Abstract
Group II introns, widely believed to be the ancestors of nuclear pre-mRNA introns, are catalytic RNAs found in bacteria, archaea, and eukaryotes. They are mobile genetic elements that move via an RNA intermediate. They retrohome to intronless alleles and retrotranspose to ectopic sites, aided by an intron-encoded protein with reverse transcriptase, maturase, and endonuclease activities. Many group II introns identified in bacteria reside on plasmid genomes rather than bacterial chromosomes, implying that plasmids are havens for these retroelements. This study demonstrates that almost one-fourth of retrotransposition events of the Ll.LtrB intron in Lactococcus lactis are into the plasmid donor. This level is more than twice that predicted based on target size and plasmid copy number relative to the chromosome. In particular, the fraction of such plasmid targeting events was elevated to more than one-third of retrotransposition events by mutation of the intron-encoded endonuclease, a situation that may resemble most bacterial group II introns, which lack the endonuclease. Target-site sequences on the plasmid are more relaxed than those on the chromosome, likely accounting for preferred integration into plasmid replicons. Furthermore, the direction of integration relative to promoters and origins of replication is consistent with group II intron retrotransposition into single-stranded DNA at replication forks. This work provides mechanistic rationales for the prevalence of group II introns in natural plasmid populations and underscores that targeting to plasmids, which are themselves mobile elements, could promote intron spread.
The self-splicing group II intron Ll.LtrB from Lactococcus lactis is a mobile retroelement. Ll.LtrB is capable of both efficient retrohoming into its cognate intronless gene (1, 2) and low-frequency retrotransposition to ectopic sites (3, 4). Both processes require a ribonucleoprotein (RNP) complex consisting of the excised intron RNA and the intron-encoded protein (IEP), LtrA (Fig. 1A), which possesses RNA maturase, reverse transcriptase, and DNA endonuclease activities (5). The excised intron recognizes the DNA target sequences through pairings of the exon binding sites (EBS1, EBS2, and δ) on the RNA with the intron binding sites (IBS1, IBS2, and δ′) on the DNA and cleaves the target sites by maturase-assisted reverse splicing. The IEP cleaves the opposite strand and initiates reverse transcription of the intron RNA by using the resulting 3′-OH end as a primer, in a target-primed reverse transcription reaction (reviewed in ref. 6; see also Fig. 5B, pathway a). The similarities in mobility mechanisms and reverse transcriptase sequences suggest that group II introns are the progenitors of non-LTR retrotransposons, which are abundant in eukaryotic genomes (7, 8).
Fig. 1.
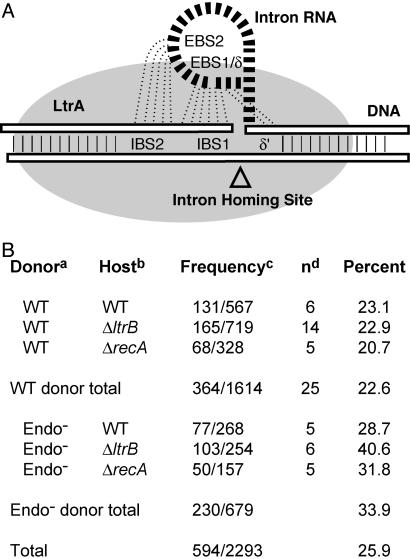
Retrotransposition into plasmid targets. (A) Schematic representation of target recognition by the RNP during retrohoming. Pairings between EBS1, EBS2, and δ of the intron RNA and IBS1, IBS2, and δ′ of the DNA target are shown. The IEP is depicted as a gray cloud. The sizes shown are not proportional to the molecular masses. (B) Frequency of retrotransposition into plasmid targets. a, WT = RIG. b, WT = NZ9800, ΔltrB = NZ9800ΔltrB, and ΔrecA = NZ9800ΔrecA. c, Plasmid events/total number of events scored. d, n = number of independent experiments.
Fig. 5.
