Abstract
Functional interactions between factors bound at multiple sites on DNA often lead to a synergistic or more-than-additive transcriptional response. We previously defined a class of peptide sequences termed synergy control motifs (SC motifs) that function in multiple regulators by selectively inhibiting synergistic activity driven from multiple but not single response elements. By studying the prototypic SC motifs of the glucocorticoid receptor, we show that SC motifs inhibit transcription per se both in cis and in trans, and that a requirement for multiple contacts with DNA renders them selective for compound response elements. Notably, SC motifs are sites for SUMOylation, and the degree of modification correlates strongly with the extent of synergy control. Recruiting SUMO to the promoter either independently or as a fusion to the glucocorticoid receptor is sufficient to recapitulate the in trans and in cis inhibition by SC motifs without apparent changes in subcellular localization. Moreover, we find that the core ubiquitin fold domain of SUMO is sufficient for inhibition and that, independently of their potential for polySUMO chain formation, SUMO-2 and SUMO-3 are more effective inhibitors than SUMO-1.
Eukaryotic transcriptional control is highly combinatorial, and the mosaic of response elements in the regulatory elements of a given gene nucleates the assembly of multiprotein complexes where various forms of functional interactions take place (1). One of the most prevalent is the more-than-additive or synergistic response resulting from the recruitment of an activator to multiple copies of a recognition site (compound response element). Despite their importance, the mechanisms that control synergistic effects are poorly understood (2).
We have identified a short regulatory motif embedded in a number of sequence-specific regulators that is both necessary and sufficient to limit their transcriptional synergy (3). Disruption of these conserved synergy control (SC) motifs selectively enhances synergistic activation at compound response elements without altering the activity driven from a single site. Structure/function analysis of nine SC motifs from the glucocorticoid (GR), mineralocorticoid, and androgen receptors as well as from ETS-1 (3) and C/EBPα (4) revealed a common core sequence (I/V-K-X-E) and the presence of proline residues within 0–3 aa from either or both ends of the core. Such sequences occur in conserved regions of many factors and, in some cases, these regions have negative regulatory functions (e.g., SP3, SREBP, c-Myb, and C/EBPε). Moreover, a human mutation (P390S) in one of the SC motifs of the androgen receptor is associated with impaired spermatogenesis (5).
A clue to the critical role of the Lys residues in SC motifs came from the identification of the consensus motif (Ψ-K-x-E/D) for the posttranslational modification by SUMO. Conjugation of this ubiquitin-like protein follows an analogous pathway to that of ubiquitination and requires dedicated E1 activating (SAE1/SAE2) and E2 conjugating (UBC9) enzymes. UBC9 interacts directly with substrates to catalyze the formation of an isopeptide bond between the C terminus of SUMO and the amino group of the target lysine. This step is facilitated by a class of E3-like proteins known as SUMO ligases (6, 7). SUMOylation is reversible, and specific isopeptidases release the SUMO moiety (8). Three mammalian SUMO isoforms have been described (SUMO-1, -2, and -3) and, although their profile of target proteins seems to be different, little is known regarding their specificity of action. SUMOylation, however, does not share with ubiquitination a direct role in targeting proteins for proteasomal degradation.
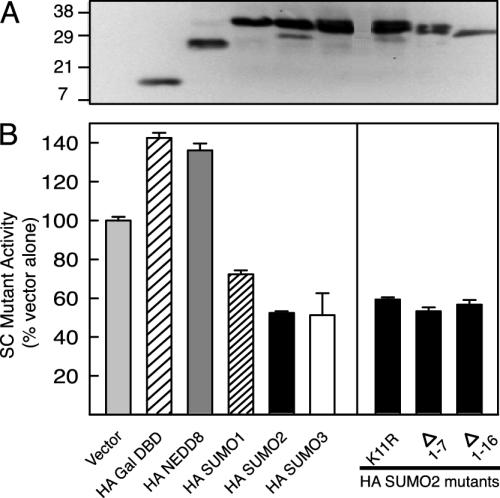
Mounting evidence indicates an important role of SUMO modification in transcriptional regulation. Several studies show that disruption of SUMO acceptor lysines in sequence-specific transcription factors results in enhanced activation (9–13). We recently demonstrated the presence of a functional SC motif in C/EBPα that functions as the main SUMOylation site. Mutation of the key Lys residue enhances transcriptional synergy and concurrently abolishes SUMOylation (4).
We initially discovered SC motifs in the N-terminal region of GR, a prototypic member of the nuclear receptor superfamily. On agonist binding to its C-terminal ligand-binding domain, GR translocates to the nucleus where a central zinc finger region tethers it to specific DNA sequences or glucocorticoid response elements (GREs) from which it regulates transcription. A potent activation function (AF-1) as well as determinants for repression are located in the N-terminal region of GR (14). The ligand-binding domain harbors a second activation function (AF-2) that operates as a ligand-dependent surface for interaction with coactivators (15). Recent work indicates that some steroid receptors, including GR, are targets of modification by SUMO-1, but this modification has been interpreted as having either positive (16) or negative (17) influence on transcription.
In this article, we have used GR as a paradigm to examine the mechanism of synergy inhibition by SC motifs and have revealed that individual SUMO isoforms play active and distinguishable roles in the control of transcriptional synergy.
