGAGAAGCCCCG-3′). Transcripts Mei1Δ58 and Mei1Δ148 were detected by RT-PCR of mutant testes by using the primers M1C-5BF and M1C-6BR.
5′ RACE. 5′ RACE was performed on C57BL/6J testes RNA by using the FirstChoice RLM-RACE kit (Ambion, Austin, TX).
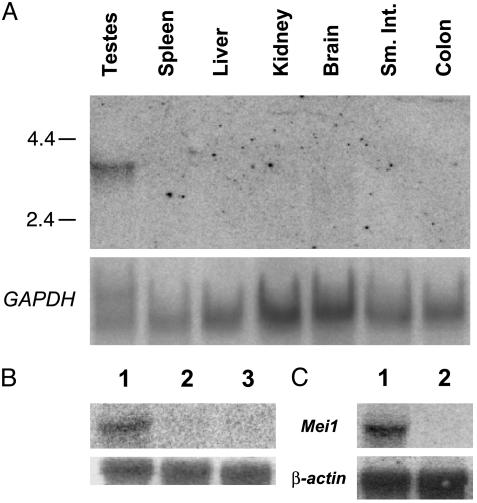
Northern Analysis. Total RNA was isolated by using the RNeasy kit (Qiagen). Twenty micrograms of total RNA was electrophoresed through formaldehyde agarose (1%) gels followed by standard capillary transfer onto a nylon membrane (Micron Separations). The blots in Fig. 3 were hybridized to PCR-generated probes corresponding either to nucleotides 1009–1500 (Fig. 3A; primers M1C-5BF and -6BR) or to nucleotides –147 to +308 (Fig. 3A; primers M1C-3F and -6R) of the Mei1 transcript, then stripped and rehybridized with either a β-actin or GAPDH probe.
Fig. 3.
Northern analysis of Mei1 expression. (A) A Mei1 cDNA probe was hybridized to RNA from the indicated tissues. The numbers to the left are sizes, in kilobases, of a size marker. The lower half is the same Northern blot rehybridized with GAPDH as a loading control. (B and C) Northern blots hybridized to a Mei1 cDNA fragment and a β-actin control. Lanes 1–3in B are wild-type testis, Mei1m1Jcs1/Mei1m1Jcs1 testis, and wild-type adult ovary, respectively. Lanes 1 and 2 in C are wild-type testis and KitW/KitW-v, respectively.
Immunocytochemistry. Spermatocytes from 2- to 3-wk-old male mice were microspread and immunolabeled as described (10). Slides were either used immediately for immunolabeling or stored at –20°C for up to 2 wk. Slides were washed three times for 10 min in 10% antibody dilution buffer (ADB) (10% horse serum/3% BSA/0.05% Triton X-100 in PBS). Primary antibody was diluted in ADB and applied to slides, which were placed in a humid chamber at 37°C for 1 h followed by washing three times for 10 min in 10% ADB, then incubating with secondary antibody for 1 h at 37°C, in the dark. Slides were then washed three times for 10 min in PBS, incubated in 4′,6-diamidino-2-phenylindole (0.2 μg/ml in PBS) for 5 min, and finally washed for 5 min in PBS. Slides were mounted in antifade (Molecular Probes) and viewed immediately or stored at 4°C in the dark for up to 1 wk. Primary antibodies were rabbit anti-RAD51 (1:500, Oncogene Research Products, La Jolla, CA); rabbit anti-H2AX (1:800, phosphorylated at Ser-139, from Upstate Biotechnology, Lake Placid, NY); and rat anti-SCP3 (1:1,000, gift from Mary Ann Handel, University of Tennessee). Secondary antibodies were Alexa 488-conjugated goat anti-rabbit IgG and Alexa 594-conjugated goat anti-rat IgG, both at 1:1,000. Slides were viewed on an Olympus (Melville, NY) BX51 microscope with epifluorescence, and digital images were collected by using an Olympus MagnaFire charge-coupled device camera.