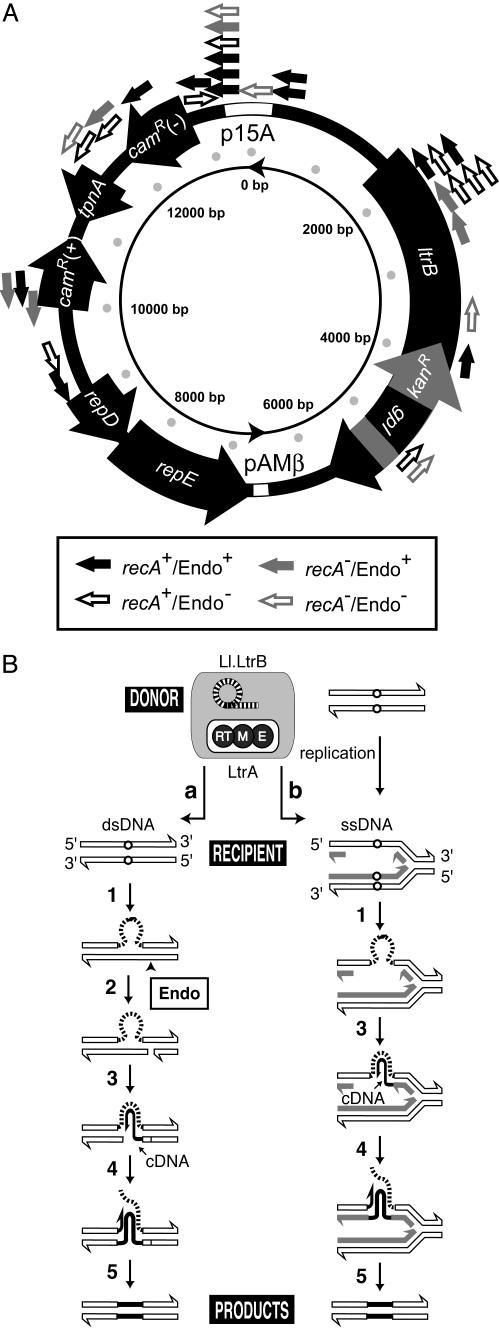
Plasmid integration sites give mechanistic insights. (A) Map of plasmid retrotransposition sites. Genes on the plasmid, ltrB, kanR interrupted by gpI (td intron), repD, repE, camR(+), and camR(–) (for Gram-negative camR), and truncated tnpA, are indicated as large arrows. The coordinates of the truncated ltrB gene are 1625–5894, group II intron 1803–5818, ltrA 2376–4185, kanR 4236–5550, group I intron 4559–4951. The small arrows outside the plasmid indicate positions and orientations of retrotransposition events identified in this study (Fig. 1B). Unidirectional plasmid replication begins at pAMβ ori and proceeds in the same direction as repE (inner circle). The recA status of the host and endonuclease (Endo) status of the plasmid for specific events are indicated by arrows as described in the rectangular box. (B) Pathways for retromobility. Retrohoming into dsDNA (2) (a) is contrasted with the ssDNA-targeted retrotransposition pathway at the replication fork as described in this and previous work (4) (b). Reaction steps are as follows: 1, reverse splicing into DNA target; 2, bottom-strand cleavage by LtrA endonuclease; 3, cDNA synthesis by LtrA reverse transcriptase; 4, removal of intron RNA and second-strand cDNA synthesis; 5, repair synthesis.
Whereas high-frequency retrohoming utilizes a double-stranded DNA (dsDNA) target and requires the endonuclease to cleave the second strand (2), rare retrotransposition in L. lactis is less dependent on endonuclease activity of LtrA and appears to frequently use single-stranded DNA (ssDNA) (4). Although the retrohoming site is recognized over ≈40 bp by base pairing with intron RNA and by protein contacts (Fig. 1 A) (5, 9, 10), retrotransposition has fewer base-pairing possibilities and protein contacts (3, 4).
Many group II introns reside in mobile elements such as transposons and plasmids, providing an additional means of intron dispersal (11–13). We report here that in addition to invading chromosomal sites, Ll.LtrB retrotransposes into many sites spread over the intron donor plasmid. Plasmid invasion is even less discriminatory than chromosomal retrotransposition and occurs more frequently than predicted based on target size. This work not only provides further mechanistic insights into group II intron retrotransposition in L. lactis but also gives clues about the preponderance of group II introns within plasmids in nature.
Materials and Methods
Retrotransposition Assay and Product Characterization. L. lactis NZ9800 and its derivatives, plasmids (pLERIG, pLERIG-Endo–), media and growth conditions, induction of the RIG-marked intron (Fig. 2A), and selection of KanR colonies were as described (4). These KanR colonies, routinely selected on plates containing 20 μg/ml kanamycin, were hybridized against a labeled 24-nt splice-junction probe to identify clones that had undergone retrotransposition (Fig. 2) (4). Hybridization and washing were carried out at 45°C. Plasmids isolated from high-intensity positive clones were transformed into Escherichia coli DH5α, tested for KanR, and sequenced.
