Abstract
We present a methodology for gene functional prediction based on extraction of physiologically relevant growth variables from all viable haploid yeast knockout mutants. This quantitative phenomics approach, here applied to saline cultivation, identified marginal but functionally important phenotypes and allowed the precise determination of time to adapt to an environmental challenge, rate of growth, and efficiency of growth. We identified ≈500 salt-sensitive gene deletions, the majority of which were previously uncharacterized and displayed salt sensitivity for only one of the three physiological features. We also report a high correlation to protein–protein interaction data; in particular, several salt-sensitive subcellular networks indicating functional modules were revealed. In contrast, no correlation was found between gene dispensability and gene expression. It is proposed that high-resolution phenomics will be instrumental in systemwide descriptions of intragenomic functional networks.
Afundamental approach in determining the cellular role of unclassified genes is to determine the consequences of gene loss. Recently, systematic approaches toward the targeted inactivation of each gene in a genome have been completed in model organisms such as the yeast Saccharomyces cerevisiae (1) and the nematode Caenorhabditis elegans (2). These initiatives have opened the door to phenomics (3), a novel field with the aim of providing systematic descriptions of phenotypic characteristics on a genomewide scale (4–10). Conventional phenotypic analysis is qualitative in nature, which should be cause for serious concern because a majority of phenotypes are expected to be marginal in nature (11). Furthermore, it is to be expected that different genes are linked to different aspects of cell physiology. Thus, the ability to distinguish different physiological features, like time to adapt to environmental challenges and kinetics or efficiency of growth, will be instrumental in a high-quality description of the cellular role of a gene.
Here we report a high-resolution quantitative analysis of salt phenotypes of the complete collection of haploid deletion strains in yeast, providing a genomewide view of the consequences of loss of individual genes under saline stress. Each strain was cultivated in isolation, providing automatically generated growth curves that make it possible to distinguish between gene deletion effects on different aspects of cellular physiology (12). There is ample information about the salt response of yeast, where the production and accumulation of the osmolyte glycerol and efficient extrusion of the sodium ion via ion pumps appear as key osmoregulatory events (13). In addition, genomewide phenotyping has earlier been performed during saline cultivation (1), making salt an ideal test case for the potency of the herepresented high-resolution phenomics to provide novel functional information during a well documented cell stress.
The gene dispensability during salt stress was compared with other systemwide datasets such as protein–protein interaction and gene expression data, providing an integrative and holistic view of salt response in yeast.
Materials and Methods
Deletion strains in the BY4741 background (1) were inoculated in 350 μl of synthetic defined (SD) medium. Two consecutive precultures were run. For experimental runs, strains (duplicates) were cultivated as reported (12) for 47 h with duplicates on separate plates. Rate of growth, efficiency of growth, and adaptation time were calculated essentially as reported (12). In each run, eight wild types were included and used for normalization of strain behavior, forming strain coefficients [logarithmic (natural logarithm) strain coefficients (LSC)]. Strain coefficients in basal SD medium (LSC basal) and in salt medium (LSC NaCl) were compared forming phenotypic indexes [logarithmic (natural logarithm) phenotypic index (LPI)]. We performed statistical tests of the null hypothesis that LPI equals 0, applying a threshold of three mean standard deviations and a two-tailed, two-sample Student's t test. These combined measures gave a significance level of α < 0.001. Statistical significance of the representation of functional classes, localization, and interaction within a category of phenotypes (e.g., rate of growth sensitivity) was tested assuming Poisson distributed data.
Details are published as Supporting Materials and Methods on the PNAS web site.
Results
Quantitative Phenomics. Analyzing the growth of all 4,711 viable isogenic haploid deletion strains from yeast in isolation, we provide systematic high-resolution growth curves for a complete mutant collection. Evaluation of 20,000 growth curves, each represented by 142 automated measurements of optical density, identified and resolved different aspects of cellular physiology (12). With high precision we quantified the evolutionary and physiologically important growth characteristics length of lagphase (time to adapt to environmental challenge), growth rate (kinetics of growth), and stationary-phase cell density (efficiency of growth). To maximize standardization and reproducibility, the quantitative growth measures in a particular environment for each strain were normalized to the average response of eight wild types included in each run, providing LSC. All three growth variables could be determined with high precision (average coefficient of variation of strain coefficients was 1.6–8.1%).