Cisplatin Treatment of Mice. Mice were injected with cisplatin as described (11). Testes were removed from animals 3 d after cisplatin treatment. Spermatocytes were collected and microspread as described above.
Quantitation of Anti-γH2AX Localization. The metamorph imaging system (Universal Imaging, Media, PA) was used to analyze immunofluorescent images of leptotene nuclei labeled with anti-γH2AX. For each genotype, 15 leptotene nuclei were selected from a single slide (prepared from age-matched animals on the same day) and digitally photographed by using identical exposure settings. With the aid of the metamorph imaging system, average pixel intensity (a digital expression of average fluorescence) was generated for each nucleus. A subsequent average of the pixel intensity of all 15 nuclei was calculated for each genotype. To compare means between samples, unpaired Student t tests were used to calculate the P value.
Results and Discussion
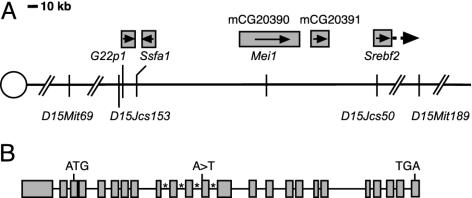
Positional Cloning of Mei1. The Mei1m1Jcs mutation was induced in CJ7 ES cells of strain 129S1/Sv (“129”). It was mapped to a 2-Mb interval between D15Mit69 and D15Mit189 on central Chr 15 by outcrossing to C57BL/6J (“B6”), intercrossing the resulting heterozygotes and genotyping affected offspring with MIT microsatellite markers polymorphic between 129 and B6 (9). Analysis of these animals with a polymorphic marker, D15Jcs50, identified a key recombinant that localized the mutation to a ≈1.6-Mb interval (based on the Celera mouse sequence) between D15Mit69 and D15Jcs50. To map Mei1m1Jcs with higher resolution, Mei1m1Jcs heterozygotes were outcrossed to Mus castaneous (CAST/Ei), and F1 carriers were intercrossed. One F2 animal inherited a chromosome recombinant between Mei1m1Jcs and D15Jcs153, located ≈3 kb proximal to G22p1 (Fig. 1). Based on genotypic and phenotypic data obtained from this recombinant, Mei1m1Jcs was positioned distal to D15Jcs153. Thus, Mei1m1Jcs was restricted to the genomic interval between D15Jcs153 and D15Jcs50 (≈250 kb).
Fig. 1.
Location and structure of Mei1. (A) A diagram of central mouse Chr-15 indicating the position of Mei1 (mCG20390) and one other gene (mCG20391) between Ssfa1 and Srebf2. Boxes above gene names represent approximate gene size, except in the case of Srebf2, which extends further distally than is shown. Arrows in boxes indicate direction of transcription. The distance between D15Mit69 and D15Mit189 is 2 Mb. (B) Schematic of the intron/exon structure of Mei1. Boxes represent exons, and asterisks mark introns whose sizes are unknown due to gaps in available sequence. The location of the Mei1m1Jcs A-to-T splice acceptor mutation is indicated.
Genomic DNA sequence and gene predictions presented in the Celera and Ensembl databases indicated that the D15Jcs153-D15Jcs50 interval contains portions of five genes: G22p1 (encoding Ku70), Ssfa1 (the mouse ortholog of human NHP2l1), two unknown genes, and the first exon of Srebf2 (Fig. 1). Because Mei1 mutant spermatocytes fail to load RAD51 onto chromosomes, implying a possible failure of DSB repair, G22p1 and Ssfa1 seemed potential candidates for Mei1. The former is required for nonhomologous end joining of DSBs, and the latter's product associates with the DNA damage checkpoint protein RAD17 (12). We therefore sequenced the coding sequences (generated by RT-PCR) and ≈1 kb upstream of each gene from mutant animals. No mutations were detected in either gene, nor was there any indication of abnormal levels of gene expression or aberrant transcript sizes.
