Abstract
The type III restriction endonuclease EcoP15I was used in isolating fragments of 26 bp from defined positions of cDNAs. We call this substantially improved variant to the conventional serial analysis of gene expression (SAGE) procedure “SuperSAGE.” By applying SuperSAGE to Magnaporthe grisea (blast)-infected rice leaves, gene expression profiles of both the rice host and blast fungus were simultaneously monitored by making use of the fully sequenced genomes of both organisms, revealing that the hydrophobin gene is the most actively transcribed M. grisea gene in blast-infected rice leaves. Moreover, SuperSAGE was applied to study gene expression changes before the so-called hypersensitive response in INF1 elicitor-treated Nicotiana benthamiana, a “nonmodel” organism for which no DNA database is available. Again, SuperSAGE allowed rapid identification of genes up- or down-regulated by the elicitor. Surprisingly, many of the down-regulated genes coded for proteins involved in photosynthesis. SuperSAGE will be especially useful for transcriptome profiling of two or more interacting organisms like hosts and pathogens, and of organisms, for which no DNA database is available.
Understanding of molecular mechanisms underlying host–pathogen interactions is of primary importance in devising strategies to control diseases. For this purpose, gene expression analysis is massively applied. One of the most powerful techniques for such gene expression analysis is serial analysis of gene expression (SAGE) as developed by Velculescu (1). Briefly, DNA (cDNA) is reverse-transcribed from mRNA isolated from defined cells, tissues, or organs. A short tag of 13–15 bp representing each expressed sequence is excised from the cDNA by use of the type IIS restriction endonuclease BsmFI, and tags from different expressed sequences are ligated for sequence analysis and assignment of the mRNA fragment to a certain gene or EST in the database. Thus, after sequencing thousands of tags, it is possible to count the number of each tag for transcripts in the sample and, further, to describe the gene expression profile in the sample.
To study the gene expression changes involved in plant host–pathogen interactions, we have applied SAGE to rice cells treated with fungal elicitor of Magnaporthe grisea, a causal pathogen of rice blast disease (2) as well as to M. grisea developing appressorium on artificial membrane (3). In both cases, SAGE revealed useful information about gene expression changes in the host and pathogen. However, the limited tag sequence size of only 13–15 bp was not always sufficient to unequivocally identify the gene from which the tag is derived. A single tag sequence frequently corresponded to several different ESTs and genomic sequences, confounding further analysis.
Here, we report of a method for the isolation of tag sequences >25 bp from defined positions of cDNAs by using the type III restriction enzyme EcoP15I. This enzyme is a member of the type III restriction endonucleases exhibiting some particular properties (4–7). Use of EcoP15I as the “tagging enzyme” dramatically improves the conventional SAGE protocol through a reliable identification of the corresponding genes and an accurate gene expression analysis. We call this improved SAGE variant involving the endonuclease EcoP15I “SuperSAGE.” By using SuperSAGE, gene expression profiles of rice and M. grisea were simultaneously studied in M. grisea-infected rice leaves. Furthermore, gene expression change was monitored in Phytophthora elicitor-treated Nicotiana benthamiana, a typical “nonmodel” organism. We demonstrate with both examples that SuperSAGE has an unprecedented power for studying global gene expression in any eukaryotic organisms with known or even unknown nucleotide sequence data.
Materials and Methods
Blast-Infected Rice Plants and Blast Inoculation. Rice plants (Oryza sativa L. cv. Norin1) susceptible to the blast fungus M. grisea race 007 were grown in an experimental field under strong infection pressure of blast fungus race 007. Leaves showing disease lesions were collected and used for this study. For blast inoculation, two rice cultivars were used, cv. Kakehashi (susceptible to M. grisea race 007) and cv. Himenomochi (resistant). These cultivars were grown in a glass house for 3 weeks after the germination of the seeds. For inoculation of blast fungus race 007, rice plants were sprayed with fungal spores suspended in 0.01% Tween 20 solution and incubated in the fully humidified chamber (13 h of light/11 h of dark) for 24 h as described (8). Leaves were harvested 10 days after the inoculation.
INF1-Treated N. benthamiana Plants. N. benthamiana plants were grown in a glass house for 6 weeks after germination of the seeds. Leaves of N. benthamiana were infiltrated with 100 nM Phytophthora infestans elicitor (INF1) prepared according to Kamoun (9). As negative control, the identical amount of water was infiltrated into the leaves (flooding treatment).
Preparation of EcoP15I Endonuclease. EcoP15I was purified from 10-g batches of Escherichia coli TG1 (pMT15) to apparent homogeneity as described by Meisel and coworkers (10).