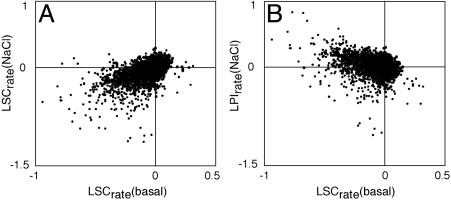
The majority of recorded growth defects in basal medium or in 0.85 M NaCl-containing medium were small (data not shown), confirming the prediction that most phenotypes are marginal in nature (11). In total, 1,415 strains displayed a significant salt growth defect (LSC < 0; α < 0.001). However, we found that many strains with a defect in growth in the presence of salt also displayed a similar defect in basal media (Fig. 1A). To distinguish the specific salt growth defect of a deletion strain from a general growth defect, LSC with and without salt were combined into a LPI providing a sensitivity measure for each strain to salt, in relation to the response of the wild type. By statistical comparison of LPIs, we found 488 deletion strains to have a significant (LPI < 0; α < 0.001) salt sensitivity for at least one of the physiological features; 38% of all sensitivity phenotypes constituted rate-of-growth defects, 12% were efficiency of growth defects, and 50% were defects in the time of adaptation. The different physiological features also constituted predominantly independent markers of growth aberrations, because in the range 50–70% of the scored phenotypes were exclusive for a single growth variable. Even among the top scores from the three growth categories, we found frequent examples of selective phenotypes (Table 1). In fact, not for a single deletion strain was a phenotype scored in all three growth categories. It is also apparent in Fig. 1B that strains with a salt-resistant phenotype (LPI > 0) displayed a strong tendency for decreased growth in basal medium. Hence, most strains scored as salt-resistant were found not to have improved salt growth per se. Rather, they displayed growth defects in basal medium and growth was not proportionally affected by the addition of salt.
Fig. 1.
Scatter plot of quantitative growth defects in basal synthetic medium, LSCrate(basal), versus growth defects in saline medium (A), LSCrate(NaCl), and salt-specific growth defects (B), LPIrate(NaCl).
Table 1. Quantitative phenotypes of the most salt-sensitive strains.
| Gene | Rate | Phenotype (LPI) efficiency | Adaptation |
|---|---|---|---|
| Cellular defense | |||
| BRO1 | -0.73 | ||
| HOG1 | -0.79 | -1.23 | |
| PBS2 | -1 | -1.27 | |
| YDR119w | -0.72 | -1.59 | |
| Cellular transport | |||
| GRR1 | -0.43 | -1.5 | |
| SRO7 | -0.95 | -1.94 | |
| TRK1 | -1.25 | ||
| Cytoskeleton | |||
| ARC18 | -0.9 | -1.23 | |
| ARP5 | -1.68 | ||
| RVS161 | -1.45 | ||
| Metabolism | |||
| SHM2 | -1.71 | ||
| Protein fate | |||
| BUL1 | -0.44 | -0.72 | |
| PEP3 | -1.61 | ||
| VPS25 | -0.54 | ||
| Protein synthesis | |||
| RPL6A | -0.57 | ||
| RPL37A | -0.2 | -1.38 | |
| YDR115w | -0.87 | ||
| Transcription | |||
| NCL1 | -0.56 | ||
| Unclassified | |||
| KRE25* | -0.47 | -1.54 | |
| PSL10 | -1.04 | -2.18 | |
| YJL184w | -1.55 | ||
| YLR097c | -1.21 | ||
| YNL047c | -0.57 | -0.51 |
The 10 most salt-sensitive deletion strains in each physiological category are listed. LPIs are indicated for physiological features that displayed statistically significant (α < 0.001) salt sensitivity. Classifications are taken from the Munich Information Center for Protein Sequences (http://mips.gsf.de/proj/yeast/CYGD/db/index.html).
Gene is spurious and the results probably reflect the partial deletion of the overlapping gene MON1.
The complete collection of growth data can be found at our yeast phenomics database, PROPHECY (http://prophecy.lundberg.gu.se). PROPHECY allows for visualization and quantitative evaluation of phenotypes with the simultaneous integration of information regarding functional class, protein–protein interaction, and subcellular localization.