Methods
Mammalian Expression Plasmids. Expression vectors for WT (p6R GR) and SC mutant (p6R GR K297R/K313R) rat GR are described in ref. 3. Exchanging an XmaI/PstI fragment containing the second SC motif in p6R GR K297R/K313R with fragments containing the indicated mutations in the second SC motif yielded all other mutants. The hemagglutinin (HA)-tagged version of WT (p6R HA GR) and SC mutant (p6R HA GR K297R/K313R) GR contain a 66-bp Acc65I/BamHI fragment encoding the amino acids MALEYPYDVPDYASRSGRGS followed by a Pro residue and GR residues 4 to 795. The Acc65I site precedes the above sequence, the HA epitope is italicized, and residues encoded by sites for XbaI and BamHI are underlined. The first 95 and 91 residues of SUMO-1 or SUMO-2, respectively, followed by residues NRLN were inserted into p6RHAGR K297R/K313R as XbaI/BamHI PCR fragments. pHAGal4 contains the same Acc65I/BamHI fragment described above followed by a BamHI/ApaI PCR product encoding the first 100 aa of Gal4 in pcDNA3.1(+). To generate SUMO GAL4 fusions, XbaI/BamHI PCR products containing SUMO-1 (1–95), SUMO-2 (1–91), or SUMO-3 (1–90) residues followed by NRLN linker amino acids or NEDD8 (1–74) residues were inserted at the same sites of pHAGal4. The point mutant and deletions of SUMO-2 were generated by PCR. pG4(SC)2 and pG4(SCmut)2 are pcDNA3 derivatives in which amino acids 289–326 of WT or K297R/K313R GR are fused directly downstream of Gal4 residues 1–100.
Reporter Plasmids. The pΔODLO reporter plasmid in which a minimal Drosophila distal alcohol dehydrogenase promoter (-33 to +55) drives the luciferase gene as well as its derivatives pΔTAT1-Luc and pΔTAT3-Luc that harbor one and three copies of a minimal GRE from the tyrosine aminotransferase (TAT) gene have been described (14). One or two Gal4 response elements (5′-CGGAGGACTGTCCTCCG-3′) were introduced upstream of the distal GRE in pΔTAT1-Luc and pΔTAT3-Luc to generate pΔ(Gal)1(TAT)1-luc, pΔ(Gal)2(TAT)1-luc, pΔ(Gal)1(TAT)3-luc, and pΔ(Gal)2(TAT)3-luc, respectively. The center-to-center distances between Gal4 response elements and between adjacent Gal4 and TAT response elements are 28 and 40 bp, respectively.
Cell Culture, Transfections, and Immunoblotting. Monkey CV-1 and COS-7 cells were maintained in DMEM (GIBCO-BRL) supplemented with 5% and 10% FBS, respectively. Cells were transfected by using Lipofectamine (Invitrogen). Unless indicated, cells received equimolar amounts of each type of expression plasmid to control for promoter effects. For functional assays, 3 × 104 cells (CV-1) were seeded into 24-well plates and transfected 24 h later with the indicated amounts of expression plasmids, 50 ng of reporter plasmid, and 50 ng of the control pCMVβgal plasmid. The total amount of DNA was supplemented to 0.3 μg per well with pBSKS(-). After 16 h, cells were exchanged into media containing 10 nM dexamethasone (Dex) or vehicle (0.1% ethanol). Cells were lysed 20 h later and luciferase and β-galactosidase activities were determined as described (14). Results represent the average ± SEM of at least three independent experiments performed in triplicate. Average values for WT GR (30 ng) in the presence of Dex from reporters containing a single or three GRE(s) were 1.3 ± 0.2 and 11.4 ± 0.8, respectively. For Western blotting, CV-1 cells (3 × 105) in 6-well plates were transfected with 3 μg of expression plasmids as above, lysed in 150 μl of 8 M urea SDS/PAGE sample buffer 36 h after transfection, and processed for immunoblotting as described below. For in vivo SUMOylation experiments, COS-7 cells (2 × 106) were seeded in charcoal-stripped medium onto 10-cm plates and transfected 24 h later with 5 μg each of the indicated receptor and HA-SUMO expression vectors. After 24 h, cells were treated with 100 nM Dex or vehicle (0.1% ethanol) and harvested 16 h later in 750 μl of buffer A (20 mM Hepes, pH 7.5/400 mM NaCl/5 mM EDTA/1 mM EGTA/5% glycerol/1% Nonidet P-40) containing 20 mM N-ethylmaleimide and protease inhibitors (Roche Diagnostics). This and subsequent steps were carried out at 4°C. Cleared lysates were treated with 40 mM DTT and immunoprecipitated (10 μl of ascites fluid for 60 min) with BuGR2 monoclonal antibody (18). Complexes were recovered (60 min) with 100 μl of 50% protein A-agarose in buffer B (buffer A containing 150 mM NaCl), washed three times with 1 ml of buffer B and resuspended in 50 μl of 2× SDS/PAGE sample buffer. Samples (15 μl) were resolved by SDS/7.5% PAGE and processed for immunoblotting with BuGR2 or HA-11 (Covance, Berkeley, CA) primary and anti-mouse IgG peroxidase conjugate (Bio-Rad) secondary antibodies. Images were captured in a Kodak Image Station 440 using Super Signal West Femto substrates (Pierce). All of the experiments were performed at least three times with similar results.
