Abstract
Retrotransposons can facilitate repair of broken chromosomes, and therefore an important question is whether the host can activate retrotransposons in response to chromosomal lesions. Here we show that Ty1 elements, which are LTR-retrotransposons in Saccharomyces cerevisiae, are mobilized when DNA lesions are created by the loss of telomere function. Inactivation of telomerase in yeast results in progressive shortening of telomeric DNA, eventually triggering a DNA-damage checkpoint that arrests cells in G2/M. A fraction of cells, termed survivors, recover from arrest by forming alternative telomere structures. When telomerase is inactivated, Ty1 retrotransposition increases substantially in parallel with telomere erosion and then partially declines when survivors emerge. Retrotransposition is stimulated at the level of Ty1 cDNA synthesis, causing cDNA levels to increase 20-fold or more before survivors form. This response is elicited through a signaling pathway that includes Rad24, Rad17, and Rad9, three components of the DNA-damage checkpoint. Our findings indicate that Ty1 retrotransposons are activated as part of the cellular response to telomere dysfunction.
LTR retrotransposons are eukaryotic mobile elements in which a cDNA copy of the RNA genome is formed by reverse transcription and integrated into a new location in the host genome. The genome of the yeast Saccharomyces cerevisiae contains ≈30 copies of the LTR-retrotransposon Ty1, most of which are competent for transposition (1). Ty1 RNA is one of the most abundant mRNA species in yeast, but only one copy of Ty1 cDNA is present for every 10,000 Ty1 transcripts, and transposition is rare (reviewed in ref. 2). Over 30 host factors that inhibit retrotransposition have been identified (3–7). Many of these regulators are previously characterized proteins with conserved roles in genome maintenance, including telomerase (6).
The telomerase holoenzyme contains a reverse transcriptase, Est2, associated with its template RNA, Tlc1 (8–10). Telomerase polymerizes the addition of simple DNA repeats onto the ends of chromosomes. In yeast, telomeres consist of ≈300 bp of TG1–3/C1–3A repeats bound by telomere-specific binding proteins. This nucleoprotein complex protects the ends of chromosomes from being recognized as double-strand breaks, thereby preventing recombination between telomeres that leads to chromosomal fusions and instability (11, 12). In the absence of telomerase, telomeres shorten with every round of DNA replication, eventually reaching a critically short length that triggers arrest in the G2/M stage of the cell cycle (13–16). Although the majority of cells cease dividing after 50–100 generations, a fraction of cells escape arrest by forming alternative telomere structures containing either tandem arrays of the subtelomeric repeat, Y′ (type I survivors), or long and heterogeneous tracts of telomeric repeat DNA (type II survivors) (14, 17, 18).
The signaling pathway that triggers G2/M arrest in response to telomere shortening has recently been defined (15, 16). The “telomere checkpoint pathway,” which is a segment of the DNA-damage checkpoint pathway (reviewed in refs. 19, 20), includes sensor proteins Rad24, Rad17, Mec3, Ddc1, and the Mec1 kinase. A second arm of the DNA-damage checkpoint pathway that includes Mre11, Rad50, Xrs2, and the Tel1 kinase as sensors is not required for the telomere checkpoint. Interestingly, DNA-damage transducer proteins Rad9 and Rad53 are also dispensable for the telomere checkpoint; however, they are activated in response to telomere erosion and required for induction of a DNA damage-response gene (15, 16).
A common feature of transposons is their responsiveness to genetic and environmental stresses (21), yet little is known about the signals that activate transposition or the pathways through which they are transduced. Although it has been suggested that DNA lesions can trigger the activation of transposition, the idea has not been addressed directly by analyzing the effects of specific DNA lesions. Here, we demonstrate that telomere erosion triggers the mobilization of Ty1 via a DNA-damage signaling pathway. Ty1 elements are activated in parallel with progressive telomere shortening in est2Δ mutants and then partially deactivated in survivors. Thus, our results provide compelling evidence that DNA lesions can trigger the activation of Ty1 retrotransposition.
Materials and Methods
Strains. All strains used in this study are derivatives of the congenic strains BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) (22). Details of strain construction are provided in Supporting Text, which is published as supporting information on the PNAS web site.
Subculturing and Transposition Assays. Diploid strains homozygous for Ty1his3AI [Δ1]-3114 and heterozygous for est2Δ or est2Δ and rad9Δ, rad24Δ, rad50Δ, or rad17Δ were sporulated, and tetrads were dissected by standard genetic methods. Spores were grown on yeast extract/peptone/dextrose (YPD) agar for 2 days at 30°C. A small portion of each spore colony was removed and grown for phenotypic analysis and PCR analysis to determine the genotype of the spore, and the remaining portion was stored at 4°C.
To subculture strains by serial dilution in broth (Fig. 1), 1-ml YPD cultures were inoculated with a spore colony and grown overnight at 30°C. Cells were counted in a hemocytometer, diluted to 4 × 104 cells per ml in YPD broth and grown for 72 h at 20°C. Subsequent subcultures (SC1 through SC9) were inoculated at 4 × 104 cells per ml into variable quantities of YPD (50–600 ml, depending on the expected viability of the cells, which was determined in pilot experiments). Cells were removed from each subculture for counting in a hemocytometer, for plating on YPD and SC-His agar to determine cell viability (number of cells plated that formed a colony) and transposition frequency (His+ prototrophs per viable cell) and for extraction of genomic DNA. Two segregants of each genotype analyzed in this manner yielded consistent results. The results of one segregant of each genotype are shown in Fig. 1.