Because Mei1m1Jcs mutants do not display any detectable somatic defects, we speculated that Mei1 might be expressed primarily, if not exclusively, in germ cells. One of the two unknown genes in the critical region, designated mCG20931 by Celera and ENSMUSG00000042028 in Ensembl, is somatically expressed based on the existence of multiple ESTs in GenBank originating from various tissues. However, the second gene was represented by a single EST (GenBank accession no. AK016514) derived from a testis cDNA library. To determine whether this gene, designated mCG20390 in the Celera database and ENSMUSG00000022467 in Ensembl, might display an expression pattern typical of a meiotic gene, RT-PCR analysis was performed by using RNA from various tissues. These experiments indicated that mCG20390 is expressed weakly in brain and strongly in testis but not detectably in spleen, adrenal gland, or liver (Fig. 2A). Northern analysis confirmed expression of mCG20390 transcripts in testis, very weak expression in brain, but no signals in spleen, liver, kidney, small intestine, or colon (Fig. 3A).
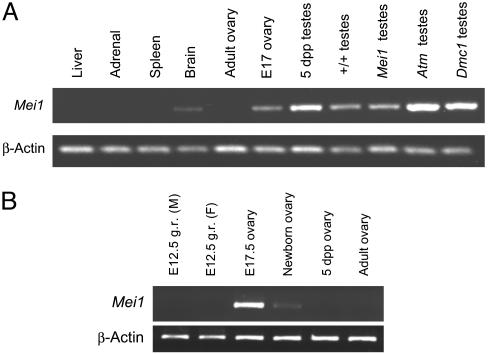
Fig. 2.
RT-PCR analysis of Mei1 expression. (A) All tissues are wild-type except for the last three lanes. “E” indicates embryonic day of gestation. (B) PCR primers were M1C-3F and M1C-6R, resulting in 455-bp amplimers. With these primers, Mei1 expression is detectable in mutant testes when extra template is used. g.r., genital ridge.
To better characterize the mCG20390 expression pattern in reproductive tissue, RT-PCR was performed on germ-cell-defective mutants and on gonads of different ages. Transcription was high in embryonic day 17 (E17) ovaries, significantly reduced in newborn ovaries, and undetectable in ovaries from 5-d-and 8-wk-old mice (Fig. 2B). The absence of transcripts in 8-wk-old ovaries was confirmed by Northern analysis (Fig. 3B), indicating that expression in females is associated with early meiotic prophase I, which occurs between ≈E14 and birth but not later meiotic stages. mCG20390 was also absent in testes of KitW-v/KitW mice, which lack germ cells (Fig. 3C). Transcripts were detectable in wild-type 5-d-old mice and in testes of DMC1- and ATM-deficient mice, which enter meiosis but do not progress past zygonema. These results suggest that, in males, mCG20390 transcription initiates in germ cells before meiotic prophase I. Based on the absence of postmeiotic expression in oocytes, the qualitatively normal levels of transcripts in Dmc1 and Atm1 mutant mice, and the absence of expression in germ-cell-deficient males, the major site of mCG20390 transcription appears to be meiotic and premeiotic germ cells. Hereafter, mCG20390 will be referred to as Mei1, which reflects conclusions of experiments described below.
The genomic organization of Mei1, comprising 24 exons and spanning ≈45 kb (Fig. 1B), was deduced by aligning the available EST sequence (AK016514) against genomic sequence obtained from the Celera database. 5′ RACE was performed, extending the 5′ sequence by 12 nt beyond that in AK016514. Comparisons of DNA sequences from RT-PCR amplicons and the Celera database revealed errors in the AK016514 sequence; the corrected transcript has a much larger (approximately three times larger) ORF.