Fig. 2.
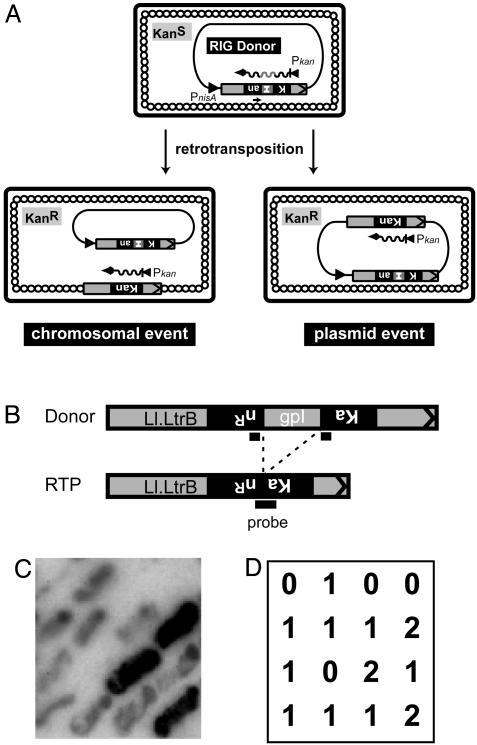
Selection and screening for plasmid retrotransposition events. (A) The pLERIG intron donor. The RIG donor contains the kanR (Kan) gene interrupted by an inverted group I intron (I). Retrotransposition can take place into either the host chromosome (Lower Left) or the donor plasmid itself (Lower Right). (B) Hybridization probe. KanR colonies were hybridized against a 24-nt kanR probe having 12 nt each of upstream and downstream sequences flanking the group I td intron (gpI) (4). Probe binding favors the joined kanR sequences in retrotransposition products over donor sequences with split of homology. (C) Representative result of hybridization. (D) Interpretation of hybridization signals in C. 0, no retrotransposition; 1, chromosomal retrotransposition; 2, plasmid retrotransposition.
Plasmid Copy Number Determination. Total cellular DNA was prepared from NZ9800ΔltrB cells containing both pLERIG (or pLERIG-Endo–) and a retrotransposed intron on its chromosome as described (4, 14). DNA was digested with EcoRI or KpnI, separated on a 0.8% agarose gel, and transferred onto a neutral Hybond membrane (Amersham Biosciences). EcoRI-digested DNA was hybridized against a 523-bp random primer [α-32P]dCTP-labeled Ll.LtrB-specific probe (probe A). KpnI-digested DNA was hybridized against a 5′-radiolabeled 28-nt probe (5′-CACGTGTTGCTTTGATTGATAGCCAAAA-3′; probe B). The probe B sequence is present once on the chromosome and once on the pLERIG backbone. Radioactivity in bands was measured on a PhosphorImager with imagequant (Molecular Dynamics), and plasmid copy number was deduced from radioactivity ratios, using sum-above-background calculations. The mass of the plasmid DNA was estimated to be 11.1% of the total DNA in a cell for pLERIG (13.5 kb × 24)/[(13.5 kb × 24) + 2,600 kb] and 12.7% for pLERIG-Endo– (13.5 kb × 28)/[(13.5 kb × 28) + 2,600 kb] for copy numbers of 24 and 28, respectively (see Results).
Results
Retrotransposition of the Ll.LtrB Group II Intron Occurs Frequently Into the Donor Plasmid. A group II intron donor with a retrotransposition indicator gene (RIG) was used (15), wherein a kanR reporter gene is inactivated by insertion of the inverted self-splicing group I td intron and placed in the antisense orientation in Ll.LtrB (Fig. 2 A) (4). This configuration places the group I intron in spliceable orientation in the Ll.LtrB transcript but not in the kanR transcript. Therefore, only after group I intron excision is an intact kanR gene generated within Ll.LtrB, reporting an RNA-mediated event. Reverse transcription and cDNA integration of the group II intron then allow direct selection of retrotransposition events via the KanR phenotype, without curing of the donor plasmid. In recent studies of retrotransposition into chromosomal sites of L. lactis NZ9800, we also observed intron invasion into the donor plasmid, pLERIG. This donor is a 13.5-kb CamR plasmid with the Ll.LtrB-RIG intron cartridge driven by the nisin promoter in L. lactis/E. coli shuttle vector pLE1 (Fig. 2 A) (4, 16).