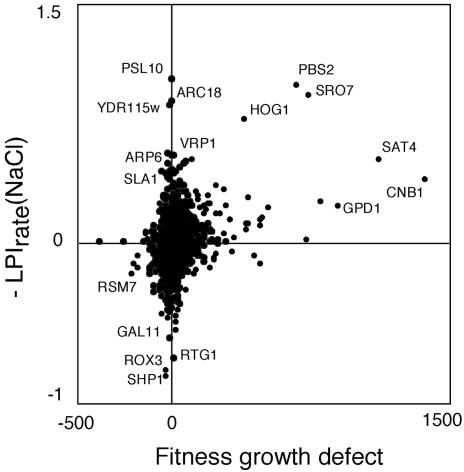
Correlation to Previously Reported Salt Growth Data. The data reported here on gene dispensability during saline growth were compared with data from a genomewide competitive fitness screen in salt utilizing the barcoding concept with corresponding microarray analysis (Fig. 2) (1). Of 57 nonauxotrophic strains scoring as significantly salt-sensitive in the barcoding/fitness experiment (fitness defects >100 in both samples at 15 generations), 31 were verified by our data as significantly salt-sensitive, using cultivation in isolation. All but three of these fitness defects were here scored as rate-of-growth phenotypes. Some genes known from earlier reports to be required for optimum growth in salt, such as HOG1, PBS2, CNB1, and SRO7, were found in both large-scale screens (Fig. 2), as were some functionally unclassified genes, such as YNR029c, YCR087w, and YGL046w. A majority of the remaining reported salt-fitness defects not scored in our screen corresponded to functionally uncharacterized ORFs. Most phenotypes identified by our microcultivation but not captured as significant by the barcoding approach were minor and presumably not detected because of the primarily qualitative nature of the fitness growth defect measure. However, some of the most severe rate phenotypes in our analysis, notably the growth defects of strains deleted for genes associated with the actin cytoskeleton (for instance ARC18, ARP6, VRP1, and SLA1) or well known salt responders such as the gimcomplex components (14) displayed reference strain behavior in the barcoding/fitness screen. Resistance phenotypes in fitness experiments have been proposed to primarily constitute artifacts (1). Nevertheless, 5 of 11 strains with significant increases in fitness, rsm7Δ, bem4Δ, ygr219wΔ, ylr261cΔ, and yor309cΔ, appeared as significantly salt-resistant in the screen reported here (all but ylr261cΔ as rate-resistant phenotypes), indicating that this assumption may be overly cautious.
Fig. 2.
Scatter plot of growth dispensability data (–LPIrate) of strains cultivated individually to strains cultivated in barcoding competition experiments [fitness growth defect after 15 generations; average of two samples (1)]. To allow direct comparison, resistant strains from the barcoding experiment were assigned a negative sign.
No correlation to fitness/barcoding data could be found for either adaptation time or efficiency of growth data (Fig. 8, which is published as supporting information on the PNAS web site). This was to be expected because the time of adaptation for most strains in a competition experiment is short compared with the period of steady-state growth and because most strains in that experimental setup never reach stationary phase. Thus, although the salt phenotypes of some heavily affected strains, such as sro7Δ, hog1Δ, and pbs2Δ, are composites of effects on more than one growth variable (Table 1 and Fig. 8), the majority of fitness defects captured by competition/fitness experiments are to be related to a slower rate of growth.
We captured 19 of the 25 deletion strains listed as hyperosmotically sensitive by MIPS based on earlier reports and present in the strain collection; 11 of these we identified as rate phenotypes (among our 210 strains with rate-of-growth defects), and the barcoding/fitness screen captured 6 [among their top 210 scores (1)].
Functional Dissection of Salt Phenotypes. The major physiological characteristics of gene dispensability during growth with salt were deduced from overrepresentation of known cellular functions (annotations and classes from MIPS, as of March 2003) among observed phenotypes. We found that saline stress affected gene deletions from a wide variety of cellular processes and thus constitutes a physiologically broad environmental stimulus (Figs. 3 and 4 and Table 1). Not surprisingly, genes implicated in yeast stress responses, a subgroup of the cell rescue/defense class, were significantly overrepresented within the rate of salt sensitivity category. Prominent in this group were the only nonredundant members of the yeast high osmolarity glycerol (hog) signal transduction pathway, HOG1 and PBS2. The strains deleted for these genes displayed some of the most severe rate- and efficiency-of-growth sensitivity phenotypes in our analysis, whereas no defects in adaptation time were observed (Table 1). Scored as rate-of-growth-sensitive but also displaying significantly increased adaptation time was the strain deleted for one of the prime final targets of the hog pathway, GPD1, which is crucial in the biosynthesis of the main yeast osmolyte glycerol (Fig. 2). Other than the production of glycerol, no major metabolic pathways were found to be nondispensable for saline growth. However, strains deleted for genes involved in the regulation of carbohydrate utilization, such as GRR1, a ubiquitin ligase implicated in glucose repression, were significantly overrepresented (α < 1e–4) as required for optimum saline rate of growth. We also observed that many of the genes with small but significant rate phenotypes in this functional subgroup, e.g., SWI3, GCR2, MIG1, and SSN8, are listed as transcriptional regulators, suggesting involvement in novel salt steady-state regulatory mechanisms.
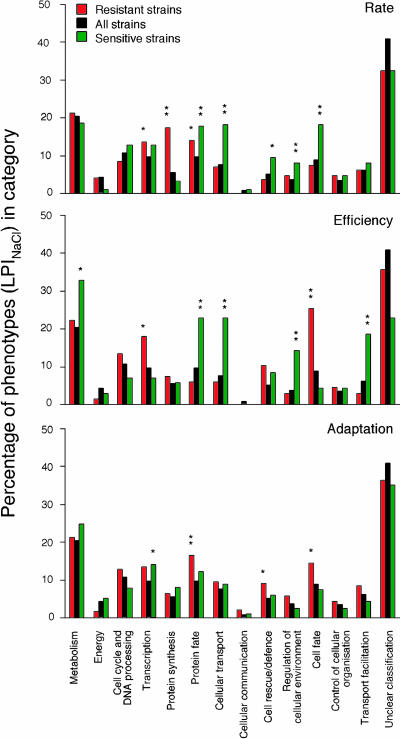
Fig. 3.