RT-PCR products generated by primer pairs situated throughout the Mei1 coding region revealed at least six alternatively spliced variants of the 5′ UTR. The first Mei1 exon maximally consists of 739 nt but contains multiple splice sites. The presence of multiple minor species of PCR products generated by primers throughout the coding region, as visualized by agarose gel electrophoresis, suggests that additional alternative splicing events occur. The largest encoded Mei1 transcript is 3,729 nt in length. The longest ORF, which is comprised of 2,682 nucleotides, begins in exon 3, terminates 136 nt upstream of a consensus polyadenylation sequence (AATAAA) in exon 24, and is predicted to encode a product 894 aa (99.26 kDa) in size. The Mei1 sequence has been deposited in GenBank (accession no. AY270177).
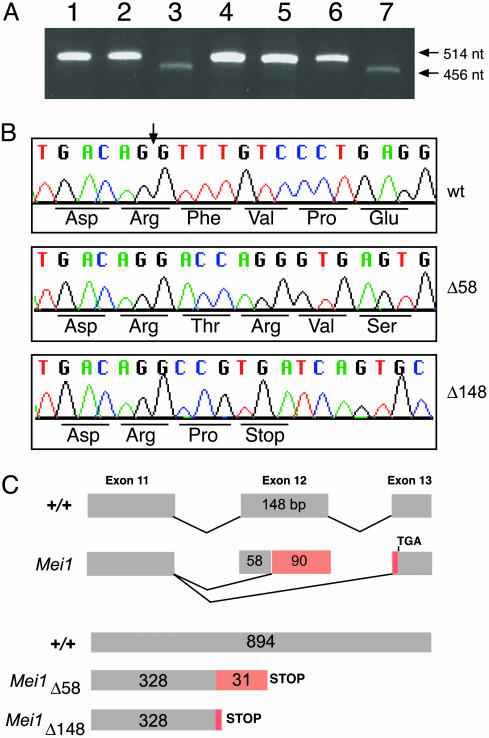
To identify possible mutations in Mei1m1Jcs mice, a series of RT-PCR reactions was performed by using primer pairs directed against various parts of the Mei1 gene. Primers flanking exons 11 and 12 generated a product of expected size (514 bp) from wild-type brain and testis cDNAs (DMC1- and ATM-deficient testis cDNA produced the same result). In contrast, two smaller transcripts, the predominant of which was 456 bp, were detected in testes and brains of Mei1m1Jcs/Mei1m1Jcs animals (Fig. 4A). The more abundant mutant transcript, designated Mei1Δ58, lacks the first 58 nt of exon 12. Notably, the wild-type genomic sequence immediately upstream of the portion of exon 12 that is retained in Mei1Δ58 features a long pyrimidine-rich tract, and the penultimate, and ultimate nucleotides of the sequence before the start of aberrant mutant sequence (A and G, respectively) form a consensus splice acceptor site. An aberrant 366-bp transcript of lesser abundance, dubbed Mei1Δ148, was also identified in mutants and entirely skips exon 12. However, Mei1Δ148 appears at low levels in wild-type samples. These mutant transcripts are diagrammed in Fig. 4C.
Fig. 4.
Splicing defects in the Mei1m1Jcs allele. (A) RT-PCR of wild-type and mutant RNAs. Lane 1, wild-type testes; lane 2, Mei1m1Jcs/+ testes; lane 3, Mei1m1Jcs/Mei1m1Jcs testes; lane 4, Atm–/– testes; lane 5, Dmc1–/– testes; lane 6, wild-type brain; lane 7, Mei1m1Jcs/Mei1m1Jcs brain. (B) Sequence analysis of wild-type and mutant transcripts. The vertical arrow indicates the beginning of exon 12. (C) Diagrams of the wild-type and mutant splicing patterns associated with exon 11. The red regions represent coding sequences and corresponding abnormal amino acids resulting from the mutant transcripts.