SuperSAGE. A flowchart of the SuperSAGE experiment is summarized in Fig. 1. From total RNA, five micrograms of mRNA were isolated by using the mRNA Purification Kit (Amersham Biosciences). This mRNA was reverse-transcribed (cDNA synthesis system, Invitrogen) to generate single-stranded cDNA by using reverse transcription-primer with the sequence 5′-CTGATCTAGAGGTACCGGATCCCAGCAGTTTTTTTTTTTTTTTTT-3′ containing the 5′-CAGCAG-3′ recognition site of the type III restriction enzyme EcoP15I. The product was converted to double-stranded cDNA and digested with NlaIII (NEB, Beverly, MA). A suspension of streptavidin magnet beads (Promega) was added to the digestion reaction and the 3′-end fragments of the cDNAs were bound to streptavidin magnetic beads. The streptavidin-bound cDNA was washed and divided into two portions in separate tubes. Two FITC-labeled linkers (1E and 2E) were prepared by annealing commercially synthesized oligonucleotides (linker-1E, FITC-5′-TACAACTAGGCTTAATACAGCAGCATG-3′ and 5′-CTGCTGTATTAAGCCTAGTTGTA-3′-NH2; linker-2E, FITC-5′-TTCTAACGATGTACGCAGCAGCATG-3′ and 5′-CTGCTGCGTACATCGTTAGAA-3′-NH2; Qiagen, Valencia, CA). The unblocked 5′ termini of linker-1E and -2E were phosphorylated by T4 polynucleotide kinase (NEB). Both linker-1E and -2E harbor the EcoP15I recognition sequence (5′-CAGCAG-3′). To each of the two tubes containing cDNA bound to magnet beads, linker-1E or -2E, respectively, were added and ligated to the cDNA ends by T4 DNA ligase (NEB). Consequently, each cDNA fragment is flanked by two inverted repeats of 5′-CAGCAG-3′. The type III restriction enzyme EcoP15I recognizes the asymmetric hexameric sequence 5′-CAGCAG-3′ and cleaves the DNA 25 nt (in one strand) and 27 nt (in the other strand) downstream of the recognition site leaving a 5′ overhang of two bases. Two unmethylated and inversely oriented recognition sites in head-to-head configuration (5′-CAGCAG-N(i)-CTGCTG-3′) are essential for efficient cleavage (7). Linker-ligated cDNA on the magnetic beads was digested with 10 units of EcoP15I in the reaction mixture (10 mM Tris·HCl, pH 8.0/10 mM KCl/10 mM MgCl2/0.1 mM EDTA/0.1 mM DTT/5 μg/ml BSA/2 mM ATP) at 37°C for 90 min (7). Released fragments after EcoP15I digestion were separated on PAGE, and the ≈69-bp “linker-tag” fragments (linker, 42 bp; tag, ≈27 bp) were visualized by FITC fluorescence on a UV transilluminator and collected from the gel. Linker-1E-tag and linker-2E-tag fragments were mixed, blunt-ended by filling-in with KOD DNA polymerase (from Thermococcus kodakaraensis strain KOD1, Toyobo, Osaka), and ligated to each other. The resulting ditags were amplified by PCR with biotinylated primers (ditag primer 1E, biotin-5′-CTAGGCTTAATACAGCAGCA-3′; ditag primer 2E, biotin-5′-TTCTAACGATGTACGCAGCAGCA-3′). After digestion of ditag PCR products with NlaIII, digested fragments were separated on PAGE and the fragment of ≈54 bp was isolated from the gel. The 54-bp fragments were concatenated by ligation. Concatemers larger than 500 bp were size-selected by PAGE, isolated, and cloned into a plasmid vector (pGEM3Z, Promega). Electrocompetent E. coli cells (DH10B, Invitrogen) were transformed with pGEM3Z harboring the concatemers by electroporation and plated on LB medium containing 100 μg/ml ampicillin, 20 μg/ml X-gal, and 0.1 mM IPTG. Plasmid inserts were amplified by colony PCR and directly sequenced with a RISA384 DNA autosequencer (Shimadzu). DNA sequences of the plasmid inserts were analyzed with the sage2000 program (supplied by Johns Hopkins University, Baltimore) for extraction of the 22-bp tags (adjacent to CATG). Although in most cases it is possible to extract a 23-bp tag sequence (leading the total size to 27 bp), filling-in reaction sometimes removes one base from the end of the fragment, reducing the tag size to 26 bp. Therefore, we decided to isolate a 26-bp sequence from each cDNA and call it the “SuperSAGE tag.”
Fig. 1.
Flowchart of SuperSAGE. For details see Materials and Methods.
cDNA Synthesis. Total RNA was extracted from blast fungus-infected rice leaves. From 1 μg of RNA of each sample, single-stranded cDNA was synthesized by using oligo(dT) primer and SuperScript II reverse transcriptase (Invitrogen). Three M. grisea genes identified by SuperSAGE were amplified by PCR with the gene-specific primers (5′-AGCTATTTTCTCACATCAGG-3′ and 5′-AATGAGTGGAACGAGAAGAG-3′ for hydrophobin, 5′-GCTTCATTGCCATCAAGCCC-3′ and 5′-GTCACCACGGATGGTGCCAG-3′ for nucleoside diphosphate kinase, and 5′-GCACAGGCTCGCTAAAATGC-3′ and 5′-TTCTCAGCCTCCTTGCTCAC-3′ for 60S ribosomal protein).