Distribution of genes in functional classes within a category of significant (α < 0.001) salt-specific phenotypes (LPI) compared to the distribution within the complete set of strains: rate resistance (271 strains), rate sensitivity (210 strains), stationary phase resistance (67 strains), stationary phase sensitivity (65 strains), adaptation time resistance (172 strains), and adaptation time sensitivity (285 strains). For lists of significant strains, see Supporting Materials and Methods. Functional classifications were taken from MIPS. *, Significant deviation in distribution (α < 0.025); **, highly significant deviation in distribution (α < 0.00125) from the complete set of strains.
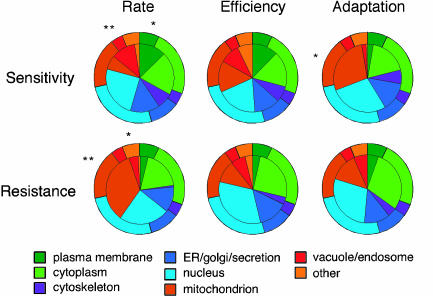
Fig. 4.
Subcellular localization of genes with significant (α < 0.001) salt-specific phenotypes. The outer pie chart indicates the subcellular distribution of all strains. The inner pie chart indicates the distribution of strains within the specified category (e.g., rate sensitivity). Subcellular distributions were taken from MIPS. *, Significant deviation in distribution (α < 0.025); **, highly significant deviation in distribution (α < 0.00125) from the complete set of strains.
Besides the osmotic effect, salt is toxic to yeast cells by disturbing ion homeostasis. Many genes implicated in ion transport, holding the functional classification cellular transport or transport facilitation and primarily localized to the plasma membrane, were enriched for rate- and efficiency-of-growth sensitivity in the screen (Figs. 3 and 4). Cation transporters, such as the potassium importer TRK1 (Table 1) and the Na+/H+ antiporter NHA1, accounted for the majority of proteins within these classes. Nondispensability of ion transporters was primarily manifested as efficiency-of-growth defects (Fig. 3), confirming the suggestion that higher maintenance costs from ion transport form a physiological basis of salt sensitivity (15). A surprising observation was the enrichment (α < 1e–7) of heavy metal transporters, such as the Cu2+ transporter PCA1 and the iron transporters FTR1 and FET3, within the category of mutants with a growth-efficiency defect.
Overrepresented in the rate-sensitivity category were genes implicated in the regulation of the cellular environment (Fig. 3). Most highly enriched of the subgroups within this class were genes regulating functions of importance in ion homeostasis. In yeast, Na+ export is primarily regulated by the calcineurin signal transduction pathway, dephosphorylating and thereby activating the zinc-finger transcription factor Crz1p, which in turn induces transcription of genes involved in Na+ extrusion (16). Strains deleted for CRZ1 or CNB1, the latter encoding the regulatory subunit of calcineurin, were among the most rate-sensitive strains in the screen. The phosphatase Ppz1p, proposed to perform functions overlapping the calcineurin pathway (16), was not required for optimum rate of growth in salt but was indispensable for a fully functional salt adaptation. Minor but significant (α < 0.001) rate phenotypes were also found for the regulatory subunits of the casein kinase II complex, CKB1 and CKB2, and for one of the subunits of the casein kinase I complex, YCK1, in agreement with a suggested role for these genes in the regulation of ion homeostasis (17).
Also, the cell fate class of genes was highly overrepresented within the rate-sensitivity category (Fig. 3). Many marginal salt phenotypes, e.g., for the clathrin light chain encoding gene CLC1 and the 14-3-3 gene BMH1, were found in the most overrepresented group of genes involved in regulating cell growth/morphogenesis. In addition, genes classified as involved in sporulation were surprisingly frequent in our screen for salt phenotypes; for instance, three of the four members of the RIM101 pathway, RIM101, RIM13, and RIM9, were found to have rate defects in salt, confirming indications linking this pathway to ion tolerance (18). Strains deleted for RIM15 or RIM16, with undefined but related roles in the regulation of sporulation, or for the kinase MCK1, which is suggested to act in parallel to the RIM101 pathway, were also detected as rate-sensitive.