Both Mei1Δ58 and Mei1Δ148 cause a frameshift of the wild-type sequence, each resulting in a predicted truncated protein about one-third the size of wild-type MEI1. Specifically, in the case of Mei1Δ148, the first 328 residues of the predicted mutant protein are encoded normally before a proline residue is substituted for Phe-329 and a stop codon (TGA) replaces Val-330. The first 328 amino acids encoded by Mei1Δ58 are also wild type, but these are followed by 31 aberrant amino acids before encountering the same stop codon that terminates Mei1Δ148 (Fig. 4 B and C).
Although Mei1Δ58 and Mei1Δ148 were detected in mutant testes by RT-PCR, Northern analysis was not sufficiently sensitive to detect Mei1 transcripts in mutant testis RNA (Fig. 3B). The reduced levels of mutant RNA might be a consequence of nonsense-mediated decay.
To identify the basis of faulty splicing in mutant cells, the intron/exon junctions of exon 12 were PCR-amplified from Mei1m1Jcs/Mei1m1Jcs genomic DNA and sequenced. The Mei1m1Jcs mutants bear an A-to-T transversion (Fig. 1B; sequencing data not shown) in the canonical AG splice acceptor sequence of Mei1 intron 11 (i.e., –2 position relative to the first base of exon 12). In contrast, the parental strain (CJ7 ES cells), C57BL/6J, and CAST/Ei genomes contained the consensus AG splice junction. Thus, the Mei1m1Jcs point mutation occurred de novo during the course of the mutagenesis experiment. Interestingly, ethylmethanesulfonate (EMS) typically causes G-to-A or C-to-T transitions in ES cells (8), flies (13), and worms (14). It is possible that EMS induces A-to-T changes at a low frequency in ES cells as in Caenorhabditis elegans (≈2% of all changes) (14). Alternatively, the Mei1m1Jcs lesion may have arisen independently during the culture of ES cells.
Searches of various databases with the predicted MEI1 amino acid sequence revealed no significant homology to any previously described protein or protein motifs. However, a human Mei1 ortholog, designated dJ821D11.1, is located between G22P1 and SREBF2 on chromosome 22q13. It is predicted to encode a protein of 884 aa that is 79.4% identical to mouse MEI1. Furthermore, tblastn searches of EST databases revealed homologs in several other vertebrates, namely bull, pig, chicken, and zebrafish. The predicted amino acid sequences of several Mei1 homologs are shown in Fig. 5.
Fig. 5.
Alignment of vertebrate MEI1 homologs. The non-mouse sequences are translated from ESTs present in sequence databases. Only that subregion for which sequence was available from all or three of the species is compared. Residues in red are identical among all three or four sequences. Areas where two or more amino acids are identical or are similar are boxed. The consensus sequence is indicated; where two of the sequences contain one amino acid and the other two contain another amino acid, the consensus sequence contains a “2.” The numbering is with respect to mouse MEI1. The zebrafish sequence is derived from two testis EST clones (faa59b01.x1/y1 and fu76e02.x1/y1) and the chicken sequence from a single EST from liver (pgl1n.pk007.e22). The chicken sequence was altered to compensate for a possible sequencing error that disrupted the reading frame. Not shown are the cow (derived from several Bos taurus testis ESTs, assembled by TIGR as TC144584) and pig (GenBank accession no. BG609787) sequences.
Searches of translated ESTs or genomic sequences of Drosophila melanogaster, C. elegans, and S. cerevisiae failed to identify any proteins resembling MEI1. Thus, MEI1 is a protein that appears to be restricted to vertebrates. As such, computational approaches would not have implicated Mei1 as a critical meiotic gene based on comparisons to model organisms such as yeast.
The sequence comparisons reveal a few highly conserved regions, the most striking being a 12-aa stretch, having the consensus QPWNRFLLFTLL, in which nine are identical among mouse, human, chicken, and zebrafish. However, as mentioned above, none correspond to any known amino acid motifs. Another notable feature of mouse MEI1 is its high leucine content (18.3%).