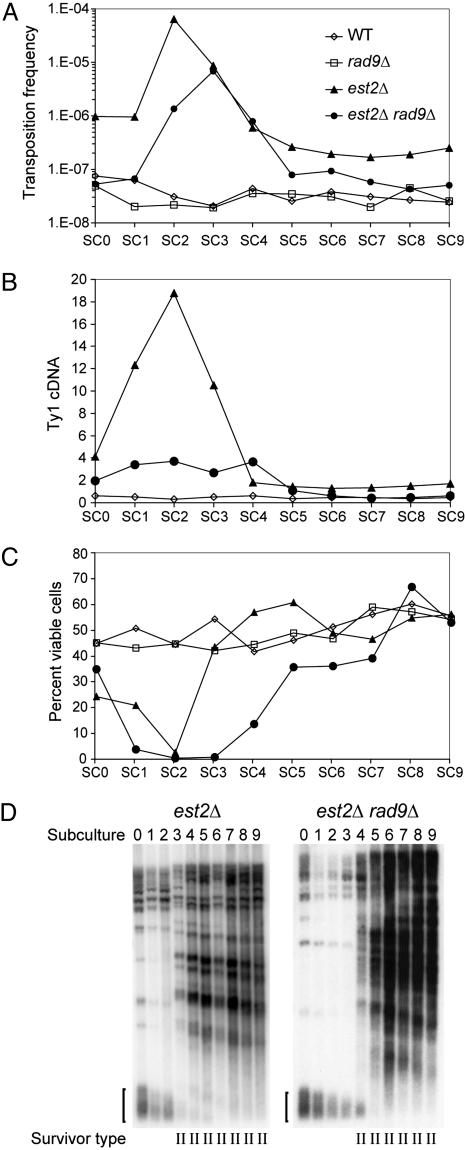
Fig. 1.
Ty1 retrotransposition and cDNA levels increase as telomeres erode and decrease when survivors form. Spore colonies of each of the genotypes indicated were subcultured by serial dilution in YPD broth at 20°C (SC0–SC9). SC0 is equivalent to ≈35 generations, and subsequent subcultures are equivalent to ≈15 generations. (A) Transposition frequency, the number of His+ prototrophs per viable cell, at each subculture. (B) The amount of Ty1 cDNA relative to genomic Ty1 element DNA at each subculture. (C) The percentage of cells that form colonies when plated on YPD agar at each subculture. (D) Southern blot of genomic DNA digested with XhoI and hybridized to a TG1–3/C1–3A DNA probe. The bracket indicates the terminal telomeric repeat tract of telomeres that contain Y′ elements, which have a conserved telomere-proximal XhoI site. Upper bands represent either telomeres lacking Y′ elements or subtelomeric DNA fragments with internal TG1–3 tracts. II, type II survivor.
Strains were subcultured on agar (Figs. 2, 3, 4) by serially restreaking spore colonies at 30°C. The spore colony (SC0) or a single colony from subculture SC1-SC4 was used to inoculate a 1-ml YPD culture, which was grown overnight at 30°C. Cultures were diluted 1:1,000 and grown for ≈72 h at 20°C. Cells were counted in a hemocytometer and then plated on YPD and SC-His agar to determine cell viability and transposition frequency, respectively.
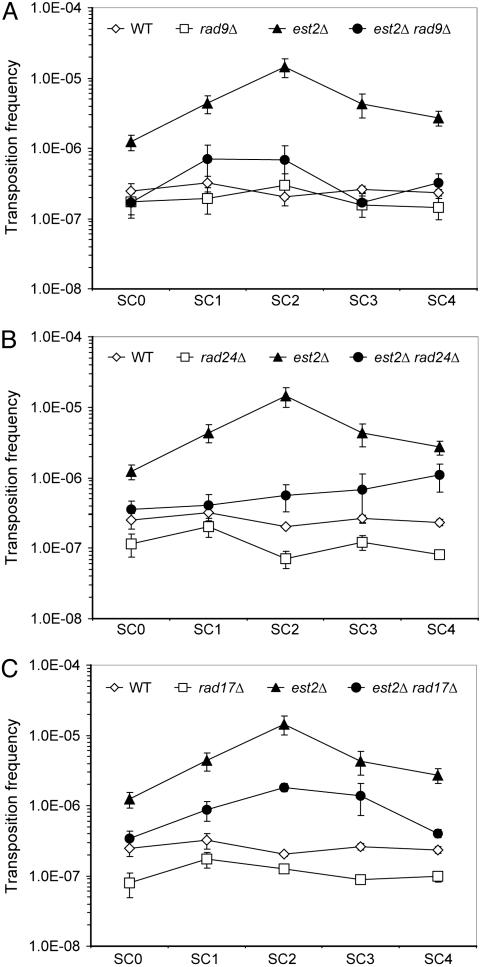
Fig. 2.
RAD9, RAD17, and RAD24 are required for the full activation of transposition during senescence of est2Δ mutants. Spore colonies were subcultured on agar at 30°C. SC0 is equivalent to ≈35 generations, and subsequent subcultures are equivalent to ≈20 generations. The average transposition frequency in three to seven segregants of each genotype at each subculture (SC0–SC4) is presented on a semilog graph. Data for wild-type and est2Δ segregants is the same in all three graphs. Error bars indicate the standard error.