When selected KanR colonies were probed with an oligonucleotide that discriminates the contiguous kanR gene from the RIG donor intron (Fig. 2 B–D), some positive clones gave a stronger signal than others in a variety of host and donor backgrounds (Figs. 1B and 2 C and D). The plasmids harvested from these strongly positive clones carried an additional copy of Ll.LtrB-RIG cassette, which had lost the td intron, indicating retrotransposition back into the plasmid.
A total of 2,293 retrotransposition events were scored in wild-type ΔltrB (lacking the chromosomal copy of the intron and some flanking exon sequence) and ΔrecA NZ9800 hosts with wild-type and endonuclease-defective pLERIG donors (Fig. 1B). With the wild-type donor, 22.6% of events were plasmid integrants, whereas with the Endo– donor, 33.9% of events were plasmid integrants. Notably, in the complete absence of endonuclease, in the ΔltrB host with the Endo– donor, 40.6% of events were into the plasmid, almost double the number when endonuclease is plentiful.
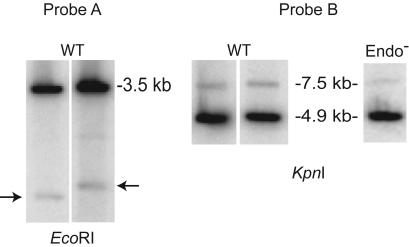
Plasmid Copy Number Relative to the Chromosome. Because plasmid integration seemed inordinately high, the plasmid copy number was determined relative to the chromosome by using two different methods. Total DNA from cells with different chromosomal retrotransposition events and pLERIG (4) was digested with EcoRI or KpnI. EcoRI-digested DNA was probed with a 32P-labeled fragment specific for a 523-bp region of Ll.LtrB present in both plasmid and chromosome (Fig. 3, probe A), whereas KpnI-digested DNA was probed with a 28-nt oligonucleotide specific for a sequence on the chromosome and on the pLERIG backbone (Fig. 3, probe B). Whereas probe A detects a plasmid fragment of 3.5 kb and variable chromosomal fragments depending on location of the chromosomal Ll.LtrB insertion, probe B detects two uniform bands in all cases (4.9 kb for plasmid and 7.5 kb for chromosome). For the wild-type donor, plasmid copy number relative to that of the chromosome was 23.6 ± 3.2 for probe A and 24.9 ± 8.4 for probe B. Based on a 13.5-kb plasmid, a 2,600-kb chromosome (17, 18), and a 24:1 plasmid:chromosome ratio, the total DNA mass of the plasmid is 11.1% of the total DNA in a cell. The fraction of retrotransposition events into the plasmid (22.6%) is therefore twice that predicted (11.1%).
Fig. 3.
Plasmid copy number determination. Probe A: Hybridization with an Ll.LtrB-specific probe. The chromosomal Ll.LtrB intron is indicated by arrows, whereas pLERIG is represented by the 3.5-kb band. Probe B: Hybridization with an oligonucleotide probe specific for chromosome and plasmid sequences. Genomic sequence corresponds to the 7.5-kb band, whereas pLERIG is represented by the 4.9-kb band. WT = pLERIG, and Endo– = pLERIG-Endo–. Although four strains with independent chromosomal Ll.LtrB intron locations were analyzed for WT, only two are shown in the figure, as pairs. Plasmid copy number was determined at least in duplicate for each strain and averaged as indicated in the text.
To ensure that the enhanced tendency toward plasmid retrotransposition when endonuclease is limiting is not due to an increased amount of plasmid, copy number was determined for the Endo– donor. This number was 28.0 ± 5.0, representing 12.7% of potential targets in a cell (Fig. 3, probe B). Therefore, the fraction of the plasmid events for the Endo– donor in a ΔltrB host (40.6%) is ≈3 times higher than predicted based on target size.
This bias toward plasmid events at 20 μg/ml kanamycin could be due to the multicopy nature of the plasmid providing better kanR expression than a single-copy insertion in the chromosome. However, selection under stringent conditions requiring more kanR expression, at 30, 40, 50, 60, 80, and 160 μg/ml kanamycin, did not increase the fraction of plasmid events (see Table 1, which is published as supporting information on the PNAS web site), arguing against this hypothesis. It is also formally possible that the use of hybridization for the selection introduced a bias, through preferential detection of multicopy events. However, control experiments to examine retrotransposition frequency without employing the probing method gave similar retrotransposition frequencies (data not shown), rendering such an explanation unlikely. We thus conclude that rather than representing some experimentally introduced biases, our results reflect a real propensity for retrotransposition into the donor plasmid.