In Vitro SUMOylation Assays and Immunofluorescence. WT and SC mutant GR proteins were translated in vitro (SP6-TNT-Quick; Promega) in the presence of [35S]methionine and 1 μM Dex with 1 μg of pSP64T-N795 or pSP64T-N795 K297E/K313E plasmids as templates (3). SUMOylation reactions (4 μl of TNT lysate) were carried out in 20 mM Hepes (pH 7.9), 50 mM KCl, 10% glycerol, 5 mM MgCl2, 0.1 mM EDTA, and 5 mM DTT as described (4) except that the amounts of GST SAE2/SAE1 and GST-Ubc9 were 2.5 and 5 μg, respectively. For immunofluorescence, CV-1 cells were seeded onto glass coverslips in 6-well plates and transfected with 1 μg of the indicated constructs. After 20 h, cells were treated with vehicle or 10 nM Dex for an additional 1 h and fixed in 100% methanol. After incubation with HA-11 primary antibody, slides were rinsed in 1× PBS and incubated with goat anti-mouse Oregon Green-labeled secondary antibody (Molecular Probes) followed by a 20-min incubation in Hoechst 33258. Images were captured in a Zeiss inverted microscope using SPOT (Diagnostic Instruments, Sterling Heights, MI) camera and software.
Results
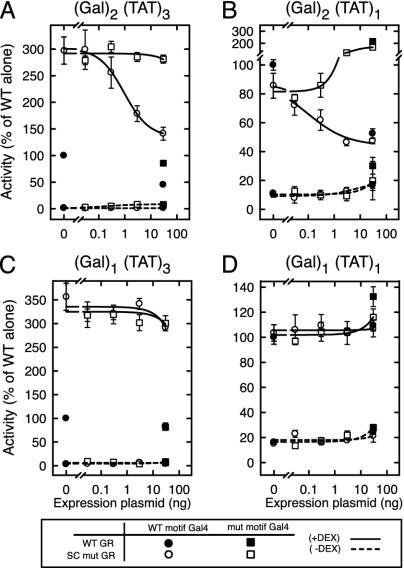
In Trans Inhibition by SC Motifs Requires Multiple Binding Sites. SC motifs are both necessary and sufficient to inhibit the synergy of the activator in which they are embedded (3). To examine whether the effect of SC motifs extends in trans to nearby bound factors, we fused a short region of GR (amino acids 289–326) encompassing both SC motifs to the Gal4 DNA-binding domain and tested its ability to inhibit GR activity at a promoter bearing two Gal4 sites upstream of three copies of a GRE from the TAT gene (Fig. 1A). As shown previously, disruption of the SC motifs in GR leads to loss of synergy control and a concomitant 3-fold enhancement of activity (leftmost open vs. filled symbols). Expression of a Gal4 DBD fusion to WT SC motifs (open circles) dose-dependently reduces the activity of SC mutant GR to levels approaching those of the WT GR. Mutations that disable the motifs eliminate this activity (open squares). The WT SC motif-Gal4 fusion also reduces WT GR activity to 50% (filled symbols). Inhibition is restricted to ligand-activated transcription because the Gal4 fusions had no effect in the absence of hormone. Western blotting confirmed expression of the fusions at comparable levels (data not shown). These results indicate that, although silent on their own, SC motifs are capable of inhibiting nearby regulators in trans.
Fig. 1.
SC motifs inhibit GR activity in trans. CV-1 cells were transfected with vectors for either WT (p6RGR) or SC mutant (p6RGR K297R/K313R) GR (30 ng) and the indicated amounts of vectors for Gal4 DBD fusions to either WT (pG4(SC)2) or mutant (pG4(SCmut)2) SC motifs of GR. The reporter plasmids used were pΔ(Gal)2(TAT)3-Luc (A), pΔ(Gal)2(TAT)1-Luc (B), pΔ(Gal)1(TAT)3-Luc (C), and pΔ(Gal)1(TAT)1-Luc (D).
The capacity of SC motifs to function in trans allowed us to dissociate the number of DNA sites contributing to activation from those contributing the SC motif function. At a reporter bearing two Gal4 sites upstream of a single GRE (Fig. 1B), agonist-induced activation by WT GR (leftmost filled symbol) is only one-tenth of that seen from three GREs (see Methods). As expected (3), mutations that disrupt the SC motifs in GR have no effect in this context (leftmost open symbol). The Gal4 fusion to the WT SC motif potently inhibited the activity of both the WT GR (single point filled circle) and SC mutant GR (open circles) by means of the two upstream Gal4 sites. In contrast, the Gal4 fusion to the mutant SC motifs failed to inhibit and instead led to an enhancement of GR activity, perhaps due to some residual activation potential of this fusion cooperating with the single GR site. These results indicate that SC motifs can inhibit activation emanating from a single nearby site. Interestingly, using reporters bearing only one Gal4 site revealed that the SC motif fusions are unable to inhibit WT or SC mutant GR activity emanating from either multiple GREs (Fig. 1C) or even a single GRE (Fig. 1D). These fusions can occupy the promoters because they effectively displaced a Gal4 VP16 fusion from the same sites (data not shown). Thus, regardless of the strength of the nearby activator, SC motifs inhibit activated transcription in trans only when recruited to multiple sites.