3′ RACE. The recovery of longer cDNA fragments downstream of the SuperSAGE tag identified in N. benthamiana was achieved with the 3′ RACE system (Invitrogen). Single-stranded cDNA was synthesized from 1 μg of total RNA by using an adapter primer containing oligo(dT) and used as template for PCR. A primer oligonucleotide complementary to the adapter–primer sequence was used for PCR in combination with the 26-bp oligonucleotides corresponding to the SuperSAGE tag sequences.
Results
Gene Expression Profiling in Blast-Infected Rice Leaves. Blast disease is a devastating rice disease caused by the ascomycete fungus M. grisea. Whole genome sequences are available for both rice (11, 12) and the blast fungus (13), so that the rice–blast fungus interaction system offers to test the utility of genomic information for understanding host–pathogen interactions.
A total of 12,119 “SuperSAGE tags” were obtained from M. grisea-infected rice leaves. The number of different tags, corresponding to the different genes, was 7,546 (Table 1). To identify these genes, the complete GenBank database comprising all available cDNA, EST, and genome sequences was searched by blast with 26-bp tag sequences as query. In most cases, only a single stretch of DNA sequence from the rice genome perfectly matched the tag sequence (see Discussion), demonstrating that the information in the 26-bp tag sequence is sufficient for identifying the gene of origin. The 10 most abundantly expressed genes thus annotated are listed in Table 1. As expected, genes for photosynthetic proteins were most abundantly expressed in rice leaves.
Table 1. The 10 most abundantly expressed genes in blast-infected rice leaves as revealed by SuperSAGE.
| Tag sequence* | Count | GenBank accession no.; encoded protein† |
|---|---|---|
| TTCGGCTTCTTCGTCCAGGCCA | 122 | D00641; chlorophyll a/b binding protein |
| GATCCGTCTCTCTGGGAGGAAT | 116 | AU172529; thiazole biosynthetic enzyme |
| GCGACGCATCGCCTTCAGCTAA | 114 | X13909; chlorophyll a/b binding protein |
| TGGTGGCTTAGCTCTACGTGTA | 111 | AU174449; glycine rich protein |
| TCGGACAAGTGCGGCAACTGCG | 94 | AF001396; metallothionein |
| TTGTAATACTCCATCAAAGAGT | 86 | D29966; catalase |
| AATTGAGTTCGCTTTGGTTATG | 78 | AF010579; glycine rich protein |
| ATGATGATATACTACACTTGAT | 58 | BE230408; photosystem II 10-kDa protein |
| GCGTCCACGCTGACCAACGTCG | 57 | BE230423; unknown protein |
| TATGTATGTACCTTAATTGTGT | 52 | D00642; chlorophyll a/b binding protein |
| Total number of tags (different tags) | 12,119 (7,546) |
Tag represented as a 22-bp sequence excluding the NIaIII site (CATG).
Rice gene bank.
Next, taking advantage of the information content in the 26-bp tag, we tried to identify the tags derived from M. grisea messages among the total tags isolated from blast-infected rice leaves. All tags isolated from infected rice leaves were applied to blast search with M.grisea draft genome sequences (http://www-genome.wi.mit.edu/cgi-bin/annotation/magnaporthe/blast_page.cgi?organismName=Magnaporthe). A total of 35 different tags matched putative genes in M. grisea genome sequences only (Table 2), but had no homologues in the rice genome. The total number of tags presumably derived from M. grisea messages was 74 representing 0.6% of the analyzed transcripts (74/12119) in blast-infected rice leaves (Table 2). The hydrophobin gene alone accounted for half of the M. grisea transcripts (38 tags), and nucleoside diphosphate kinase and 60S ribosomal protein genes contributed two tags each (Table 2). The rest of the tags were encountered only once. To see whether the M. grisea genes identified by SuperSAGE are really expressed in blast-infected rice leaves, RT-PCR was carried out on cDNA from M. grisea-infected leaves (cv. Norin 1) as template, using gene specific primers for hydrophobin, nucleoside diphosphate kinase, and 60S ribosomal protein genes. Specific PCR amplification was observed for messages from all three genes (Fig. 2A). As mentioned, cv. Kakehashi is susceptible to M. grisea race 007, and cv. Himenomochi is resistant. When RT-PCR for the mRNAs from the three genes was carried out with template cDNAs isolated from rice leaves of (i) cv. Kakehashi mock-inoculated with water, (ii) cv. Kakehashi inoculated with M. grisea race 007, (iii) cv. Himenomochi mock-inoculated, and (iv) cv. Himenomochi inoculated with M. grisea race 007, PCR products of expected sizes were only observed in ii, i.e., the susceptible cultivar inoculated with the compatible M. grisea race. This is solid proof that transcripts of the three M. grisea genes identified by SuperSAGE actually derived from M. grisea messages in the infected rice leaves, again demonstrating the power of SuperSAGE for the simultaneous resolution of gene expression in two or more interacting organisms such as hosts and pathogens.