We also report a high number of salt-resistance phenotypes (LPI > 0). The most striking salt-resistance feature in the screen was the significant overrepresentation of strains deleted for genes involved in protein synthesis (Fig. 3). This observation correlates well with the salt repression of ribosomal genes reported from expression profiling experiments (19). However, close examination revealed that strains deleted for protein synthesis genes were not salt-resistant per se. Rather, they were found to have growth defects in basal medium but displayed similar growth behavior as the reference strain during saline growth (see Fig. 1), suggesting that although undisturbed protein synthesis is essential during conditions of optimal growth in basal medium, this process is no longer rate limiting when cells proliferate at a lower rate because of environmental stress. Taking all three growth variables into account, salt-dependent loss of growth-limiting properties formed the basis for ≈70% of the salt-resistance phenotypes, among them the highly overrepresented class of proteins localized to the mitochondria (Fig. 4). Some of these effects may be due to specific osmotic remediability (20); however, the vast majority can be observed also to apply to nonosmotic environmental stresses (J.W., E.E., L.F., and A.B., unpublished work) and thus constitute general effects.
Revealing Salt-Sensitive Functional Modules. To test the hypothesis that deletion strains corresponding to physically interacting proteins also share similar patterns of growth behavior, we correlated the salt growth data reported herein to data from global protein–protein interaction studies (21–25). Indeed, within each of the three growth variables we found a significant (α < 0.00125) enrichment of interactions. This overrepresentation ranged from 2.3-fold for adaptation-sensitivity phenotypes to 8.5-fold for growth-efficiency phenotypes. For some of the functionally unknown genes this integrative approach provided functional clues. For instance, Yhl002wp interacts with and was required for optimal salt adaptation in a similar manner as Doa1p, supporting a role for this protein in ubiquitin-dependent proteolysis. Similarly, the strain deleted for Ypl068cp displayed similar rate resistance as and interacts physically with Ecm15p, indicating involvement in cell wall biogenesis.
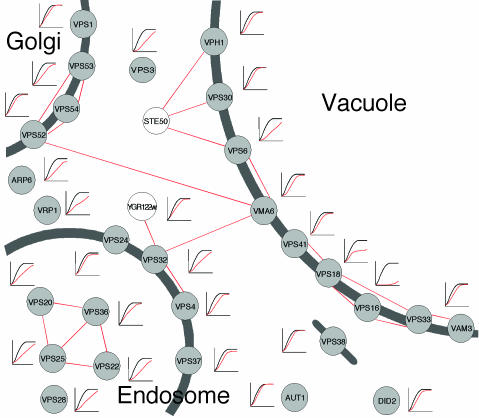
A high frequency of interactions was especially pronounced among the highly enriched (rate) class of nondispensable genes encoding proteins localized to the vacuole/endosome (Fig. 4). All but one of these vacuolar/endosomal proteins are classified as involved in the transport of proteins to the vacuole. Within this class of proteins, which constituted the most highly enriched (13-fold) subgroup in the screen, the reported physical interactions (21–25) were found to link the endosome, Golgi, and vacuole together in a salt-sensitive module (Fig. 5). For instance, Vps41p, Vps16p, and Vps18p have all been reported to interact physically, as have Vps52p, Vps54p, and Vma6p. Of particular interest was the interaction module of Vps20p, Vps25p, Vps22p, and Vps36p, because deletions of these largely uncharacterized proteins were found to have similar, severe growth defects in salt (Fig. 5), suggesting a common function in the targeting of proteins to the vacuole. Also interesting was the uncharacterized protein Ygr122wp, which has earlier been reported as physically linked to the VPS gene network via Vps32p. Vps32p was here found to be nondispensable in salt for optimum rate of growth as all of the other endosomal components (Fig. 5), providing evidence for a function of this gene in the vacuolar targeting of proteins.
Fig. 5.
Physical interactions, subcellular localization, and salt growth of genes involved in protein targeting to the vacuole. Red lines indicate reported physical interactions (21–25). Growth curves of genes exhibiting significant sensitivity (α < 0.001) to salt for at least one growth variable are displayed on logarithmic scale (OD = 0.05–5.0) over 47 h. Red curves indicate deletion strains and black curves indicate a representative reference strain. Gray circles represent genes earlier classified as involved in the targeting of proteins to the vacuole. Subcellular localizations were taken from the Saccharomyces Genome Database (http://genome-www.stanford.edu/Saccharomyces). Genes with unknown subcellular localization are shown as cytosolic.