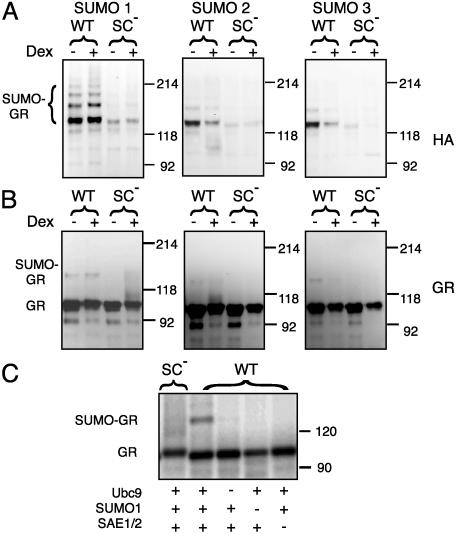
SC Motifs Are Sites for SUMO Modification. SC motifs can be viewed as a subset of the more general consensus site for posttranslational modification by SUMO, and Tian et al. (17) have shown that GR can be modified by SUMO-1. We therefore examined the ability of all three mammalian SUMO isoforms to modify GR in vivo. As seen in Fig. 2A, when WT GR is coexpressed with HA-SUMO-1, we detect a major HA-immunoreactive ≈147 kDa band in GR immunoprecipitates corresponding to mono-SUMOylated GR as well as a ladder of at least three additional multiply SUMOylated slower migrating forms. The 147-kDa band also is visible as a minor (4–6% of the unmodified GR) immunoreactive species (Fig. 2B). Similar results were obtained in the case of SUMO-2 and SUMO-3 except that forms above the diSUMOylated species are not detectable in the HA blot. Although the exact topology and composition of GR forms bearing multiple SUMO moieties remain to be defined, both SC motifs can be SUMO-modified (see below), and although SUMO-1 is thought not to form chains, it is possible that higher-order species contain endogenously expressed SUMO isoforms capable of forming chains. Agonist stimulation leads to a down-regulation of GR levels to ≈70% and a parallel reduction in the SUMO-2/3-modified forms. This effect is less pronounced in the case of SUMO-1, suggesting a modest enhancement of SUMO-1 modification by the agonist. Other doses or times of exposure to Dex yielded similar results (data not shown). The levels of SUMO conjugates were undetectable or severely reduced for the SC mutant form of GR (K297R/K313R), indicating that disruption of the SC motifs essentially eliminates the ability of GR to be modified by SUMO. A similar pattern of modification was observed for the Gal4 fusions used in Fig. 1 (data not shown). In a reconstituted SUMOylation reaction with purified recombinant components and in vitro-translated GR as substrate, we detected a slower migrating species corresponding to SUMO-1-modified GR only with the WT but not the SC mutant GR (Fig. 2C). Conjugation depended on the presence of SUMO-1 and the E1 (SAE2/SAE1) and E2 (Ubc9) activities. Similar results were obtained by using purified GST fusions to a small region (amino acids 289–326) encompassing both SC motifs (data not shown). Taken together, these results show that the SC motifs in GR are the main sites for conjugation by all three SUMO isoforms and suggest that this modification plays a key role in SC motif function.
Fig. 2.
The SC motifs in GR are sites for SUMO modification. COS-7 cells were transfected with vectors for WT (p6RGR) or SC mutant (p6RGR K297R/K313R) GR- and pCDNA3-based vectors for HA-tagged SUMO-1, -2, and -3. After a 16-h incubation with either vehicle or Dex, GR immunoprecipitates were resolved by SDS/PAGE and processed for immunoblotting by using anti-HA (A) or BuGR (B) antibodies. (C) In vitro SUMOylation reactions were performed as described in Methods by using in vitro-transcribed and -translated [35S]-labeled WT or SC mutant (K297E/K313E) GR as substrates in the presence of the indicated components.
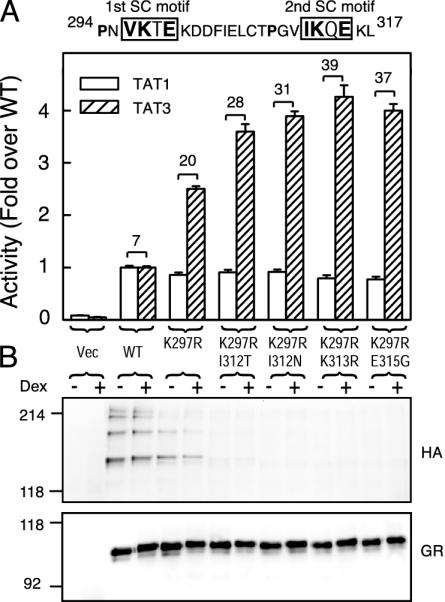
SC Motif Function Correlates with SUMO Modification. If SUMO modification is a prerequisite for SC motif function, then mutations that disrupt SUMO modification at positions other than the acceptor lysines are predicted also to affect synergy control. As can be seen in Fig. 3, replacement of lysine 297 in the first motif by arginine leads to a substantial reduction in the higher-order SUMO-modified forms. This mutation inactivates the first SC motif and leads to a partial loss of synergy control because it does not alter TAT1 activity but causes a 2.5-fold enhancement at TAT3. A similar mutation in the second motif yielded identical results (data not shown). In the context of an inactive first motif, replacing Ile-312 at the first position of the second motif by threonine or asparagine reduces GR SUMOylation to very low levels and causes a further loss of synergy control (TAT3/TAT1 = 28 and 31, respectively). Substitution with valine, which is a common residue at this position in SC motifs, alters neither SUMOylation nor synergy control (data not shown). Substitution of Glu-315 with glycine at the critical fourth position prevents SUMO modification and enhances synergistic activation to levels comparable to that of the K313R mutation. Similar results were obtained for SUMO-2 and -3 (data not shown). The positive correlation between SUMOylation and synergy control strongly implies that SUMO modification of SC motifs is required for their function.