Table 2. Some M. grisea expressed genes in blast-infected rice leaves as revealed by SuperSAGE.
| Tag sequence* | Count | GenBank accession no.; putative protein† |
|---|---|---|
| CGATCACGAGGGGATGATGGTG | 38 | L20685; hydrophobin |
| TCAGACACAGGCTGTACAAGGC | 2 | Nucleoside-diphosphate kinase |
| TCACGTTTAGAAAGGCGACCCG | 2 | 60S ribosomal protein |
| TTGCCCGTATGTACATAAACAA | 1 | BM865406; NADH-ubiquinone oxidoreductase |
| CAATTGGTGTTTCTTTGGGTTT | 1 | AF056625; poly-ubiquitin |
| TCGTCTGTGGCTTCAGTTGCTG | 1 | Unknown protein |
| ACGAGCTGATGCGCAAGGATGG | 1 | ABC transporter |
| Total number of tags presumably derived from M. grisea | 74 |
Tags represented as a 22-bp sequence excluding the NIaIII site (CATG).
Putative protein was deduced from cDNA sequence and genomic sequence of M. grisea that matched the corresponding tag.
Fig. 2.
(A) RT-PCR of M. grisea genes encoding hydrophobin, nucleoside diphosphate kinase, and 60S ribosomal protein, from cDNA template prepared from M. grisea-infected rice cv. Norin 1. (B) RT-PCR of M. grisea genes from cDNA templates prepared from rice cv. Kakehashi and cv. Himenomochi either mock-inoculated (–) or inoculated with M. grisea race 007 (+).
Gene Expression Profiling of INF1 Elicitor-Treated N. benthamiana, a Nonmodel Organism. On recognition of invading pathogens, plants exert their resistance in multiple front lines (14). One of the remarkable responses is the so-called hypersensitive response (HR) that involves apoptosis-like cell death occurring around the foci of pathogen invasion. HR functions in the containment of the pathogens and avoids further spread of the disease. INF1, a secreted protein of P. infestans, is a well characterized elicitor causing HR in incompatible hosts like N. benthamiana (9, 15). We are interested in the molecular mechanisms of INF1-mediated HR (16) generally and gene expression changes during HR in particular and therefore applied SuperSAGE to INF1-treated N. benthamiana. For this species, no genomic DNA sequence is available and only 55 EST/cDNA sequences were published in GenBank as of June 2003. Therefore, N. benthamiana represents a typical nonmodel organism.
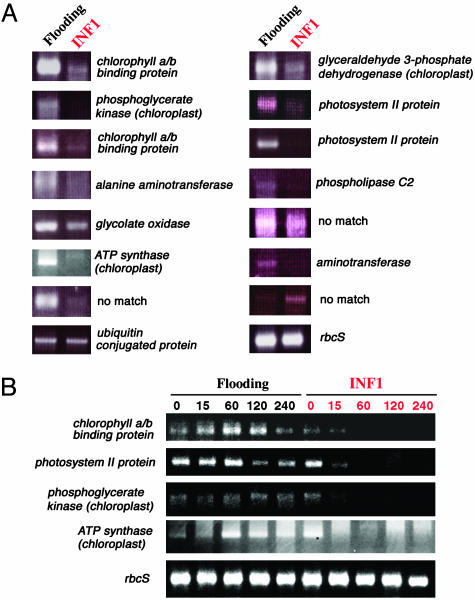
N. benthamiana leaves were infiltrated with water (flooding as negative control) or 100 nM INF1 elicitor (9) by a needleless syringe. One hour after the infiltration, leaves were harvested and used for SuperSAGE. HR usually becomes visible 24–48 h after the infiltration. A total of 5,095 and 5,089 tags were isolated from flooding- and INF1-treated leaves, respectively. SuperSAGE tags differentially represented in the differentially treated plants were identified, and most of them showed lower representation in INF1-treated leaves as compared to the control. We randomly selected 14 tags, and cDNA fragments downstream of the tags were recovered by 3′ RACE with PCR primers corresponding to the tag sequences (Table 3). For all of them, partial cDNA fragments containing a polyA tail were easily recovered. Of 14 cDNA sequences, 11 showed significant homology to known protein genes of higher plants. Many of the genes down-regulated after INF1 treatment encode chloroplast-localized and photosynthesis-related proteins. We confirmed the expression change in these genes by 3′ RACE with 26 bp-tag primers. In a separate experiment, RNA was isolated from N. benthamiana leaves 60 min after flooding or INF1 treatment. The mRNA was reverse-transcribed and used as template for RT-PCR (3′ RACE using the 26-bp tag primer). For all of the genes tested, the expression changes already detected by SuperSAGE were fully reproduced by RT-PCR (Fig. 3A), demonstrating the accuracy of transcripts profiling by SuperSAGE. Note that the constitutive expression of the genes for ribulose bisphosphate carboxylase small subunit (rbcS) and ubiquitin-conjugated protein as revealed by SuperSAGE was also confirmed by RT-PCR. The kinetics of down-regulated genes after INF1 treatment was probed with RT-PCR (3′ RACE using the 26-bp tag primer). Expression of genes encoding chlorophyll a/b binding protein, photosystem II protein, phosphoglycerate kinase, and ATP synthase started to decrease 15 min after INF1 infiltration, and after 60 min expression it was completely silenced, whereas the rbcS gene was almost constantly expressed under both treatment (Fig. 3B). We conclude that the 26-bp tag primers can directly be applied to 3′ RACE kinetics of gene expression.