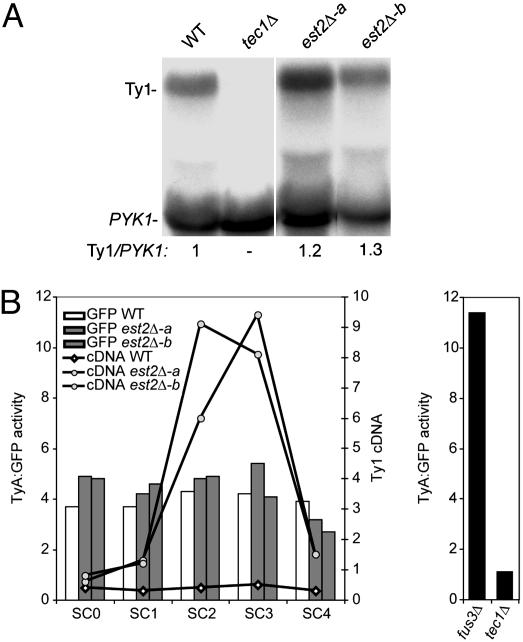
Fig. 3.
Ty1 RNA and TyA protein levels are not increased during senescence in est2Δ mutants. (A) Northern blot analysis of total RNA from one EST2 and two est2Δ segregants (-a and -b) at SC2 and a tec1Δ strain as a negative control. The blot was hybridized to Ty1 and PYK1 probes sequentially. The ratio of Ty1 RNA to the loading control, PYK1 RNA, is indicated below each lane. (b) Graphical representation of total Ty1 cDNA levels and TyA:GFP activities in each culture. Bars indicate the relative fluorescence (TyA:GFP activity) attributable to the Ty1-GFP-3566 element (Left) in SC0 through SC4 subcultures of EST2 and est2Δ segregants, as well as in single cultures of tec1Δ and fus3Δ segregants. Diamonds and circles indicate the levels of Ty1 cDNA relative to genomic Ty1 elements in each subculture (Right).
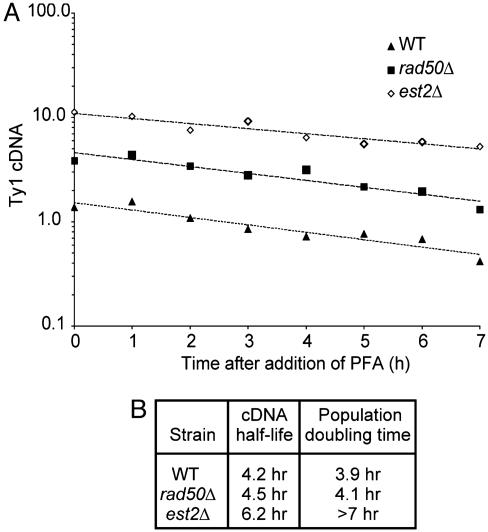
Fig. 4.
Ty1 cDNA half-life and population doubling time of SC2 cultures of wild-type (WT), rad50Δ, and est2Δ segregants. (A) The level of Ty1 cDNA relative to genomic Ty1 DNA against time after addition of the reverse transcriptase inhibitor, PFA. A best-fit line was generated for each data set. (B) Table showing the half-life of the Ty1 cDNA (determined from the slope of the best-fit line) and the population-doubling time of each culture.
Southern Blot Analyses. Southern blot analysis of Ty1 cDNA was performed as described (6). Details of quantifying Ty1 cDNA relative to Ty1 genomic DNA are provided in Supporting Text. The structure of telomeres was determined by Southern blot analysis using the method of Teng et al. (17). Telomeric C1–3A/TG1–3 repeat DNA was synthesized by PCR, using plasmid pCA75, a gift from V. Zakian (Department of Molecular Biology, Princeton University, Princeton), as a template, and labeled by random priming (23).
Northern Blot Analysis. Total RNA was extracted from cells in SC2 cultures grown at 20°C for 72 h by the hot phenol method (23). RNA was treated with glyoxal, subjected to electrophoresis in 1% agarose, and transferred to a nylon membrane. Blots were probed with 32P-labeled RNA generated by in vitro transcription of pGEM-TYA1 (24) and pGEM-PYK1 (25).
TyA:GFP Activity Assay. Congenic wild-type, est2Δ, fus3Δ, and tec1Δ segregants containing either the marked Ty1-GFP-3566 or unmarked Ty1–3566 element were obtained by tetrad dissection. Wild-type and est2Δ spore colonies were subcultured on YPD agar at 30°C, and SC0 through SC4 colonies and single colonies of fus3Δ and tec1Δ strains were used to seed YPD cultures, which were grown for ≈72 h at 20°C. Cultures were diluted in YPD broth to a density of 0.5 OD600 units and grown at 20°C to a density of 1.0 OD600 units. Cells were diluted in water to 0.03 OD600 units. Flow cytometry of 10,000 cells was carried out by using a FACScan (Becton Dickinson). To correct for differences in the average cell size among strains of different genotypes, the mean fluorescence per particle in each strain harboring Ty1-GFP-3566 was divided by the mean fluorescence per particle in a congenic strain harboring Ty1–3566.