Fig. 3.
SUMO modification correlates with extent of synergy control. (A) CV-1 cells were transfected with pΔTAT1-luc or pΔTAT3-luc reporters and 30 ng of expression vectors for either WT (p6RGR) or SC mutant forms of GR containing a K297R mutation in the first SC motif alone or in combination with the indicated mutations in the second motif. The number above the brackets represents the TAT3/TAT1 activity ratio. The sequence above represents GR amino acids 294–317 containing the two SC motifs. (B) COS-7 cells cotransfected with HA-SUMO-1 and the GR vectors used in A were processed as in Fig. 2.
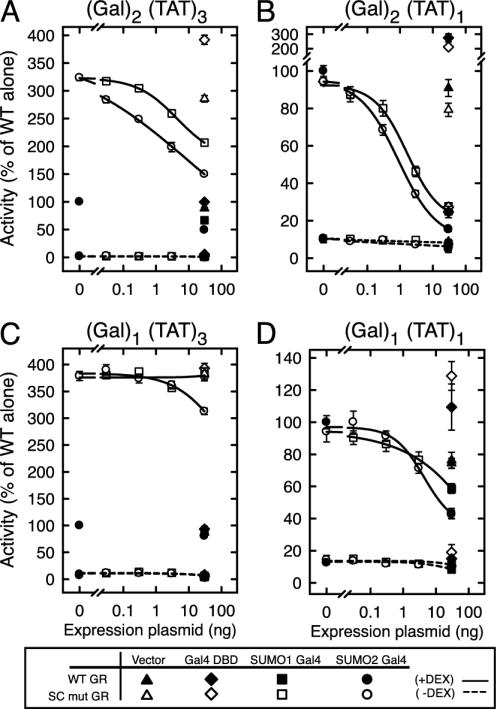
A Main Role of SC Motifs Is to Recruit SUMO to Promoters. To test more directly whether a main role of SC motifs is to recruit SUMO to the promoter, we generated noncleavable Gal4 DBD fusions to SUMO-1 or SUMO-2 and examined their ability to inhibit GR-activated transcription in the same promoter contexts examined in Fig. 1. In the (Gal)2(TAT)3 context (Fig. 4A), expression of fusions to SUMO-1 or SUMO-2 resulted in a dose-dependent inhibition of agonist-bound SC mutant GR. Notably, the SUMO-2 fusion was significantly more potent than SUMO-1 and inhibited activity to lower levels (50% vs. 70% of the SC mutant alone). Cotransfection of an empty vector or expression of the Gal4 DBD alone did not inhibit GR activity. This in trans inhibition also was observed for WT GR. Comparable expression of the fusions was confirmed by Western blot (data not shown and Fig. 5A). At the (Gal)2(TAT)1 promoter (Fig. 4B), recruitment of SUMO-1 or -2 caused a profound inhibition of both WT and SC mutant GR with the same order of potency as in Fig. 4A. Thus, like SC motifs, recruitment of SUMO to the promoter can inhibit activity emanating from a single nearby site. At promoters harboring a single Gal4 site, only the highest concentration of the SUMO-2 fusion weakly inhibited activity from three GREs (Fig. 4C). In contrast, both SUMO-1 and -2 can inhibit significantly the activity emanating from a single GRE (Fig. 4D). The stronger effects of the SUMO fusions compared to SC motifs may be attributable to a higher SUMO presence at the promoters, because only a small fraction of the SC motif fusions is SUMO-modified (≈4%; data not shown). As for the SC motif fusions, SUMO inhibited activated transcription much more effectively when bound to multiple sites. Taken together, these results show that SUMO is sufficient to recapitulate the in trans effects of SC motifs and that there is a strong dependence on recruitment to multiple sites on DNA to elicit their inhibitory effects.
Fig. 4.
SUMO inhibits GR activity in trans. CV-1 cells were transfected with expression vectors for WT (p6R GR) or SC mutant (p6RGR K297R/K313R) GR (30 ng) and the indicated amounts of empty (pCDNA3) or Gal4 DBD fusion expression vectors to either HA-tagged SUMO-1 or SUMO-2 or the HA epitope alone. The reporter plasmids were the same as in Fig. 1.
Fig. 5.
Specific inhibition by SUMO isoforms does not require their N-terminal region. (A) Expression of Gal4 DBD fusions in CV-1 cells was confirmed by immunoblotting for the HA epitope as described in Methods. (B) CV-1 cells were transfected with 30 ng of expression vectors for WT (p6RGR) or SC mutant (p6RGR K297R/K313R) GR and 30 ng of pCDNA3.1(+)-based vectors for the indicated Gal4 DBD fusions. The reporter was pΔ(Gal)2(TAT)3-Luc.