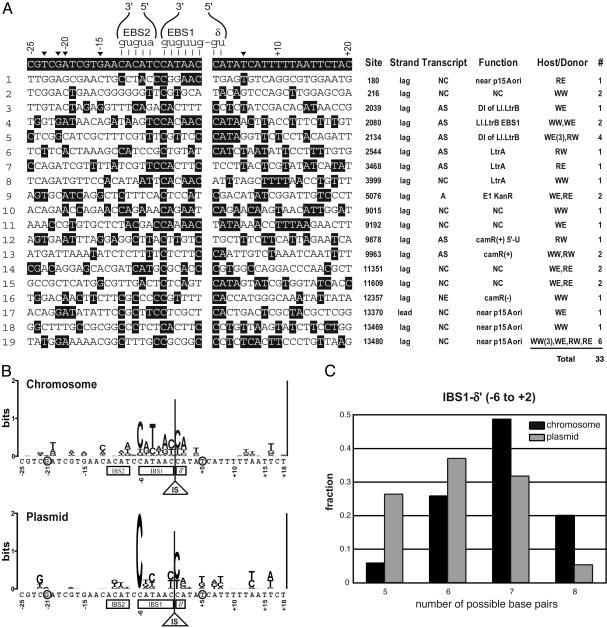
The Nature of Plasmid Retrotransposition Events. To gain insights into plasmid target preference and into the mechanism of retrotransposition, 33 independent plasmid retrotransposition events were sequenced and analyzed (Fig. 4A). Neither the distal 5′ nucleotides, which are recognized by LtrA to promote DNA unwinding during retrohoming (–23T, –21G, –20A, and –15G), nor +5T in the 3′ region, which is recognized by LtrA to promote the second strand cleavage (9, 10, 19), are conserved in either plasmid or chromosomal target sites (Fig. 4 A and B).
Fig. 4.
Retrotransposition sites. (A) Plasmid retrotransposition sites aligned with the homing-site sequence. (Left) Base-pairing interactions between the intron RNA (EBS1, EBS2, and δ) and target DNA (IBS1, IBS2, and δ′) during retrohoming are shown. Important residues for LtrA recognition in retrohoming, –23T, –21G, –20A, –15G, and +5T, are indicated by black arrowheads. (Right) Sites on the plasmid, strand (lag = lagging, lead = leading), transcript (S, sense; AS, antisense; NC, noncoding; NE, not expressed; A, ambiguous because of bidirectional transcription), gene function, host/donor genotypes, and number of independent events (#). Gene function: NC, noncoding; DI, ribozyme domain 1; E1, exon 1; camR(+), Gram-positive camR gene; 5′-U, 5′ untranslated. Host: W, wild-type NZ9800; R, recA–. Donor: W, wild-type pLERIG; E, pLERIG-Endo–.(B) Nucleotide conservation is represented by a Logos display (2 bits = 100%) (29). The conservation of the chromosomal sites is from ref. 4. (C) Statistical analysis of base-pairing potential of integration sites. Nucleotides that can base pair with the intron were counted for each site for the IBS1-δ′ region (–6 to +2). The number of sites having the indicated number of base-pairing possibilities was divided by the total number of events, as indicated in the text. G-T and U-G pairs were counted, although only nucleotides that are identical to the homing site are highlighted in A. The anticipated value for random sequence is 4.0 bp, because C and T are allowed at –6 to maintain base pairing; likewise, A and G at –5, C and T at –4, A and G at –2 and –3, C and T at –1, C and T at +1, and A and G at +2; thus, the number of base pairs would be (0.5 × 8 positions) = 4. Gray bars, plasmid; black bars, chromosome.
We next compared base pairing possibilities in IBS2, IBS1, and δ′ in plasmid and chromosomal sites. The IBS2 of plasmid and chromosomal target-site populations (positions –12 to –8) resembles random sequences (data not shown), suggesting that the IBS2–EBS2 contacts are of negligible importance for target recognition, in contrast with the importance of the IBS2–EBS2 interaction during retrohoming (9, 20, 21). On the other hand, IBS1 (–6 to –1) and δ′ (+1 and +2), which together comprise a continuous duplex with intron RNA, maintain significant base pairing possibilities. In a statistical analysis examining the number of base-pairing possibilities between the intron and different targets, the distributions of plasmid (19 sites) and chromosomal (47 sites; ref. 4) populations are significantly different in the IBS1–δ′ stretch (Fig. 4C). For this 8-nt stretch, the chromosomal population has a complementarity peak at 7 bp and the plasmid population has a peak at 6 bp (Fig. 4C), where the peak for random sequences is anticipated at 4 bp (legend of Fig. 4C). Whereas one-fourth of plasmid integration sites have 5-bp complementarity, this is so for only 5% of chromosomal sites. Moreover, one-fifth of chromosomal sites have a perfect match (8 bp), whereas only one of 19 plasmid sites (5%) has absolute complementarity. The differences in these distributions suggest that target selection for plasmid invasion is more relaxed than that for chromosomal invasion. Despite generally lower discrimination in targeting to IBS1 sequences, –6C, which is the most conserved among the chromosomal target sites, is absolutely conserved among the plasmid sites.