Ty1 cDNA Stability. The half-life of Ty1 cDNA was measured essentially as described in ref. 26. The population doubling time was calculated from the number of cells per volume culture at every time point, as determined by counting cells in a hemocytometer. Details are provided in Supporting Text.
Results
Telomere Shortening Triggers the Activation of Ty1 Transposition in est2Δ Mutants. To investigate the possibility that telomere dys-function triggers the activation of Ty1 transposition, we determined whether transposition is activated in parallel with progressive telomere shortening in est2Δ mutants. To measure transposition, we used a yeast strain containing a chromosomal Ty1 element marked with the retrotransposition indicator gene, his3AI (27). Cells that sustain transposition of the marked element are detected as His+ prototrophs. This assay is specific for retrotransposition, because the formation of a functional HIS3 gene absolutely requires splicing and reverse transcription of the Ty1his3AI transcript.
A diploid strain heterozygous for est2Δ and homozygous for Ty1his3AI-3114 was sporulated to obtain congenic est2Δ Ty1his3AI-3114 and EST2 Ty1his3AI-3114 spores in which telomeres were wild type in length. Spore colonies were transferred to YPD broth (SC0) and grown at 20°C, which is a permissive temperature for retrotransposition, and then serially subcultured to allow progressive telomere shortening to occur in the est2Δ strains (SC1–SC9). In EST2 cells, levels of transposition (Fig. 1 A) and Ty1 cDNA (Fig. 1B) remained relatively constant throughout 10 serial subcultures (SC0–SC9, Fig. 1 A), as did cell viability (Fig. 1C) and telomere length (data not shown). In contrast, Ty1his3AI transposition in the est2Δ mutant was increased ≈14-fold in the original culture (SC0) and the first subculture (SC1), and >1,000-fold in SC2 (Fig. 1 A). Transposition decreased in SC3 through SC5, and then stabilized at levels ≈5-fold higher than in the EST2 strain. Correspondingly, the level of Ty1 cDNA relative to genomic DNA increased substantially from SC0 to SC2, reaching levels 38-fold higher than those in the EST2 strain (Fig. 1B). A substantial increase in cDNA as telomeres erode is seen consistently in analyses of est2Δ segregants (e.g., see Fig. 3B). The increase in cDNA relative to genomic DNA was not caused by degradation of genomic DNA (Fig. 6, which is published as supporting information on the PNAS web site). After SC2, Ty1 cDNA levels dropped quickly and then stabilized in SC5 through SC9. The stimulation and subsequent reduction of retrotransposition and cDNA were correlated with loss and recovery of cell viability, which reached its nadir at SC2 and then recovered to wild-type levels by SC5 (Fig. 1C). Similarly, activation of Ty1 transposition was linked to the progressive shortening of terminal telomeric repeats in est2Δ mutants from SC0 to SC2 (Fig. 1D). In SC3, when transposition levels began to decline, heterogeneous telomeric repeat tracts appeared, indicating the emergence of type II survivors (18). Therefore, Ty1 retrotransposition and Ty1 cDNA levels increased in parallel with telomere shortening and loss of viability in senescing est2Δ mutants, and subsequently declined when survivors form. Ty1 retrotransposition and cDNA levels remained modestly elevated in survivors, despite the recovery of cell viability to wild-type levels. These results, together with those of additional experiments in which transposition was assayed at shorter intervals of population growth (Fig. 7, which is published as supporting information on the PNAS web site), indicate that loss of telomere function, rather than the absence of telomerase, is the trigger for the activation of Ty1 in telomerase-negative mutants.
A Rad9-Dependent Signaling Pathway Activates Ty1 in est2Δ Mutants. The activation of transposition in response to a DNA lesion suggests that a DNA-damage signaling pathway may be involved in activating transposition. Rad9 is a central component of the DNA-damage checkpoint pathway. We compared levels of Ty1 retrotransposition and cDNA during senescence of an est2Δ rad9Δ spore to that of an est2Δ spore. Similar to the est2Δ mutant, Ty1his3AI retrotransposition in the est2Δ rad9Δ mutant increased progressively as viability dropped and telomeres shortened and then declined when survivors formed, indicating that deleting RAD9 does not abolish the transpositional response to telomere erosion (Fig. 1 A, C, and D). However, the peak transposition frequency of the est2Δ rad9Δ mutant at SC3 was only 11% of that of the est2Δ mutant at SC2 (Fig. 1 A), and Ty1 cDNA was reduced 7-fold (Fig. 1B). In contrast, transposition and cDNA levels were equivalent in a wild-type strain and a rad9Δ mutant. These findings indicate that the mobilization of Ty1 elements in response to senescence in est2Δ mutants partially requires Rad9, implicating the involvement of a DNA-damage signaling pathway.
DNA-Damage Checkpoint Proteins Required for Ty1 Activation. To further characterize the signaling pathway that triggers transposition, we performed epistasis analysis using mutant spores that were subcultured by serial restreaking on YPD agar. Single colonies were grown at 30°C, a nonpermissive temperature for transposition, and then shifted to 20°C to measure transposition. Shifting from the nonpermissive to permissive temperature facilitated analysis of multiple segregants of each genotype.