Table 3. Differentially expressed genes in INF1 vs. flooded leaves of N. benthamiana (1 h after the treatment).
| Number of tags
|
|||
|---|---|---|---|
| Tag sequence* | Flooding | INF1 | Gene product† |
| TTTTCTATGTTCGGATTCTTTG | 17 | 8 | Chlorophyll a/b binding protein |
| AGGAATAGAGGGCAAGGTGCTC | 11 | 6 | Phosphoglycerate kinase (chloroplast) |
| GGCTTTTGCCACTAACTTTGTA | 14 | 4 | Chlorophyll a/b binding protein |
| GAGCAATATGAAGACCACAGAG | 11 | 3 | Alanine aminotransferase |
| GCTCTTGAAGAGGTTGTGAAAG | 11 | 3 | Glycolate oxidase |
| GGCAACAATGCTCTAGAGAAAG | 10 | 2 | ATP synthase (chloroplast) |
| CCTAGCTATTGACTACTGAAGT | 10 | 2 | No match |
| GTTAAGGTTATTGCTTGGTATG | 7 | 2 | Glyceraldehyde 3-phosphate dehydrogenase (chloroplast) |
| TTTCCTTGACGATCACTCTTGG | 7 | 2 | PhotosystemII 23-kDa protein |
| GTGATTCCCGACGTAGCCGAAG | 6 | 1 | PhotosystemII protein |
| TTGCAACTTCTAGTCAATGACT | 16 | 4 | Phospholipase C2 |
| ATGGCCAAGTAATTTCACCATC | 6 | 1 | No match |
| AACTCATTAGAGACTCGAAGGG | 6 | 0 | Amino transferase |
| CAACACGAGCACGCACCTCTCT | 0 | 7 | No match |
| TGCGGGATTCGGTGGTGCCGGA | 5 | 7 | Ubiquitin-conjugated protein |
| TTCGGGTGCACTGATGCCACTC | 32 | 33 | Ribulose bisphosphate carboxylase small subunit |
| Total number of tags | 5,095 | 5,089 | |
Tags represented as a 22-bp sequence excluding the NIaIII site (CATG).
Encoded proteins were deduced by blast search with 3′ RACE fragment recovered by using a 26-bp tag primer.
Fig. 3.
(A) 3′ RACE-PCR of genes identified by SuperSAGE to be differentially expressed in flooded and INF1-treated N. benthamiana leaves. Oligonucleotides corresponding to the 26-bp tag sequences were used as PCR primers. (B) Expression kinetics of four N. benthamiana genes encoding chlorophyll a/b binding protein, photosystem II protein, phosphoglycerate kinase, and ATP synthase after flooding and INF1 treatments as revealed by 3′ RACE-PCR. Numbers above indicate minutes after treatment.
Discussion
Response of rice cells to M. grisea fungal elicitor has previously been studied by conventional SAGE (2). After analyzing >10,000 tags for elicitor-treated and control rice cells, 139 unique tags were identified as elicitor-inducible, of which 96 tags were assigned with putative gene names after consulting EST and genomic DNA databases. However, many 13-bp tags had matches to multiple positions of the genome so that decisive identification of the gene was difficult. A similar difficulty was encountered in SAGE analysis of M. grisea that was treated with cAMP to induce appressorium on artificial membrane (3). As such, simultaneous monitoring of gene expression of rice and M. grisea in blast-infected rice leaves is not possible with conventional SAGE. SuperSAGE, on the other hand, extracts a 26-bp tag from each cDNA and allows the assignment of each tag to rice or M. grisea genome without ambiguity. This opens a possibility to directly study the gene expression of two organisms at the foci of interaction. SuperSAGE revealed that hydrophobin is the most highly expressed M. grisea gene in rice leaves (Table 2). The same gene was identified as highly inducible by cAMP treatment (3) and corroborates the finding that hydrophobin is required for appressorium formation (17).
The INF1 protein of P. infestans induces a HR including cell death in the host and is responsible for non-host resistance of N. benthamiana to P. infestans (9). The molecular mechanisms of HR induction by INF1 have not yet been clarified. Therefore, we used SuperSAGE to identify the genes that are up- or down-regulated before HR induction in INF1-treated N. benthamiana plants. Because water-infiltrated leaves were used as control, wound-inducible genes such as PR protein genes (18) are not represented. Therefore, we expected to find only genes specifically induced or repressed by the INF1 elicitor signal.