Notably, for retrotransposition into expressed regions on plasmids, the introns were inserted in the antisense orientation (7 of 7), as for chromosomal events (Figs. 4A and 5A). Additionally, almost all (18 of 19) retrotransposition sites are on the strand that serves as a template for lagging-strand DNA synthesis during plasmid replication of the unidirectional θ mode (22). Other noteworthy observations are that about one-half of the plasmid target sites are in nontranscribed regions (Figs. 4A and 5A), whereas almost all of the chromosomal sites are within genes, and that there is a retrotransposition hot-spot at plasmid residue 13480 and another at 2134.
It is striking that, whereas the intron was inserted within the donor intron in 10 of the plasmid events, no such intron-into-intron events were isolated on the chromosome of intron-containing hosts. It would therefore appear that the “recipient” Ll.LtrB sites need to be on the plasmid if they are to be frequently targeted.
Discussion
Retrotransposition of a group II intron in L. lactis resulted in a disproportionally high number of events into the plasmid donor, relative to the chromosome. A detailed characterization of integration sites and intron orientation reinforces the model that the group II intron retrotransposes preferentially into ssDNA molecules in its natural L. lactis host (4), and provides mechanistic rationales for the prevalence of group II introns in plasmids in natural populations.
Target Specificity Differences on Plasmid Relative to Chromosomal Sequences. Target site discrimination is somewhat lower for plasmid than for chromosomal sequences. This is particularly true for deoxynucleotides involved in base-pairing with EBS1 of the RNA, which are most stringently required for retrotransposition (Fig. 4 A–C) (3, 4). Although it is unknown why plasmid integration is less discriminatory, this somewhat relaxed specificity is likely an important factor in the preferential integration into the plasmid target.
Residue –6C in IBS1 is invariant among all of the plasmid integration sites (Fig. 4 A and B). In addition, positions +1 and +2 (δ′) are fairly well conserved. These residues are likely important for intron-target base-pairing to form a complex that can initiate reverse splicing (5). Regardless, this specificity at –6C and δ′ is consistent with the concept that the most discriminatory contacts are those that flank the intron insertion site, in accord with the group II RNP being active on intronless but not intron-containing alleles.
The Nature of Plasmid Integrants Favors a ssDNA Replication-Coupled Retrotransposition Pathway. Retrotransposition events are distributed around pLERIG, with cold-spots in the essential replication region (repD, repE, and pAMβ ori), and hot-spots at 13480, close to the E. coli plasmid origin (p15A ori), and at 2134, corresponding to a site in domain I of Ll.LtrB (Fig. 5A). Frequent integration at 13480 could be ascribed to the ability of the stretch from –6 to +1 to perfectly base pair with the intron (allowing G-U pairs) and/or to bases –23T, –21G, –20A, and +5T involved in IEP contacts being conserved. Alternatively, although the p15A ori is not functional for replication in L. lactis, a proximal DNA structure might facilitate integration in this region. This may also be the case for the hotspot at 2134, which has neither nucleotides favorable for protein contacts nor significant base-pairing possibilities.
The integration sites on the donor plasmid highlight several important points relating to the mechanism of retrotransposition. Although the strong bias toward the antitranscription orientation and nontranscribed regions (Fig. 4 A and C) may be due to selection for KanR events (4), they preclude the use of an RNA target (3) with the RIG selection system. These results indicate that retrotransposition occurred via reverse splicing into DNA, as for chromosomal events.
The bias toward retrotransposition into the template for the lagging-strand plasmid DNA synthesis is stronger than that for chromosomal events: all but one insertion site reside on the lagging template of the plasmid (Figs. 4A and 5A). Because retrotransposition into the leading-strand template generates a direct repeat of the intron, these products might have been counterselected via recombination-mediated deletion. However, such recombination events are rare (see, for example, ref. 23) and therefore unlikely to account for the observed lagging-strand bias. Instead, the bias may reflect the intron favoring invasion of ssDNA intermediates during plasmid replication, followed by reverse transcription initiated with the 3′ end of a nascent Okazaki fragment (Fig. 5B, pathway b) (4).