Seven EST2 Ty1his3AI-3114 and six est2Δ Ty1his3AI-3114 spore colonies were subcultured on agar, and the frequency of Ty1his3AI-3114 transposition in individual segregants of each genotype was averaged at each subculture. As seen in the previous experiment, the transposition frequency increased substantially between SC0 and SC2 in est2Δ mutants (Fig. 2 A). The increase in transposition was consistently lower than that of est2Δ segregants grown continuously at 20°C in the prior experiments (Figs. 1 A and 7), because the accumulation of His+ events is reduced when cultures are shifted from the nonpermissive to the permissive temperature. However, transposition was still elevated 75-fold, on average, in est2Δ SC2 cultures compared with EST2 SC2 cultures. (The highest frequency of transposition occurred at SC2 in all six est2Δ segregants.)
In three est2Δ rad9Δ mutants, the average transposition frequency at SC2 was 5% of that in est2Δ mutants (Fig. 2 A). In contrast, the average transposition frequency in rad9Δ mutants at all subcultures was 76% of that in wild-type segregants. Consistent with the findings of the liquid subculturing experiment (Fig. 1 A), these results indicate that Rad9 is required to stimulate transposition to high levels in response to telomere dysfunction.
Rad24 and Rad17 function as sensors in both the DNA damage and telomere checkpoint pathways; we therefore asked whether deletion of either gene blocked the activation of transposition during senescence. In six est2Δ rad24Δ mutants tested, the progressive increase in Ty1his3AI transposition that occurs in SC0 through SC2 of est2Δ mutants was largely abolished (Fig. 2B). The average transposition frequency in est2Δ rad24Δ segregants at SC2 was only 4% of that in est2Δ strains. Notably, transposition increased in SC3 and SC4 of est2Δ rad24Δ segregants, but this stimulation of transposition occurred after survivors formed (data not shown). In addition, the average transposition frequency in three est2Δ rad17Δ segregants at SC2 was reduced to 12% of that in the est2Δ segregants (Fig. 2C). In contrast, the average transposition frequency in rad24Δ and rad17Δ strains was 43% and 47% that in wild-type strains, respectively. A role for Rad24 in the activation of transposition in response to telomere erosion is further supported by the finding that deleting RAD24 significantly reduced the level of Ty1 cDNA in est2Δ mutants during senescence (Fig. 8, which is published as supporting information on the PNAS web site). Overall, these data indicate that Rad9, Rad24 and Rad17 are components of the signaling pathway that mobilizes Ty1 elements in response to telomere erosion. Although the absence of Rad24, Rad17, or Rad9 alone did not completely abrogate the function of the Ty1 activation pathway, partial loss of function in the absence of Rad24 or Rad9 is also observed for the DNA-damage checkpoint and telomere checkpoint pathways (16, 28, 29).
Rad50 functions as a sensor in the DNA damage pathway but not the telomere checkpoint pathway. Therefore, we hypothesized that Rad50 is not required for the activation of Ty1 by dysfunctional telomeres. Ty1 transposition is elevated in rad50Δ mutants (5), which could complicate the interpretation of epistasis analysis with the est2Δ mutation. Therefore, we analyzed est2Δ and est2Δ rad50Δ mutants at the peak of senescence, when the effect of an est2Δ mutation on transposition should be substantially greater than that of a rad50Δ mutation. The average transposition frequency in nine est2Δ rad50Δ mutants subcultured to SC2 was 1.6-fold higher than that in nine est2Δ segregants at SC2 (Table 1, which is published as supporting information on the PNAS web site). Thus, deletion of RAD50 does not reduce transposition in an est2Δ mutant, but may increase it slightly. Moreover, the average transposition frequency in est2Δ rad50Δ segregants was 10-fold higher than that of four rad50Δ mutants (Table 1). Therefore, Rad50 is not required for activating transposition in response to telomere erosion in est2Δ mutants, which is consistent with activation occurring via the telomere checkpoint pathway.
Telomere Erosion Triggers an Increase in Ty1 cDNA. To determine whether telomerase-negative mutants are affected at a posttranscriptional step of retrotransposition during senescence, two est2Δ segregants and one EST2 segregant were subcultured on YPD agar. The est2Δ segregants were senescent at SC2 and formed survivors at either SC3 or SC4 (data not shown). Northern analysis of RNA from SC2 cultures revealed that Ty1 RNA was increased 1.3-fold or less in the est2Δ mutants relative to the EST2 strain (Fig. 3A). Because small increases in RNA of endogenous Ty1 elements are correlated with proportional increases in transposition (30), the minor increase in Ty1 RNA levels cannot be the cause of the dramatic increase in transposition and cDNA in senescing est2Δ mutants.
Each spore analyzed in this experiment also harbored a chromosomal Ty1-GFP-3566 element, in which the ORF of green fluorescent protein (GFP) is fused to the TYA1 ORF. The Ty1-GFP-3566 element is responsive to characterized regulators of Ty1 expression, because TyA:GFP activity was diminished in a tec1Δ mutant (Fig. 3B), in which Ty1 transcription is reduced (31), and was increased in a fus3Δ mutant, in which TyA protein stability is enhanced (30). The TyA:GFP activity in SC0 through SC4 cultures of two est2Δ segregants was similar to that in the EST2 subcultures (Fig. 3B, bar graph). In contrast with TyA:GFP activity, Ty1 cDNA levels were elevated more than 20-fold in the est2Δ SC2 or SC3 cultures (Fig. 3B, line graph). We have also confirmed that total TyA levels are not increased in est2Δ mutants by Western analysis (data not shown). Taken together, these data demonstrate that Ty1 cDNA is increased at a posttranslational level in senescing est2Δ mutants.