Rescue of RAD51 Chromosome-Binding Defect by Cisplatin. In normal spermatocytes, RAD51 is immunocytochemically detected as foci along meiotic chromosomes, beginning at the leptotene stage. By midpachynema, when synapsis is complete, RAD51 foci are restricted to just a few per autosome and are associated with the synaptonemal complex. Previous studies showed that RAD51 foci are absent from meiotic chromosomes of Mei1m1Jcs/Mei1m1Jcs spermatocytes (9). This could be due to a failure to direct RAD51 to the sites of DNA repair, absent or aberrant meiotic DSB formation, or a defect in recombinational repair of DSBs. To address these possibilities, the localization of key meiotic molecules was investigated in mutant and normal spermatocytes, with and without induction of lesions that require recombinational repair.
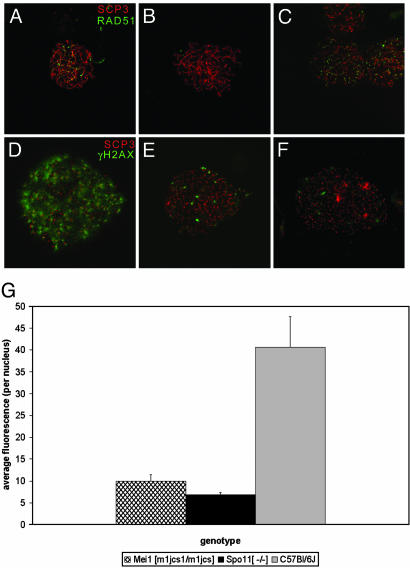
To investigate whether lack of RAD51 binding is due to an inability to direct RAD51 to the sites of recombination or DNA repair in meiocytes, exogenous DNA damage was induced by administering cisplatin to Mei1m1Jcs/Mei1m1Jcs mice. Cisplatin induces inter-and intrastrand crosslinks that require recombinational repair (15, 16) and has been shown to increase recombination in fungi, flies, and mice (11, 17, 18). In addition, cisplatin treatment restores RAD51 localization in SPO11-deficient DSB-lacking spermatocytes, resulting in a partial rescue of that phenotype (4). Cisplatin treatment of Mei1m1Jcs/Mei1m1Jcs males restored RAD51 localization in zygotene spermatocytes (Fig. 6 B and C). The localization pattern was consistent with that seen in zygotene spermatocytes of normal untreated littermates (Fig. 6A). As in normal littermates, the most advanced spermatocytes from cisplatin-treated mutants no longer showed RAD51 foci (not shown). This may reflect a premature loss of RAD51 foci, or RAD51 localization may be following its normal route, whereby autosomal foci are lost by late pachynema (a stage not reached in untreated mutants). The latter possibility would imply that cisplatin-treated Mei1m1Jcs/Mei1m1Jcs spermatocytes exhibit stages beyond zygonema, in the “developmental” if not chromosomal sense.
Fig. 6.
γH2AX and RAD51 localization in cisplatin-treated and untreated Mei1m1Jcs/Mei1m1Jcs spermatocytes. RAD51 foci (green) are absent from Mei1m1Jcs/Mei1m1Jcs zygotene spermatocytes (B) but appear after cisplatin treatment (C). A wild-type untreated control is shown in A. Localization of γH2AX antibody (green) in Mei1m1Jcs/Mei1m1Jcs leptotene (E) spermatocytes is greatly reduced compared with wild type (D) and comparable to that of Spo11–/– spermatocytes (F). In all cases, spreads were colabeled with anti-SCP3 (red). To quantitate the data represented in D–F, average fluorescence across each of 15 anti-γH2AX labeled nuclei (n = 15) was calculated for each genotype by using the metamorph imaging system (G). The amount of γH2AX in Mei1m1Jcs/Mei1m1Jcs and Spo11–/– spermatocyte nuclei was reduced 4-fold when compared with that of normal spermatocytes (P < 0.0001). Unpaired Student t tests and ANOVA were used to calculate P values. Fig. 7 shows, for each genotype, three images for each of five nuclei: anti-SCP3 staining alone, anti-γH2AX alone, and merged.