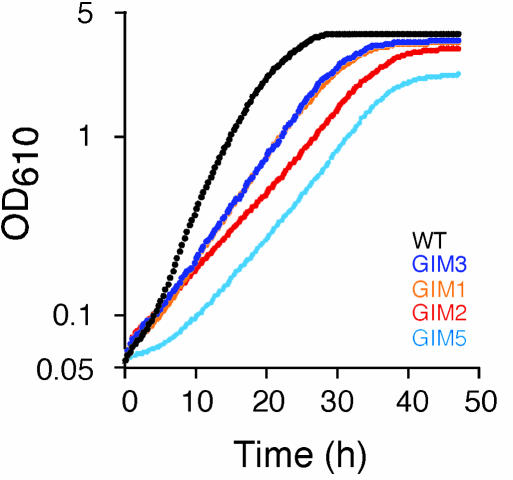
Significant functional overrepresentation within the rate sensitivity category was observed for genes involved in the determination of protein fate (Fig. 3). Some of the most severe salt phenotypes within this class were observed for strains deleted for four of the five components of the cytoskeleton-associated Gim complex, GIM1, GIM2, GIM3, and GIM5 [GIM4 was not found to be salt-sensitive, in agreement with earlier indications (14)]. Sequence analyses identify GIM1 and GIM3 as phylogenetically related (52% similarity), suggesting a possible functional overlap; gim1Δ and gim3Δ were found to have identical and minor growth defects in salt, distinct from the growth behavior of gim2Δ and gim5Δ, supporting this assumption (Fig. 6). The importance of the cytoskeleton in the salt response was also reflected in the salt sensitivity of many actin-associated components. Of particular interest was the high frequency of adaptation defects observed in this functional class, e.g., ARC18 and ARP5 (Table 1). A structural component of the actin cytoskeleton, the Arp2/3 complex, connects the previously mentioned Gim complex, the Rvs161/Rvs167 complex involved in actin polarization, and several genes previously only tentatively linked to the actin cytoskeleton, such as END5, SAC6, CHS5, and SAP155, into a network of synthetic lethality (26). Some of the most severe adaptation and rate defects in our screen were observed for strains deleted for genes in this network, such as arc18Δ, end5Δ, and rvs161Δ, whereas many others, such as sac6Δ, chs5Δ, and sap155Δ, displayed minor but significant salt growth defects. Defects in actin repolarization have been demonstrated to form the molecular basis of the rvs161Δ osmotic sensitivity (27), a gene deletion with a pronounced adaptation defect (Table 1), providing indications of the mechanisms underlying the adaptation time salt sensitivity of actin-associated genes.
Fig. 6.
Salt growth of strains deleted for genes in the gim complex as determined by 142 automated measurements of cell density. The salt sensitivity of gim4Δ was not significantly different from the wild type and is not displayed.
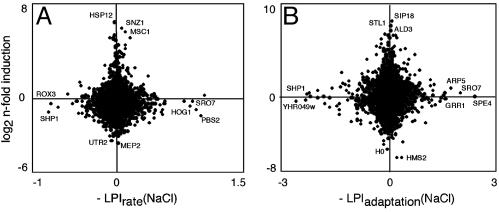
Gene Dispensability and Gene Regulation Are Not Correlated. Recent reports indicate no or a very limited qualitative overlap between expression data during the adaptation phase and gene dispensability experiments performed under steady-state salt growth (1, 28). In addition, steady-state salt gene expression was recently reported not to correlate to rate-of-growth salt phenotypes (H. Alipour, E.E., T. Ferea, P. Mostad, J. Norbeck, O.N., and A.B., unpublished work). We extend the comparison to also include expression data derived from salt shock adaptation experiments (19) to be compared with adaptation time phenotypes (Fig. 7). In agreement with earlier reports, we found no correlation at all for either steady-state growth (linear correlation, r2 = 0.0003) or adaptation-phase data (no correlation at 15, 30, 45, or 120 min; linear correlation at 60 min, r2 = 0.0004). There are ample extreme examples of this lack of correlation, such as SRO7 and STL1 (Fig. 7B).
Fig. 7.
Quantitative comparison of salt phenotypes (–LPI) displayed by strains cultivated in isolation to expression profiling data. (A) Steady-state rate of growth salt phenotype versus steady-state salt induction (average of five samples; H. Alipour, E.E., T. Ferea, P. Mostad, J. Norbeck, O.N., and A.B., unpublished work). (B) Adaptation-time salt phenotype versus adaptation-phase salt induction at 60 min [average of two samples (19)]. Correlation to adaptation-phase salt-induction data at 15, 30, 45, and 120 min was investigated in a similar manner (data not shown). No correlation was found.