Ty1 cDNA Synthesis Is Stimulated in est2Δ Mutants. To determine whether Ty1 cDNA is stabilized by the Ty1 activation pathway that is triggered by telomere erosion, we measured the half-life of Ty1 cDNA after treatment of cells with the reverse transcriptase inhibitor, phosphonoformic acid (PFA) (26). We included rad50Δ mutants, which also exhibit a posttranslational increase in transposition (5), in our analysis because of their intermediate transposition phenotype. Cultures of wild-type, rad50Δ, and est2Δ cells at SC2 were treated with 200 μg/ml PFA, and then cellular DNA was extracted immediately or from 1 to 7 h afterward. The level of unintegrated Ty1 cDNA relative to genomic Ty1 DNA at each time point was determined by Southern blotting, and the half-life was determined from a best-fit line of the cDNA levels plotted against time (Fig. 4A).
The half-life of Ty1 cDNA was 4.2 h in the wild-type strain, 4.5 h in the rad50Δ mutant, and 6.2 h in the est2Δ mutant. Because Ty1 cDNA appeared to decay at such a slow rate in all three strains, we measured the population-doubling time of each culture (Fig. 4B). Surprisingly, the population-doubling time was comparable to the respective Ty1 cDNA half-life in each strain. The same result was obtained by using a different wild-type strain (GRF167; data not shown). Hence, the apparent reduction in Ty1 cDNA relative to genomic Ty1 DNA in cells grown in PFA can be attributed to that fact that genomic DNA is replicating in doubling cells, whereas Ty1 cDNA is not. Hence, Ty1 cDNA is stable in wild-type yeast, and therefore, the elevated levels of Ty1 cDNA in est2Δ mutants cannot be caused by an increase in cDNA stability. These findings indicate that telomere erosion stimulates the synthesis of Ty1 cDNA.
Discussion
The mobility of retrotransposons is a double-edged sword for the host genome. Retrotransposons can inactivate genes, activate or alter the regulation of adjacent genes, patch together broken chromosome ends (32–34), and provide dispersed regions of homology that facilitate chromosomal rearrangements (35, 36). Although the mutations that they cause are often neutral or deleterious, retrotransposons impart evolutionary plasticity to the genome, thereby contributing to the ability of cells to adapt to new environments (37). One way of balancing the beneficial and deleterious effects of retrotransposons is for eukaryotic cells to link their mobilization to genomic insults, when their activity can promote repair or adaptive rearrangements (38). The results presented here provide strong evidence that Ty1 retrotransposons are activated as a specific response to DNA lesions.
In est2Δ spores segregated from a heterozygous est2Δ/EST2 diploid, retrotransposition of a chromosomal Ty1his3AI element and total Ty1 cDNA levels increased in parallel with progressive loss of telomeric repeats and escalating growth arrest, and then partially decreased when survivors formed and viability increased (Figs. 1 and 2). At the peak of senescence in est2Δ segregants, the frequency of Ty1his3AI transposition was increased as much as 75-fold when cells were subcultured at 30°C and then shifted to 20°C (Fig. 2), and several hundred-fold when cells were grown continuously at 20°C (Figs. 1 A and 7). Correspondingly, Ty1 cDNA levels were increased 20-fold or more when telomere erosion was most severe (Figs. 1B, 3B, and 8). These findings demonstrate that loss of telomerase is not directly responsible for the major increase in transposition, but that progressive loss of telomere function triggers the mobilization of Ty1 elements, just as it triggers activation of the telomere checkpoint.
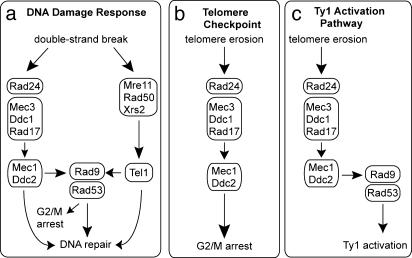
Rad24, Rad17, and Rad9, but not Rad50, are required for the full stimulation of Ty1 cDNA synthesis and transposition that is triggered by telomere dysfunction. The same requirements were found for growth arrest in response to telomere dysfunction, except that Rad9 played a minor role (15, 16). Therefore, we propose a model in which Ty1 is mobilized through the telomere checkpoint pathway, except that signaling through Rad9, and probably the Rad53 kinase with which Rad9 interacts, is critical for activation of transposition (Fig. 5). The significant reduction in transposition but not growth arrest in est2Δ rad9Δ mutants suggests that prolonging the G2/M phase of the cell cycle is not in itself sufficient to mobilize Ty1 elements to the full extent. Instead, Rad9/Rad53 might induce the expression or activity of a factor that promotes retrotransposition.
Fig. 5.
Model for the mobilization of Ty1 elements through a branch of the DNA-damage checkpoint pathway triggered by telomere erosion. This model is based on our findings and the established roles of components of the DNA-damage response and telomere checkpoint pathways (15, 16, 19, 20). (a) DNA damage can trigger arrest at the G2/M checkpoint and induction of DNA repair through either the Rad24/Mec1 arm or the MRX/Tel1 arm of the pathway. (b) The telomere checkpoint pathway utilizes some components of the DNA-damage checkpoint pathway to arrest growth. (c) Ty1 transposition is triggered by telomere erosion through the telomere checkpoint pathway, but, unlike G2/M arrest, activation of transposition requires Rad9 and probably Rad53.