Surprisingly, several photosynthetic genes were repressed by INF1 as early as 1 hour after treatment (Table 3). A few previous reports indeed relate photosynthesis with HR in higher plants. Allen et al. (19) showed that mastoparan (G-protein activator)-induced HR in Asparagus mesophyll cells was light-dependent, suggesting the involvement of photosynthesis. Seo et al. (20) reported that chloroplast gene ftsH was down-regulated before HR caused by tobacco mosaic virus (TMV) infection to resistant tobacco plant. The down-regulation of the ftsH gene is reminiscent of the repression of photosynthetic genes in N. benthamiana. FtsH may be involved in the degradation of chloroplast D1 proteins that are damaged by reactive oxygen species (ROS) produced by an imbalance in the photosystem II (PSII) reaction (21). Any decrease in FtsH protein concentration in TMV-infected leaves is supposed to inhibit PSII activity and cause HR. In INF1-treated N. benthamiana leaves, genes for PSII proteins were remarkably repressed. We hypothesize that PSII activity is lost, and this in turn causes ROS accumulation in chloroplast, inducing HR in leaf tissue. The coordinated down-regulation of several photosynthetic genes after INF1 treatment strongly suggests the involvement of common transcriptional regulatory factors responding to INF1 in N. benthamiana.
The use of the type III restriction endonuclease EcoP15I as a tagging enzyme is the key feature of SuperSAGE that enabled the isolation of 26-bp tags. Among all of the functionally related type III restriction endonucleases, EcoP15I has the cleavage site most distant from its recognition site (http://rebase.neb.com/rebase). For the use of EcoP15I as the tagging enzyme, we modified both the linker and oligo(dT) primer structure. Because EcoP15I requires two unmethylated inversely oriented 5′-CAGCAG-3′ sites in the target DNA for digestion (7), this recognition site was incorporated in the linker and oligo(dT) sequences. Another important modification concerns the purification step of the linker-tag fragments. We labeled the linker fragment with FITC, so that the linker-tag can easily be visualized under UV and isolated from other fragments after digestion of linker-cDNA with EcoP15I. This modification increased the yield of linker-tag fragments and resulted in the robustness of the technique. These two essential modifications made SuperSAGE more informative and technically less demanding than the original SAGE. Recently, a so-called LongSAGE protocol was developed that allows isolation of longer tag fragment than does SAGE (22). With the use of the type IIS restriction enzyme MmeI as the “tagging enzyme,” it is possible to recover 19- to 21-bp tags. However, MmeI digestion produces protrusions of two bases at the 3′ end. To ensure the random association of tags to form “ditags,” the 3′ protrusion has to be removed. This results in the reduction of the tag sequence length to 17–19 bp, which in turn reduces the information content in the tag sequence. Therefore, in the LongSAGE protocol (22), the MmeI-generated ends are not polished and the fragments with 3′ protrusions are ligated. This, however, implies that a linkertag fragment ligates itself only with another linker-tag fragment harboring the compatible 3′ end. This inevitably entails that ditags formed after ligation are no more the products of a random association of tags. Theoretically, this procedure skews the representation of each tag in resulting ditags, and the final result of LongSAGE may not faithfully reflect the abundance of expression of each gene. For this reason, the current LongSAGE procedure is only used to annotate the 13-bp tag sequences obtained by the conventional SAGE protocol.
The advantage of SuperSAGE over the conventional SAGE and LongSAGE is twofold. Firstly, the information content of a 26-bp SuperSAGE tag fragment is appreciably higher than a 19- to 21-bp tag obtained by LongSAGE or the 13-bp tag of conventional SAGE. Table 4 shows the summary of blast search for 30 rice SAGE tags against the whole GenBank database. For each tag fragment, its size was set to 15 bp (conventional SAGE), 18 bp (LongSAGE with blunting treatment), 20 bp (LongSAGE), and 26 bp (SuperSAGE) and used as query for blast search. The number of species that harbored DNA sequences showing perfect match to the given tag sequence was counted for each tag, and its average and maximum values across 30 tags were calculated. The 26-bp SuperSAGE tag has matches to only 1.1 species on average and 2 species maximum. This indicates that a SuperSAGE tag identifies a gene out of the entire DNA sequences deposited in the complete GenBank database. This power of gene identification is not available for conventional SAGE (15-bp tag) or for LongSAGE with blunting treatment (18-bp tag). For species without deposited DNA sequences in databases, the 26-bp SuperSAGE tag could immediately be used as a specific 3′ RACE primer to rapidly recover a longer cDNA fragment. This fragment in turn could then successfully be used for blast search and annotation of the tags. Previously, several PCR techniques were reported to recover cDNA fragment from 13- to 15-bp SAGE tags (23, 24). However, it appeared always difficult to determine the appropriate conditions for a specific amplification of cDNAs from each gene because the SAGE tag primers were too short. Secondly, to ascertain accurate gene expression profiling, the ends of linker-tag fragments generated by SuperSAGE are blunt-ended to ensure the random association of two tags to form ditags. As confirmed by RT-PCR, reliable gene expression profiles can be generated by SuperSAGE in INF1-challenged N. benthamiana leaves. In the LongSAGE protocol with 19- to 21-bp tags, this blunting treatment is not applicable. To further test the reliability of transcript profiling obtained by SuperSAGE, we performed SuperSAGE and conventional SAGE by using the same RNA sample isolated from rice suspension-cultured cells (see Table 5 and Fig. 5, which are published as supporting information on the PNAS web site). Of the 50 most highly represented SuperSAGE tags, 37 tags found corresponding 13-bp tags in the 50 most highly represented SAGE tags, and only 10 tags showed statistically significant difference in representation between SuperSAGE and SAGE data. Such discrepancies between SuperSAGE and SAGE data could be caused mainly by the two possibilities: (i) a 13-bp SAGE tag was actually derived from two or more different gene transcripts, which were separately represented in different SuperSAGE tags, and (ii) the longer SuperSAGE tag has higher chance of incorporating mutations/sequence errors during experiment than the short SAGE tag (25). In the computational extraction of tags from sequence data, the sage2000 program removes the duplicated ditags (ditag comprising of the same combination of tags) from consideration. This “duplicated ditag removal” is performed to minimize the effect of PCR bias (1). In this process, if a mutation or sequencing error occurs in duplicated ditags, these ditags are no more recognized as “duplicated” and these tags will participate in tag counting. This potentially could affect the data. However, the error caused by this process is random and will not cause a systematic bias in the data. The discrepancy of the data caused by the first possibility is attributable to the lower resolution power of conventional SAGE. The second possibility would be minimized by using a high-fidelity DNA polymerase in the PCR step. Regardless of some discrepancy, we conclude that SuperSAGE data are overall compatible with the conventional SAGE data and providing faithful gene expression profiles.