Discussion
The tailor-made deletion strain collection of all genes in the yeast genome (1) allows standardized determination of the consequences of gene loss in this model system on an unprecedented scale. Quantitative assessment of gene dispensability may be achieved by cultivation of strains either in competition, relying on the estimation of relative fitness of barcoded deletion strains (1, 29), or individually, allowing the determination of absolute growth variables (12). Individual microcultivation of strains is here shown to allow the precise, quantitative assessment of gene dispensability and to resolve independent and physiologically relevant growth variables yielding information directly interpretable as gene by environment interactions. The importance of resolving different physiological features is clearly apparent for signal transduction components identified to be of importance because of prolonged time of salt adaptation (Ppz1p), slower rate of salt growth (casein kinase II, HOG, RIM101, and calcineurin pathways) and lower efficiency of salt growth (HOG pathway). Surprisingly, the HOG pathway that displays phosphorylation cascade activation during early salt adaptation (13) reveals its importance only during later phases of the growth cycle. Also, for downstream targets of these signaling pathways, functional importance in different physiological contexts was observed, where actin components and actin-associated complexes revealed salt importance mainly during adaptation, while deletions in ion transport systems displayed lower efficiency of salt growth. It is clear that high-quality descriptions of gene/protein functionality will require high-resolution phenotypic profiling that can resolve different aspects of cell life.
The extraction of functional information from phenotypic data of deletion strains is complicated by both functional redundancy for the deleted gene (30) and by the overall metabolic plasticity of the cellular machinery. This cellular robustness is reflected in both the ≈80% nonessential genes (1) as well as in the almost exclusively minor effects on growth that we report for the 4,711 viable yeast deletion strains (Fig. 9, which is published as supporting information on the PNAS web site). We found that marginal quantitative salt effects were physiologically highly relevant and can reveal relevant responses even for redundant components, making the strategy proposed here superior to qualitative phenotypic screens. Importantly, we identified in a single genomewide analysis all major cellular features known to be important for growth in the presence of salt, e.g., glycerol production, ion homeostasis, cytoskeleton organization, signaling pathways, and vacuolar protein transport. Despite the fact that salt stress constitutes one of the most well characterized cellular stimulus for yeast, we identified several cellular pathways not known to be required for optimal saline growth and extended the list of genes with functional importance during saline growth to include ≈500 genes. Some of these defects are expected to be mechanistically indirect in nature via links to primary osmoregulatory features, as exemplified by the salt sensitivity caused by improper transportation and incorporation of the sodium transporter Ena1p by aberrant vesicle docking in the SRO7 deletion (31) or by gene knockouts in other components of vesicle transportation (Fig. 5). High-resolution phenomics thus provides important information about the design of functional intragenomic networks.
Integrating phenotypic data with functional annotation, localization, protein–protein interaction, and synthetic lethality data, we found salt-important genes/proteins to form physiologically relevant functional networks. This allowed the tentative prediction of cellular roles for previously uncharacterized genes and put functionally known genes into new perspective, information that will be instrumental in systemwide descriptions of the cell machinery. However, we found no overlap between gene dispensability and gene-regulation data. Gene redundancy or posttranslational regulation may explain this important observation; however, it also raises the question as to whether gene transcript regulatory mechanisms are optimized to each and every condition by evolution to the extent generally believed.
Automated phenotypic profiling of strains cultivated individually thus allowed for exhaustive and physiologically precise screening of mutant collections. We suggest that it will constitute a valuable approach in the emerging field of phenomics, which is gaining momentum by the construction of genomewide deletion collections in other important model organisms such as C. elegans, Arabidopsis thaliana, and mouse.
Supplementary Material
Acknowledgments
Comments from colleagues have been instrumental during the course of this work and we would like, in particular, to express our gratitude to L. Adler, G. Bjursell, A. Farewell, S. Hohmann, M. Krantz, and P. Sunnerhagen for valuable comments and suggestions on the manuscript. This work was supported by the Swedish Foundation for Strategic Research (SSF).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: LPI, logarithmic (natural logarithm) phenotypic index; LSC, logarithmic (natural logarithm) strain coefficients; MIPS, Munich Information Center for Protein Sequences.