Our data show that Ty1 RNA levels are not appreciably elevated in telomerase-negative mutants (Fig. 3A). Hence, it is not likely that the mobilization of Ty1 elements results from derepression of a putative subset of Ty1 elements that are normally silenced, because this would increase total Ty1 RNA levels. Likewise, the levels of a TyA:GFP fusion protein expressed from a chromosomal element were not significantly increased as telomeres eroded, indicating that the dramatic increase in Ty1 cDNA in est2Δ mutants occurs by a posttranslational mechanism. Several other hypertransposition mutants exhibit a posttranslational increase in Ty1 cDNA, which has been attributed to increased cDNA stability (7, 26). However, the half-life of Ty1 cDNA is at least 90 min in all wild-type strains tested to date, which is at odds with the idea that Ty1 cDNA is degraded. Moreover, by comparing the Ty1 cDNA half-life to the population doubling time, we found that Ty1 cDNA is not degraded in wild-type strains. Therefore, the increase in cDNA in est2Δ mutants cannot be due to increased cDNA stability. Our findings suggest that Ty1 cDNA is protected from nucleases, perhaps within cytoplasmic VLPs or within the preintegration complex.
Ty1 elements are activated at a transcriptional level by DNA damaging agents (39–41). In contrast, we found that telomere erosion activates transposition at the level of cDNA synthesis, clearly differentiating the response to telomere erosion from the response to mutagens. Interestingly, treatment of cells with DNA-damaging agents stimulates the environmental stress response (ESR), which involves changes in the expression of >900 genes (42). The ESR is also activated by a wide variety of environmental stresses, including heat shock and osmotic stress (43). In contrast, the transcriptional response to deletion of telomerase (TDR) involves only a subset of the ESR, and also includes the activation of a unique set of genes (44). Differences in the ESR and TDR might account for the induction of Ty1 element expression in response to DNA-damaging agents but not dysfunctional telomeres.
The activation of Ty1 in response to telomere dysfunction through a conserved DNA-damage signaling pathway suggests that increased Ty1 cDNA synthesis and retrotransposition are part of the cellular response to genome damage. The ability of Ty1 elements to mediate adaptively favorable mutations is consistent with this hypothesis (37). Remarkably, Ty1 elements have recently been shown to be associated with genomic “fragile” sites, where replication pauses and chromosomes are more likely to break (45). Hence, the increased levels of cDNA that are induced in response to chromosomal lesions may be used for recombination with chromosomes that are broken within fragile retrotransposon sequences, thereby promoting healing of double strand breaks by cDNA-mediated gene conversion. This use of cDNA would prevent ectopic recombination between chromosomal Ty1 elements, which can lead to potentially deleterious genome rearrangements.
Our findings show that the cellular response to telomere erosion provides a window of opportunity for retrotransposons to propagate in the host genome. The ubiquitous distribution of retrotransposons and the conserved nature of DNA-damage response pathways in eukaryotes hint at the possibility that this phenomenon is not confined to Saccharomyces.
Supplementary Material
Acknowledgments
This article is dedicated to our courageous colleague and friend Vicky Derbyshire. We thank the Wadsworth Center Molecular Genetics Core for oligonucleotide synthesis and the Immunology Core for flow cytometry; M. Bryk, D. Gottschling, V. Lundblad, and V. Zakian for plasmids; and K. Derbyshire, R. Morse, M. Bryk, D. Edgell, and P. Maxwell for critical review of the manuscript. This work was supported by National Institutes of Health Grant GM52072 (to M.J.C.).
Abbreviations: SC, subculture; YPD, yeast extract/peptone/dextrose; PFA, phosphonoformic acid.
References
- 1.Curcio, M. J. & Garfinkel, D. J. (1994) Genetics 136, 1245–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Voytas, D. F. & Boeke, J. D. (2001) in Mobile DNA II, eds. Craig, N., Craigie, R., Gellert, M. & Lambowitz, A. (Am. Soc. Microbiol., Washington, DC), pp. 631–662.