This model of replication-coupled integration into ssDNA during retrotransposition is further supported by the following two observations. First, those residues required for unwinding of duplex DNA (–23T, –21G, –20A, and –15G) (9, 10, 19) are not highly conserved in the plasmid targets. Second, the plasmid was preferred for retrotransposition in all host–donor combinations, and such plasmid events were enriched in the complete absence of endonuclease (Fig. 1B, ΔltrB, Endo– donor). Because the endonuclease is required for integration into dsDNA, the preponderance of events in its absence favors a ssDNA-targeted pathway (Fig. 5B), consistent with the intron's ability to reverse-splice into ssDNA in vitro (4).
The Propensity of Group II Introns to Reside in Plasmids. The bias toward retrotransposition into plasmids (Fig. 1B) is likely related to the report that at least 17 of the 36 documented full-length group II introns identified in bacteria reside on plasmids rather than chromosomes (13), despite the size differential of these replicons. Although the nature and mode of replication of the intron-containing plasmids are not known, group II intron distribution contrasts with that of bacterial group I introns, which invade dsDNA. No group I introns have been reported on natural plasmids, with almost all residing in chromosomes or bacteriophage genomes (24).
One should remain open to the possibility that plasmid-targeting reflects a cis preference, particularly because transcription and translation are coupled. Although this explanation has not been eliminated, it is not easily reconciled with the increased propensity to integrate into plasmids when endonuclease is absent (Fig. 1B). Interestingly, most bacterial group II introns lack the endonuclease domain (13). Therefore, especially in the absence of the second strand cleavage, the intron seems to prefer plasmid molecules, possibly also accounting for the prevalence of group II introns in natural plasmid populations. Regardless, the low target specificity described above would tend to allow promiscuous reverse splicing into plasmid genomes.
Accessibility of the replication fork to the intron RNP would also be an important factor for retrotransposition via the ssDNA-targeted pathway (Fig. 5, pathway b). Whereas chromosomal replisomes remain at the center of the cell and the replicated DNAs are released toward each cell pole (25, 26), plasmids lacking partitioning systems are distributed randomly in cytosolic spaces (27). Because pLERIG lacks a partitioning system, it is likely distributed and replicated in the cytosolic space, where the intron RNP would reside, rather than in the nucleoid. We therefore speculate that the single-stranded lagging-strand template on this plasmid is, as possibly also in some natural plasmids, more accessible than that on the chromosome, thereby favoring integration. Another possible explanation for plasmid preference is density of replication forks, given the greater number of forks per unit length of DNA on plasmids than on the chromosome.
The observation of lagging-strand bias provides hints on the mechanism of retrotransposition during conjugation, in which the lagging strand is synthesized in the recipient cell (28), and may explain the frequent residence of the group II intron in conjugative plasmids. It indeed seems advantageous for group II introns to reside on plasmids that are themselves mobile. Such plasmids can travel between different bacterial cells, thereby facilitating the spread of group II introns. The preferential targeting of plasmid replicons, especially conjugative plasmids, might be a consequence of the strategic choice of group II introns to survive in a bacterial world under pressure to minimize genome size.
Supplementary Material
Acknowledgments
We thank M. Gilson for sequencing pLEIItd+KR″, the parent of pLERIG, S. Lawrence for technical assistance, L. Conlan, C. Coros, J. Curcio, D. Edgell, A. Lambowitz, D. Mills, P. Perlman, and D. Smith for critical readings of the manuscript, M. Landthaler for helpful discussions, and M. Carl and J. Dansereau for help with the manuscript and figures, respectively. The Wadsworth Center Molecular Genetics Core Facility is acknowledged for DNA sequencing and oligonucleotide synthesis. This work was supported by National Institutes of Health Grants GM39422 and GM44844 (to M.B.).
Abbreviations: dsDNA, double-stranded DNA; EBS, exon binding site; IBS, intron binding site; RNP, ribonucleoprotein; ssDNA, single-stranded DNA.