Because these experiments indicate that the absence of RAD51 binding in mutant animals is not due to defects in directing or loading RAD51 to sites of recombinational repair, we revisited the possibility, previously investigated by Libby et al. (9), that genetically induced DSBs may not occur normally in Mei1m1Jcs/Mei1m1Jcs meiocytes. γH2AX early in meiosis is a marker of chromatin associated with DSBs (19). In mice, this is supported by the observation that γH2AX is severely decreased in leptotene spermatocytes lacking SPO11, an enzyme that creates DSBs (20). Similarly, the pattern and degree of γH2AX distribution in Mei1m1Jcs/Mei1m1Jcs spermatocytes was drastically reduced relative to normal littermates (P < 0.0001, Fig. 6 D, E, and G; see also Fig. 7, which is published as supporting information on the PNAS web site, and which shows additional images of immunostained meiotic nuclei), suggesting that Mei1m1Jcs/Mei1m1Jcs spermatocytes do not form normal levels of DSBs. γH2AX was slightly higher in Mei1m1Jcs/Mei1m1Jcs vs. Spo11–/– spermatocytes (Figs. 6 E–G and 7), but it should be pointed out that the genetic backgrounds of the Mei1m1Jcs and Spo11–/– mice were not identical (the former being nearly congenic in C57BL/6J, and the latter being a mix between strain 129S1/Sv and C57BL/6J). The residual levels of γH2AX in both mutants may be due to DSBs formed by late-replicating DNA (19) or chromosome breakage unrelated to recombination induction.
The meiotic phenotypes of Mei1m1Jcs/Mei1m1Jcs and Spo11–/– mice are very similar. They both exhibit meiotic arrest with identical sexually dimorphic features; lack zygotene RAD51 foci, which can be restored with cisplatin treatment; and are presumably deficient in DSB formation, as assessed by γH2AX. In S. cerevisiae, at least 15 gene products are required for the initiation of SPO11-catalyzed DSBs. In yeast, ablation of any one of these genes creates phenotypes that are very similar to spo11 strains. Some of these genes, including REC102, REC104, and MER2, have no known vertebrate homologs. It is possible that different vertebrate genes act to achieve the same functions, and Mei1 may be one example of such a gene.
Supplementary Material
Acknowledgments
We thank T. Ashley for demonstrating the microspread protocol and for helpful discussions, M. A. Handel (University of Tennessee, Knoxville) for the anti-SCP3 antibody, D. Pittman (Medical College of Ohio, Toledo) for testis RNA from Atm and Dmc1 mutants, Maria Jasin (Sloan–Kettering Memorial Hospital, New York) for ES cells containing the Spo11 targeted allele, and J. Barker (The Jackson Laboratory) for the KitW/KitW-v mice. We also appreciate helpful comments on the manuscript from J. Eppig, M. A. Handel, and B. Richards-Smith. This work was supported by National Institutes of Health Grant GM45415 (to J.C.S.) and Cancer Center Grant CA34196 (to The Jackson Laboratory).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ES, embryonic stem; DSB, double-strand break; γH2AX, H2AX phosphorylation.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY270177).
See Commentary on page 15287.