Specificity and Features of SUMO Involved in Inhibition. To explore the specificity of inhibition by SUMO, we compared the activity of Gal4 DBD fusions to all three SUMO isoforms and to NEDD8, a distantly related ubiquitin-like protein at the (Gal)2(TAT)3 reporter (Fig. 5B). In contrast to the inactive NEDD8 fusion, SUMO-2 and -3 inhibited SC mutant GR to comparable levels and, as shown above, SUMO-1 was consistently less effective. The basis for the difference between SUMO-1 and SUMO-2/3 may be related to the potential for the latter to form chains. SUMO-2 and -3 but not SUMO-1 contain a SUMOylation motif in their N-terminal region and SUMO chain formation at Lys-11 of SUMO-2 has been demonstrated in vitro (19). This region in the NMR structure of SUMO-1 is disordered (20). Mutating Lys-11 to Arg or deletion of half (Δ1–7) or the entire (Δ1–16) N-terminal region of SUMO-2, however, did not alter its ability to inhibit. Because all forms were expressed at relatively comparable levels (Fig. 5A), the results indicate that in trans inhibition is specific to the SUMO family and that the critical determinants map to the central ubiquitin fold core.
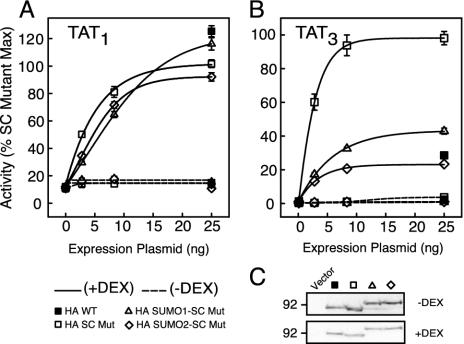
Colinear Fusion of SUMO to GR Inhibits Transcriptional Synergy Without Detectable Alterations in Subcellular Localization. The previous experiments indicate that SUMO mimics the in trans effects of SC motifs. To test whether SUMO recapitulates the function of SC motifs in cis, we compared the activity of WT to SC mutant forms of GR bearing noncleavable forms of SUMO-1 or SUMO-2. All forms displayed comparable activities from a single site (Fig. 6A) whereas at a promoter harboring three GREs, the SC mutant displayed its characteristic enhanced activity compared to WT GR. Notably, the unrestrained synergy of the SC mutant was essentially abrogated by fusing SUMO-2 to its N terminus and substantially reduced by the SUMO-1 fusion (Fig. 6B). The same fusions to WT GR also displayed a selective reduction of activity at TAT3 (data not shown), and all forms were expressed at comparable levels (Fig. 6C). Thus, consistent with the in trans effects, SUMO fusions complement the mutations in SC motifs, with SUMO-2 being more effective than SUMO-1.
Fig. 6.
Colinear fusion of SUMO inhibits GR synergy. CV-1 cells were transfected with the indicated amounts of p6R-based vectors for the expression of WT or SC mutant GR (K297R/K313R) fused to HA alone, HA-SUMO-1, or SUMO-2 and 100 ng of either pΔTAT1-Luc (A) or pΔTAT3-Luc (B) reporters. Average activity values for SC mutant GR (25 ng) in the presence of Dex were 4.5 ± 0.2 (A) and 78.1 ± 6.7 (B). (C) Expression of GR variants in CV-1 cells was confirmed by immunoblotting for the HA epitope as described in Methods.
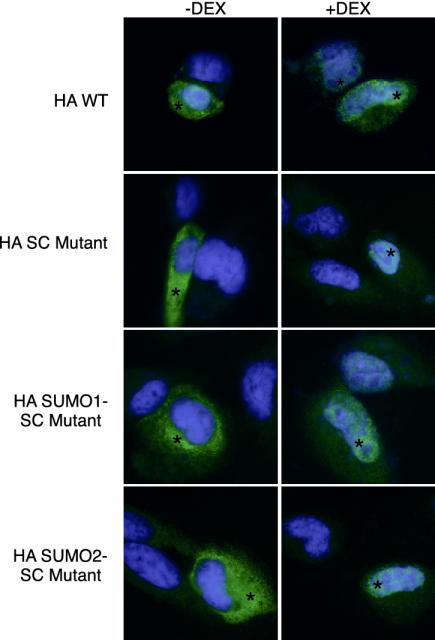
SUMO modification of certain proteins correlates with their partition to subnuclear domains such as PML bodies (21). We find, however, that there are no discernable differences in the ligand-dependent shift of WT and SC mutant GR from a cytoplasmic to a finely grained nuclear distribution (Fig. 7). Because the low stochiometry of modification (Fig. 2) could obscure the detection of SUMO-dependent alterations in localization, we examined the behavior of the SUMO fusions where the entire population of receptors bears the SUMO sequences. These forms, however, distribute similarly to the WT receptor (Fig. 7). Thus, appending SUMO to GR does not appear to target it to subnuclear domains discernible at this level of spatial resolution.
Fig. 7.
Disruption of SC motifs or colinear fusion to SUMO does not alter the subcellular localization of GR. CV-1 cells were transfected with the same expression plasmids as in Fig. 6. After 1 h of Dex (10 nM) or vehicle treatment, cells were fixed in 100% methanol and stained with anti-HA primary and Oregon Green-labeled secondary antibodies. After Hoechst 33258 (Molecular Probes) staining, slides were processed for immunofluorescence digital camera imaging as described in Methods. Representative fields are shown, and transfected cells are marked with an asterisk.