References
- 1.Mills, D. A., Manias, D. A., McKay, L. L. & Dunny, G. M. (1997) J. Bacteriol. 179, 6107–6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cousineau, B., Smith, D., Lawrence-Cavanagh, S., Mueller, J. E., Yang, J., Mills, D., Manias, D., Dunny, G., Lambowitz, A. M. & Belfort, M. (1998) Cell 94, 451–462. [DOI] [PubMed] [Google Scholar]
- 3.Cousineau, B., Lawrence, S., Smith, D. & Belfort, M. (2000) Nature 404, 1018–1021, and correction (2001) 414, 84. [DOI] [PubMed] [Google Scholar]
- 4.Ichiyanagi, K., Beauregard, A., Lawrence, S., Smith, D., Cousineau, B. & Belfort, M. (2002) Mol. Microbiol. 46, 1259–1271. [DOI] [PubMed] [Google Scholar]
- 5.Matsuura, M., Saldanha, R., Ma, H., Wank, H., Yang, J., Mohr, G., Cavanagh, S., Dunny, G. M., Belfort, M. & Lambowitz, A. M. (1997) Genes Dev. 11, 2910–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belfort, M., Derbyshire, V., Cousineau, B. & Lambowitz, A. (2002) in Mobile DNA II, eds. Craig, N., Craigie, R., Gellert, M. & Lambowitz, A. (Am. Soc. Microbiol., Washington, DC), pp. 761–783.
- 7.Zimmerly, S., Guo, H., Perlman, P. S. & Lambowitz, A. M. (1995) Cell 82, 545–554. [DOI] [PubMed] [Google Scholar]
- 8.Eickbush, T. H. (1999) Curr. Biol. 9, R11–R14. [DOI] [PubMed] [Google Scholar]
- 9.Mohr, G., Smith, D., Belfort, M. & Lambowitz, A. M. (2000) Genes Dev. 14, 559–573. [PMC free article] [PubMed] [Google Scholar]
- 10.Singh, N. N. & Lambowitz, A. M. (2001) J. Mol. Biol. 309, 361–386. [DOI] [PubMed] [Google Scholar]
- 11.Michel, F. & Ferat, J.-L. (1995) Annu. Rev. Biochem. 64, 435–461. [DOI] [PubMed] [Google Scholar]
- 12.Martínez-Abarca, F. & Toro, N. (2000) Mol. Microbiol. 38, 917–926. [DOI] [PubMed] [Google Scholar]
- 13.Dai, L., Toor, N., Olson, R., Keeping, A. & Zimmerly, S. (2003) Nucleic Acids Res. 31, 424–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leenhouts, K. J., Kok, J. & Venema, G. (1991) Appl. Environ. Microbiol. 57, 2562–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Curcio, M. J. & Garfinkel, D. J. (1991) Proc. Natl. Acad. Sci. USA 88, 936–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mills, D. A., Choi, C. K., Dunny, G. M. & McKay, L. (1994) Appl. Environ. Microbiol. 60, 4413–4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Bourgeois, P., Lautier, M., van den Berghe, L., Gasson, M. J. & Ritzenthaler, P. (1995) J. Bacteriol. 177, 2840–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rauch, P. J. & de Vos, W. M. (1992) J. Bacteriol. 174, 1280–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong, J. & Lambowitz, A. M. (2003) EMBO J. 22, 4555–4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aizawa, Y., Ziang, Q., Lambowitz, A. M. & Pyle, A. M. (2003) Mol. Cell 11, 795–805. [DOI] [PubMed] [Google Scholar]
- 21.Zhong, J., Karberg, M. & Lambowitz, A. M. (2003) Nucleic Acids Res. 31, 1656–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bruand, C., Le Chatelier, E., Ehrlich, S. D. & Janniere, L. (1993) Proc. Natl. Acad. Sci. USA 90, 11668–11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lovett, S. T., Drapkin, P. T., Sutera, V. A., Jr., & Gluckman-Peskind, T. J. (1993) Genetics 135, 631–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edgell, D. R., Belfort, M. & Shub, D. A. (2000) J. Bacteriol. 182, 5281–5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lemon, K. P. & Grossman, A. D. (1998) Science 282, 1516–1519. [DOI] [PubMed] [Google Scholar]
- 26.Lemon, K. P. & Grossman, A. D. (2000) Mol. Cell 6, 1321–1330. [DOI] [PubMed] [Google Scholar]
- 27.Niki, H. & Hiraga, S. (1997) Cell 90, 951–957. [DOI] [PubMed] [Google Scholar]
- 28.Wilkins, B. & Lanka, E. (1993) in Bacterial Conjugation, ed. Clewell, D. B. (Plenum, New York), pp. 105–136.
- 29.Schneider, T. D. & Stephens, R. M. (1990) Nucleic Acids Res. 18, 6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.