- 3.Conte, D., Barber, E., Banerjee, M., Garfinkel, D. J. & Curcio, M. J. (1998) Mol. Cell. Biol. 18, 2502–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee, B. S., Lichtenstein, C. P., Faiola, B., Rinckel, L. A., Wysock, W., Curcio, M. J. & Garfinkel, D. J. (1998) Genetics 148, 1743–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rattray, A. J., Shafer, B. K. & Garfinkel, D. J. (2000) Genetics 154, 543–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scholes, D. T., Banerjee, M., Bowen, B. & Curcio, M. J. (2001) Genetics 159, 1449–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sundararajan, A., Lee, B. S. & Garfinkel, D. J. (2003) Genetics 163, 55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singer, M. S. & Gottschling, D. E. (1994) Science 266, 404–409. [DOI] [PubMed] [Google Scholar]
- 9.Lingner, J., Hughes, T. R., Shevchenko, A., Mann, M., Lundblad, V. & Cech, T. R. (1997) Science 276, 561–567. [DOI] [PubMed] [Google Scholar]
- 10.Lendvay, T. S., Morris, D. K., Sah, J., Balasubramanian, B. & Lundblad, V. (1996) Genetics 144, 1399–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blackburn, E. H. (2000) Nat. Cell Biol. 408, 53–56. [Google Scholar]
- 12.Hackett, J. A., Feldser, D. M. & Greider, C. W. (2001) Cell 106, 275–286. [DOI] [PubMed] [Google Scholar]
- 13.Lundblad, V. & Szostak, J. W. (1989) Cell 57, 633–643. [DOI] [PubMed] [Google Scholar]
- 14.Lundblad, V. & Blackburn, E. H. (1993) Cell 73, 347–360. [DOI] [PubMed] [Google Scholar]
- 15.Enomoto, S., Glowczewski, L. & Berman, J. (2002) Mol. Biol. Cell 13, 2626–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ijpma, A. S. & Greider, C. W. (2003) Mol. Biol. Cell 14, 987–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teng, S. C. & Zakian, V. A. (1999) Mol. Cell. Biol. 19, 8083–8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teng, S. C., Chang, J., McCowan, B. & Zakian, V. A. (2000) Mol. Cell 6, 947–952. [DOI] [PubMed] [Google Scholar]
- 19.Zhou, B. B. & Elledge, S. J. (2000) Nature 408, 433–439. [DOI] [PubMed] [Google Scholar]
- 20.Rouse, J. & Jackson, S. P. (2002) Science 297, 547–551. [DOI] [PubMed] [Google Scholar]
- 21.Capy, P., Gasperi, G., Biemont, C. & Bazin, C. (2000) Heredity 85, 101–106. [DOI] [PubMed] [Google Scholar]
- 22.Brachmann, C. B., Davies, A., Cost, G. J., Caputo, E., Li, J., Hieter, P. & Boeke, J. D. (1998) Yeast 14, 115–132. [DOI] [PubMed] [Google Scholar]
- 23.Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A. & Struhl, K. (1993) Current Protocols in Molecular Biology (Wiley, New York).
- 24.Curcio, M. J., Hedge, A. M., Boeke, J. D. & Garfinkel, D. J. (1990) Mol. Gen. Genet. 220, 213–221. [DOI] [PubMed] [Google Scholar]
- 25.Curcio, M. J. & Garfinkel, D. J. (1992) Mol. Cell. Biol. 12, 2813–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, B. S., Bi, L., Garfinkel, D. J. & Bailis, A. M. (2000) Mol. Cell. Biol. 20, 2436–2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Curcio, M. J. & Garfinkel, D. J. (1991) Proc. Natl. Acad. Sci. USA 88, 936–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lydall, D. & Weinert, T. (1995) Science 270, 1488–1491. [DOI] [PubMed] [Google Scholar]
- 29.de la Torre-Ruiz, M. A., Green, C. M. & Lowndes, N. F. (1998) EMBO J. 17, 2687–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Conte, D. & Curcio, M. J. (2000) Mol. Microbiol. 35, 415–427. [DOI] [PubMed] [Google Scholar]
- 31.Laloux, I., Dubois, E., Dewerchin, M. & Jacobs, E. (1990) Mol. Cell. Biol. 10, 3541–3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore, J. K. & Haber, J. E. (1996) Nature 383, 644–646. [DOI] [PubMed] [Google Scholar]
- 33.Teng, S. C., Kim, B. & Gabriel, A. (1996) Nature 383, 641–644. [DOI] [PubMed] [Google Scholar]
- 34.Morrish, T. A., Gilbert, N., Myers, J. S., Vincent, B. J., Stamato, T. D., Taccioli, G. E., Batzer, M. A. & Moran, J. V. (2002) Nat. Genet. 31, 159–165. [DOI] [PubMed] [Google Scholar]
- 35.Dunham, M. J., Badrane, H., Ferea, T., Adams, J., Brown, P. O., Rosenzweig, F. & Botstein, D. (2002) Proc. Natl. Acad. Sci. USA 99, 16144–16149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Umezu, K., Hiraoka, M., Mori, M. & Maki, H. (2002) Genetics 160, 97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilke, C. M. & Adams, J. (1992) Genetics 131, 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McClintock, B. (1984) Science 226, 792–801. [DOI] [PubMed] [Google Scholar]
- 39.Staleva Staleva, L. & Venkov, P. (2001) Mutat. Res. 474, 93–103. [DOI] [PubMed] [Google Scholar]
- 40.Rolfe, M., Spanos, A. & Banks, G. (1986) Nature 319, 339–340. [Google Scholar]
- 41.Bradshaw, V. A. & McEntee, K. (1989) Mol. Gen. Genet. 218, 465–474. [DOI] [PubMed] [Google Scholar]
- 42.Gasch, A. P., Huang, M., Metzner, S., Botstein, D., Elledge, S. J. & Brown, P. O. (2001) Mol. Biol. Cell 12, 2987–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gasch, A. P., Spellman, P. T., Kao, C. M., Carmel-Harel, O., Eisen, M. B., Storz, G., Botstein, D. & Brown, P. O. (2000) Mol. Biol. Cell 11, 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nautiyal, S., DeRisi, J. L. & Blackburn, E. H. (2002) Proc. Natl. Acad. Sci. USA 99, 9316–9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cha, R. S. & Kleckner, N. (2002) Science 297, 602–606. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.