Discussion
SUMO Is Essential for SC Motif Function. Mechanisms that regulate transcriptional synergy are not well understood, yet they are likely to be essential for controlling patterns of gene expression, particularly during cell-specific function and development. We have characterized a class of regulatory motifs in multiple transcription factors that selectively inhibits synergistic activation. SC motifs overlap with the consensus sequence (Ψ-K-x-E/D) for posttranslational modification by SUMO proteins. Our data clearly show that the critical lysines in the SC motifs of GR are targets for SUMO modification and extend the SUMO-1 findings of Tian et al. (17) to include all three SUMO isoforms. Together with the findings that the functionally confirmed SC motifs in the androgen receptor (22) and C/EBPα (4) are also SUMO acceptor sites, these studies strongly implicate SUMO modification in the function of SC motifs. The list of transcription factors subject to SUMO modification is expanding rapidly and, for many, the modification site conforms to our definition of SC motifs. Disruption of acceptor lysine residues is associated with a concomitant enhancement in activity (4, 9–12, 17). Although the analysis has been mostly at compound response elements, it is likely that these constitute additional examples of synergy control.
We propose that the transcriptional effects of SC motifs are caused by recruitment of SUMO because the features in SC motifs required for function also are required for SUMO modification, and the degree of SUMOylation correlates with the extent of synergy control (Fig. 3). More directly, recruitment of SUMO to the promoter is sufficient to recapitulate the in trans inhibition of SC motifs (Fig. 4), and fusing SUMO in cis to a receptor lacking functional SC motifs inhibits synergistic activation without affecting activity from a single site (Fig. 6). Together with the recent results for transcription factors Sp3 (11) and p300 (23), our data clearly support a direct inhibitory function for SUMO. Although the relatively low stochiometry of SUMOylation we and others observe in vivo may reflect our inability to preserve this modification, it is also possible that SUMOylation is a transient licensing event that is sufficient to initiate, but not necessarily maintain, inhibition.
Notably, inhibition is specific to SUMO, as the structurally similar but divergent ubiquitin-like protein NEDD8 (24) is inactive (Fig. 5). Interestingly, both the SUMO in trans and in cis results indicate that SUMO-2/3 are more potent inhibitors than SUMO-1. This important functional difference between SUMO isoforms affords an additional layer of regulation and may contribute to the transcriptional response to stress during which SUMO-2/3 conjugation seems to be enhanced (25). Our mutational analysis of SUMO-2 suggests that the key inhibition/specificity features map to the surface of the ubiquitin-like core region. We are currently identifying such determinants. Although SUMO itself can account for the basic properties of SC motifs, competing lysine-directed modifications could play additional regulatory roles. Thus, although disruption of the SC motif in C/EBPα does not prevent ubiquitination (4), acetylation of SUMO acceptor sites has been described (26).
The Basis for Selective Inhibition at Multiple Sites. A defining feature of SC motifs is their selective inhibitory effects at compound but not single sites. Our further characterization shows that their effects are not a consequence of selective targeting of synergy per se because recruitment of SC motifs can counteract a nearby activator bound to either a single or multiple sites. However, in accord with their effects in cis, inhibition in trans also requires multiple contacts with the DNA. From our data, we extend our previous model (3) and propose that SUMO-modified SC motifs at compound response elements create a multivalent recognition surface for a general inhibitory factor(s). The strong dependence on multiple sites for SC motif function may be attributed to a relatively low affinity of the factor(s) for SUMO coupled to the substoichiometric SUMO modification of SC motifs. Interestingly, our data imply that multiple contacts with DNA are more important than the number of SC motifs. Thus, even though GR has two SC motifs, they are silent at a single GRE, whereas a single functional SC motif in GR is sufficient to inhibit activity from multiple GREs. This property is analogous to the synergistic behavior of activation domains because multimerizing the VP16 activation domains in Gal4VP16 does not increase activity at a single site but multimerizing Gal4 sites does (27). Thus, SC motifs control synergy by functioning as highly synergistic inhibitory modules. Although the inhibitory factor responsible for SC motif function remains to be defined, inhibitory proteins such as histone deacetylases recently have been shown to interact with SUMO (23).
The reversible SUMO modification of SC motifs provides a versatile regulatory mechanism to control the output of transcriptional regulatory complexes. As suggested by the enhancement in SUMO-2/3 modification in response to stress (25), it is likely that signaling pathways can impinge on transcriptional responses by regulating the activity of factors that enhance or reverse conjugation. In the case of GR and other nuclear receptors (17, 22), the fact that SUMOylation does not depend strictly on ligand binding suggests that such pathways may supply an independent regulatory input to modulate ligand-based control mechanisms. Like phosphorylation, SUMO modification is well suited to function in multiple contexts to modify protein function and yield a rich array of regulatory consequences.
Acknowledgments
We are thankful to Drs. Lalitha Subramanian and Sergey Chupreta for many useful discussions. This work was supported by United States Public Health Service Grant DK61656-01, National Institutes of Health Grant P60 DK20572, and American Heart Association Scientist Development Grant AHA0130559.
Abbreviations: SC motif, synergy control motif; GR, glucocorticoid receptor; GRE, glucocorticoid response element; HA, hemagglutinin; TAT, tyrosine aminotransferase; Dex, dexamethasone.