Table 4. Summary of blast search of 30 rice SAGE tags for the entire body of GenBank data.
| Tag size, bp
|
||||
|---|---|---|---|---|
| 26 | 20 | 18 | 15 | |
| Average no. of species with DNA sequences perfectly matching the tag | 1.1 | 1.2 | 1.8 | 5.0 |
| Maximum no. of species with DNA sequences perfectly matching the tag | 2 | 4 | 7 | 9 |
The functions of the genes identified by SuperSAGE as described above can also be studied by using the virus-induced gene silencing (VIGS) system (26). Because SuperSAGE tags are likely to be derived from the 3′ end of the transcripts, the 3′ UTR sequence in the 3′ RACE fragment can easily be cloned into an appropriate virus vector to trigger gene-specific VIGS. The combination of SuperSAGE and VIGS should therefore serve as a rapid and high-throughput functional genomics tool in higher plants (Fig. 4).
Fig. 4.
Application of SuperSAGE for functional genomics of eukaryotic organisms.
RNA interference (RNAi) is currently the most efficient tool for knock-down of specific genes in animals. For example, in C. elegans, thousands of genes were systematically targeted by RNAi and phenotypes of these knock-down lines studied (27). Recently, it was shown that RNAi can also be induced by the introduction of a short synthetic oligo RNA (21–20 mer) into mammalian cells (28). Therefore, our 26-bp SuperSAGE tag sequences strongly recommend themselves for direct use in RNAi, so that the knock-down lines showing any phenotypes can be generated without any knowledge about the genes. We trust that the combination of SuperSAGE and short oligo RNA-mediated RNAi will be a future high throughput gene function analysis system.
Oligonucleotides or cDNA microarrays are excellent tools to simultaneously analyze gene expression in numerous samples at a given time (29). The 3′ RACE products generated with 26-bp SuperSAGE tag primers should serve as highly gene-specific fragments suitable for spotting on the microarrays. It should be also possible to use the 26-bp SuperSAGE tag sequences directly for spotting onto oligonucleotide arrays (30). So, not only in its present format, but also in combination with other techniques, SuperSAGE promises to be a valuable addition to the repertoire of methodologies for functional genomics.
Supplementary Material
Acknowledgments
We dedicate this work to the memory of the late Professor Dr. Jeff Schell (Max Planck Institute for Plant Breeding, Cologne, Germany), who inspired our research tremendously. We thank Petra Mackeldanz for help with enzyme preparation and Prof. K. Kinzler (Johns Hopkins University, Baltimore) for the provision of sage analysis software. G.K. thanks his colleagues in the Biocenter, University of Frankfurt, for leaving him space after his retirement. R.T. thanks the Alexander von Humboldt Foundation for Fellowship IV-7121-1028559 and the Science and Technology Agency (Japan) for Fellowship ID 499072. Work in Iwate was supported in part by the Research for the Future Program of the Japan Society for the Promotion of Science. Work in Berlin was supported by the Deutsche Forschungsgemeinschaft (KR1293/1-3), Fonds der Chemischen Industrie, and the Humboldt University School of Medicine.
Abbreviations: HR, hypersensitive response; SAGE, serial analysis of gene expression.