References
- 1.Giaever, G., Chu, A. M., Ni, L., Connelly, C., Riles, L., Veronneau, S., Dow, S., Lucau-Danila, A., Anderson, K., Andre, B., et al. (2002) Nature 418, 387–391. [DOI] [PubMed] [Google Scholar]
- 2.Kamath, R. S., Fraser, A. G., Dong, Y., Poulin, G., Durbin, R., Gotta, M., Kanapin, A., Le Bot, N., Moreno, S., Sohrmann, M., et al. (2003) Nature 421, 231–237. [DOI] [PubMed] [Google Scholar]
- 3.Gerlai, R. (2002) Trends Neurosci. 25, 506–509. [DOI] [PubMed] [Google Scholar]
- 4.Bennett, C. B., Lewis, L. K., Karthikeyan, G., Lobachev, K. S., Jin, Y. H., Sterling, J. F., Snipe, J. R. & Resnick, M. A. (2001) Nat. Genet. 29, 426–434. [DOI] [PubMed] [Google Scholar]
- 5.Birrell, G. W., Giaever, G., Chu, A. M., Davis, R. W. & Brown, J. M. (2001) Proc. Natl. Acad. Sci. USA 98, 12608–12613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fleming, J. A., Lightcap, E. S., Sadis, S., Thoroddsen, V., Bulawa, C. E. & Blackman, R. K. (2002) Proc. Natl. Acad. Sci. USA 99, 1461–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan, T. F., Carvalho, J., Riles, L. & Zheng, X. F. (2000) Proc. Natl. Acad. Sci. USA 97, 13227–13232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ross-Macdonald, P., Coelho, P. S., Roemer, T., Agarwal, S., Kumar, A., Jansen, R., Cheung, K. H., Sheehan, A., Symoniatis, D., Umansky, L., et al. (1999) Nature 402, 413–418. [DOI] [PubMed] [Google Scholar]
- 9.Que, Q. Q. & Winzeler, E. A. (2002) Funct. Integr. Genomics 2, 193–198. [DOI] [PubMed] [Google Scholar]
- 10.Ni, L. & Snyder, M. (2001) Mol. Biol. Cell 12, 2147–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thatcher, J. W., Shaw, J. M. & Dickinson, W. J. (1998) Proc. Natl. Acad. Sci. USA 95, 253–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warringer, J. & Blomberg, A. (2003) Yeast 20, 53–67. [DOI] [PubMed] [Google Scholar]
- 13.Hohmann, S. (2002) Microbiol. Mol. Biol. Rev. 66, 300–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geissler, S., Siegers, K. & Schiebel, E. (1998) EMBO J. 17, 952–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watson, T. G. (1970) J. Gen. Microbiol. 64, 91–99. [DOI] [PubMed] [Google Scholar]
- 16.Cyert, M. S. (2001) Annu. Rev. Genet. 35, 647–672. [DOI] [PubMed] [Google Scholar]
- 17.Glover, C. V., III (1998) Prog. Nucleic Acid Res. Mol. Biol. 59, 95–133. [DOI] [PubMed] [Google Scholar]
- 18.Lamb, T. M. & Mitchell, A. P. (2003) Mol. Cell. Biol. 23, 677–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Causton, H. C., Ren, B., Koh, S. S., Harbison, C. T., Kanin, E., Jennings, E. G., Lee, T. I., True, H. L., Lander, E. S. & Young, R. A. (2001) Mol. Biol. Cell 12, 323–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hawthorne, D. C. & Friis, J. (1964) Genetics 50, 829–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gavin, A. C., Bosche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., Schultz, J., Rick, J. M., Michon, A. M., Cruciat, C. M., et al. (2002) Nature 415, 141–147. [DOI] [PubMed] [Google Scholar]
- 22.Ho, Y., Gruhler, A., Heilbut, A., Bader, G. D., Moore, L., Adams, S. L., Millar, A., Taylor, P., Bennett, K., Boutilier, K., et al. (2002) Nature 415, 180–183. [DOI] [PubMed] [Google Scholar]
- 23.Ito, T., Tashiro, K., Muta, S., Ozawa, R., Chiba, T., Nishizawa, M., Yamamoto, K., Kuhara, S. & Sakaki, Y. (2000) Proc. Natl. Acad. Sci. USA 97, 1143–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito, T., Chiba, T., Ozawa, R., Yoshida, M., Hattori, M. & Sakaki, Y. (2001) Proc. Natl. Acad. Sci. USA 98, 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uetz, P., Giot, L., Cagney, G., Mansfield, T. A., Judson, R. S., Knight, J. R., Lockshon, D., Narayan, V., Srinivasan, M., Pochart, P., et al. (2000) Nature 403, 623–627. [DOI] [PubMed] [Google Scholar]
- 26.Tong, A. H., Evangelista, M., Parsons, A. B., Xu, H., Bader, G. D., Page, N., Robinson, M., Raghibizadeh, S., Hogue, C. W., Bussey, H., et al. (2001) Science 294, 2364–2368. [DOI] [PubMed] [Google Scholar]
- 27.Balguerie, A., Bagnat, M., Bonneu, M., Aigle, M. & Breton, A. M. (2002) Eukaryot. Cell 1, 1021–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Birrell, G. W., Brown, J. A., Wu, H. I., Giaever, G., Chu, A. M., Davis, R. W. & Brown, J. M. (2002) Proc. Natl. Acad. Sci. USA 99, 8778–8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giaever, G., Shoemaker, D. D., Jones, T. W., Liang, H., Winzeler, E. A., Astromoff, A. & Davis, R. W. (1999) Nat. Genet. 21, 278–283. [DOI] [PubMed] [Google Scholar]
- 30.Steinmetz, L. M., Sinha, H., Richards, D. R., Spiegelman, J. I., Oefner, P. J., McCusker, J. H. & Davis, R. W. (2002) Nature 416, 326–330. [DOI] [PubMed] [Google Scholar]
- 31.Wadskog, I. (2003) Ph.D. thesis (Göteborg Univ., Göteborg, Sweden), p. 63.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.