References
- 1.Yamamoto, K. R., Darimont, B. D., Wagner, R. L. & Iniguez-Lluhi, J. A. (1998) Cold Spring Harbor Symp. Quant. Biol. 63, 587-598. [DOI] [PubMed] [Google Scholar]
- 2.Carey, M. (1998) Cell 92, 5-8. [DOI] [PubMed] [Google Scholar]
- 3.Iñiguez-Lluhí, J. A. & Pearce, D. (2000) Mol. Cell. Biol. 20, 6040-6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Subramanian, L., Benson, M. D. & Iñiguez-Lluhí, J. A. (2003) J. Biol. Chem. 278, 9134-9141. [DOI] [PubMed] [Google Scholar]
- 5.Hiort, O., Holterhus, P. M., Horter, T., Schulze, W., Kremke, B., Bals-Pratsch, M., Sinnecker, G. H. & Kruse, K. (2000) J. Clin. Endocrinol. Metab. 85, 2810-2815. [DOI] [PubMed] [Google Scholar]
- 6.Kotaja, N., Vihinen, M., Palvimo, J. J. & Janne, O. A. (2002) J. Biol. Chem. 277, 17781-17788. [DOI] [PubMed] [Google Scholar]
- 7.Kagey, M. H., Melhuish, T. A. & Wotton, D. (2003) Cell 113, 127-137. [DOI] [PubMed] [Google Scholar]
- 8.Best, J. L., Ganiatsas, S., Agarwal, S., Changou, A., Salomoni, P., Shirihai, O., Meluh, P. B., Pandolfi, P. P. & Zon, L. I. (2002) Mol. Cell 10, 843-855. [DOI] [PubMed] [Google Scholar]
- 9.Bies, J., Markus, J. & Wolff, L. (2002) J. Biol. Chem. 277, 8999-9009. [DOI] [PubMed] [Google Scholar]
- 10.Kim, J., Cantwell, C. A., Johnson, P. F., Pfarr, C. M. & Williams, S. C. (2002) J. Biol. Chem. 277, 38037-38044. [DOI] [PubMed] [Google Scholar]
- 11.Ross, S., Best, J. L., Zon, L. I. & Gill, G. (2002) Mol. Cell 10, 831-842. [DOI] [PubMed] [Google Scholar]
- 12.Sapetschnig, A., Rischitor, G., Braun, H., Doll, A., Schergaut, M., Melchior, F. & Suske, G. (2002) EMBO J. 21, 5206-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirano, Y., Murata, S., Tanaka, K., Shimizu, M. & Sato, R. (2003) J. Biol. Chem. 278, 16809-16819. [DOI] [PubMed] [Google Scholar]
- 14.Iñiguez-Lluhí, J. A., Lou, D. Y. & Yamamoto, K. R. (1997) J. Biol. Chem. 272, 4149-4156. [DOI] [PubMed] [Google Scholar]
- 15.Bledsoe, R. K., Montana, V. G., Stanley, T. B., Delves, C. J., Apolito, C. J., McKee, D. D., Consler, T. G., Parks, D. J., Stewart, E. L., Willson, T. M., et al. (2002) Cell 110, 93-105. [DOI] [PubMed] [Google Scholar]
- 16.Le Drean, Y., Mincheneau, N., Le Goff, P. & Michel, D. (2002) Endocrinology 143, 3482-3489. [DOI] [PubMed] [Google Scholar]
- 17.Tian, S., Poukka, H., Palvimo, J. J. & Janne, O. A. (2002) Biochem. J. 367, 907-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gametchu, B. & Harrison, R. W. (1984) Endocrinology 114, 274-279. [DOI] [PubMed] [Google Scholar]
- 19.Tatham, M. H., Jaffray, E., Vaughan, O. A., Desterro, J. M., Botting, C. H., Naismith, J. H. & Hay, R. T. (2001) J. Biol. Chem. 276, 35368-35374. [DOI] [PubMed] [Google Scholar]
- 20.Bayer, P., Arndt, A., Metzger, S., Mahajan, R., Melchior, F., Jaenicke, R. & Becker, J. (1998) J. Mol. Biol. 280, 275-286. [DOI] [PubMed] [Google Scholar]
- 21.Muller, S., Matunis, M. J. & Dejean, A. (1998) EMBO J. 17, 61-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poukka, H., Karvonen, U., Janne, O. A. & Palvimo, J. J. (2000) Proc. Natl. Acad. Sci. USA 97, 14145-14150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Girdwood, D., Bumpass, D., Vaughan, O. A., Thain, A., Anderson, L. A., Snowden, A. W., Garcia-Wilson, E., Perkins, N. D. & Hay, R. T. (2003) Mol. Cell 11, 1043-1054. [DOI] [PubMed] [Google Scholar]
- 24.Whitby, F. G., Xia, G., Pickart, C. M. & Hill, C. P. (1998) J. Biol. Chem. 273, 34983-34991. [DOI] [PubMed] [Google Scholar]
- 25.Saitoh, H. & Hinchey, J. (2000) J. Biol. Chem. 275, 6252-6258. [DOI] [PubMed] [Google Scholar]
- 26.Braun, H., Koop, R., Ertmer, A., Nacht, S. & Suske, G. (2001) Nucleic Acids Res. 29, 4994-5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ellwood, K., Huang, W., Johnson, R. & Carey, M. (1999) Mol. Cell. Biol. 19, 2613-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]