References
- 1.Velculescu, V. E., Zhang, L., Vogelstein, B. & Kinzler, K. W. (1995) Science 270, 484–487. [DOI] [PubMed] [Google Scholar]
- 2.Matsumura, H., Nirasawa, S., Kiba, A., Urasaki, N., Saitoh, H., Ito, M., Kawai-Yamada, M., Uchimiya, H. & Terauchi, R. (2003) Plant J. 33, 425–434. [DOI] [PubMed] [Google Scholar]
- 3.Irie, T., Matsumura, H., Terauchi, R. & Saitoh, H. (2003) Mol. Gen. Genet. 270, 181–189. [DOI] [PubMed] [Google Scholar]
- 4.Dryden, D. T. F., Murray, N. E. & Rao, D. N. (2001) Nucleic Acids Res. 29, 3728–3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janscak, P., Sandmeier, U., Szczelkun, M. D. & Bickle, T. A. (2001) J. Mol. Biol. 306, 417–431. [DOI] [PubMed] [Google Scholar]
- 6.Mücke, M., Reich, S., Moncke-Buchner, E., Reuter, M. & Krüger, D. H. (2001) J. Mol. Biol. 312, 687–698. [DOI] [PubMed] [Google Scholar]
- 7.Meisel, A., Bickle, T. A., Krüger, D. H. & Schroeder, C. (1992) Nature 355, 467–469. [DOI] [PubMed] [Google Scholar]
- 8.Tada, T., Kanzaki, H., Norita, E., Uchimiya, H. & Nakamura I. (1996) Mol. Plant–Microbe Interact. 9, 758–759. [Google Scholar]
- 9.Kamoun, S., van West, P., Vleeshouwers, V. G., de Groot, K. E. & Govers, F. (1998) Plant Cell 10, 1413–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meisel, A., Mackeldanz, P., Bickle, T. A., Krüger, D. H. & Schroeder, C. (1995) EMBO J. 14, 2958–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goff, S. A., Ricke, D., Lan, T. H., Presting, G., Wang, R., Dunn, M., Glazebrook, J., Sessions, A., Oeller, P., Varma, H., et al. (2002) Science 296, 92–100. [DOI] [PubMed] [Google Scholar]
- 12.Yu, J., Hu, S., Wang, J., Wong, G. K., Li, S., Liu, B., Deng, Y., Dai, L., Zhou, Y., Zhang, X., et al. (2002) Science 296, 79–92. [DOI] [PubMed] [Google Scholar]
- 13.Martin, S. L., Blackmon, B. P., Rajagopalan, R., Houfek, T. D., Sceeles, R. G., Denn, S. O., Mitchell, T. K., Brown, D. E., Wing, R. A. & Dean, R. A. (2002) Nucleic Acids Res. 30, 121–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maleck, K. & Dietrich, R. A. (1999) Trends Plant Sci. 4, 215–219. [DOI] [PubMed] [Google Scholar]
- 15.Kamoun, S., Lindqvist, H. & Govers, F. (1997) Mol. Plant–Microbe Interact. 10, 1028–1030. [DOI] [PubMed] [Google Scholar]
- 16.Sharma, P. C., Ito, A., Shimizu, T., Terauchi, R., Kamoun, S. & Saitoh, H. (2003) Mol. Gen. Genet. 269, 583–591. [DOI] [PubMed] [Google Scholar]
- 17.Talbot, N. J., Ebbole, D. J. & Hamer, J. E. (1993) Plant Cell 5, 1575–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grosset, J., Marty, I., Chartier, Y. & Meyer, Y. (1990) Plant Mol. Biol. 15, 485–496. [DOI] [PubMed] [Google Scholar]
- 19.Allen, L. J., MacGregor, K. B., Koop, R. S., Bruce, D. H., Karner, J. & Bown, A. W. (1999) Plant Physiol. 119, 1233–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seo, S., Okamoto, M., Iwai, T., Iwano, M., Fukui, K., Isogai, A., Nakajima, N. & Ohashi, Y. (2000) Plant Cell 12, 917–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spetea, C., Hundai, T., Lohmann, F. & Andersson, B. (1999) Proc. Natl. Acad. Sci. USA 96, 6547–6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saha, S., Sparks, A. B., Rago, C., Akmaev, V., Wang, C. J., Vogelstein, B., Kinzler, K. W. & Velculescu, V. E. (2002) Nat. Biotechnol. 20, 508–512. [DOI] [PubMed] [Google Scholar]
- 23.van den Berg, A., van der Leij, J. & Poppema, S. (1999) Nucleic Acids Res. 27, e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen, J. J., Rowley, J. D. & Wang, S. M. (2000) Proc. Natl. Acad. Sci. USA 97, 349–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pleasance, E. D., Marra, M. A. & Jones, S. J. M. (2003) Genome Res. 13, 1203–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baulcombe, D. C. (1999) Curr. Opin. Plant Biol. 2, 109–113. [DOI] [PubMed] [Google Scholar]
- 27.Kamath, R. S., Fraser, A. G., Dong, Y., Poulin, G., Durbin, R., Gotta, M., Kanapin, A., Le Bot, N., Moreno, S., Sohrmann, M., et al. (2003) Nature 421, 231–237. [DOI] [PubMed] [Google Scholar]
- 28.Yu, J. Y., DeRuiter, S. L. & Turner, D. L. (2002) Proc. Natl. Acad. Sci. USA 99, 6047–6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Churchill, G. A. (2002) Nat. Genet. 32, 490–495. [DOI] [PubMed] [Google Scholar]
- 30.Hacia, J. G., Woski, S. A., Fidanza, J., Edgemon, K., Hunt, N., McGall, G., Fodor, S. P. & Collins, F. S. (1998) Nucleic Acids Res. 26, 4975–4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.