References
- 1.Pittman, D., Cobb, J., Schimenti, K., Wilson, L., Cooper, D., Brignull, E., Handel, M. A. & Schimenti, J. (1998) Mol. Cell 1, 697–705. [DOI] [PubMed] [Google Scholar]
- 2.Yoshida, K., Kondoh, G., Matsuda, Y., Habu, T., Nishimune, Y. & Morita, T. (1998) Mol. Cell 1, 707–718. [DOI] [PubMed] [Google Scholar]
- 3.Baudat, F., Manova, K., Yuen, J. P., Jasin, M. & Keeney, S. (2000) Mol. Cell 6, 989–998. [DOI] [PubMed] [Google Scholar]
- 4.Romanienko, P. J. & Camerini-Otero, R. D. (2000) Mol. Cell 6, 975–987. [DOI] [PubMed] [Google Scholar]
- 5.Kneitz, B., Cohen, P. E., Avdievich, E., Zhu, L., Kane, M. F., Hou, H., Jr., Kolodner, R. D., Kucherlapati, R., Pollard, J. W. & Edelmann, W. (2000) Genes Dev. 14, 1085–1097. [PMC free article] [PubMed] [Google Scholar]
- 6.Edelmann, W., Cohen, P. E., Kneitz, B., Winand, N., Lia, M., Heyer, J., Kolodner, R., Pollard, J. W. & Kucherlapati, R. (1999) Nat. Genet. 21, 123–127. [DOI] [PubMed] [Google Scholar]
- 7.Cressman, V., Backlund, D., Avrutskaya, A., Leadon, S., Godfrey, V. & Koller, B. (1999) Mol. Cell. Biol. 19, 7061–7075. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 8.Munroe, R. J., Bergstrom, R. A., Zheng, Q. Y., Libby, B., Smith, R., John, S. W. M., Schimenti, K. J., Browning, V. L. & Schimenti, J. C. (2000) Nat. Genet. 24, 318–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Libby, B. J., De La Fuente, R., O'Brien, M. J., Wigglesworth, K., Cobb, J., Inselman, A., Eaker, S., Handel, M. A., Eppig, J. J. & Schimenti, J. C. (2002) Dev. Biol. 242, 174–187. [DOI] [PubMed] [Google Scholar]
- 10.Peters, A. H., Plug, A. W., van Vugt, M. J. & de Boer, P. (1997) Chromosome Res. 5, 66–68. [DOI] [PubMed] [Google Scholar]
- 11.Hanneman, W. H., Legare, M. E., Sweeney, S. & Schimenti, J. C. (1997) Proc. Natl. Acad. Sci. USA 94, 8681–8685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang, M. S., Sasaki, H., Campbell, M. S., Kraeft, S. K., Sutherland, R., Yang, C. Y., Liu, Y., Auclair, D., Hao, L., Sonoda, H., et al. (1999) J. Biol. Chem. 274, 36544–36549. [DOI] [PubMed] [Google Scholar]
- 13.Sega, G. A. (1984) Mutat. Res. 134, 113–142. [DOI] [PubMed] [Google Scholar]
- 14.Anderson, P. (1995) Methods Cell Biol. 48, 31–58. [PubMed] [Google Scholar]
- 15.De Silva, I. U., McHugh, P. J., Clingen, P. H. & Hartley, J. A. (2002) Nucleic Acids Res. 30, 3848–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McHugh, P. J., Sones, W. R. & Hartley, J. A. (2000) Mol. Cell. Biol. 20, 3425–3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vogel, E. W. & Zijlstra, J. A. (1987) Mutat. Res. 182, 243–264. [DOI] [PubMed] [Google Scholar]
- 18.Katz, A. J. (1987) Environ. Mol. Mutagen. 10, 197–203. [DOI] [PubMed] [Google Scholar]
- 19.Hunter, N., Valentin Borner, G., Lichten, M. & Kleckner, N. (2001) Nat. Genet. 27, 236–238. [DOI] [PubMed] [Google Scholar]
- 20.Mahadevaiah, S. K., Turner, J. M., Baudat, F., Rogakou, E. P., de Boer, P., Blanco-Rodriguez, J., Jasin, M., Keeney, S., Bonner, W. M. & Burgoyne, P. S. (2001) Nat. Genet. 27, 271–276. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.