Abstract
Positive-strand RNA viruses are the largest virus class and include many pathogens such as hepatitis C virus and the severe acute respiratory syndrome coronavirus (SARS). Brome mosaic virus (BMV) is a representative positive-strand RNA virus whose RNA replication, gene expression, and encapsidation have been reproduced in the yeast Saccharomyces cerevisiae. By using traditional yeast genetics, host genes have been identified that function in controlling BMV translation, selecting BMV RNAs as replication templates, activating the replication complex, maintaining a lipid composition required for membrane-associated RNA replication, and other steps. To more globally and systematically identify such host factors, we used engineered BMV derivatives to assay viral RNA replication in each strain of an ordered, genome-wide set of yeast single-gene deletion mutants. Each deletion strain was transformed to express BMV replicase proteins and a BMV RNA replication template with the capsid gene replaced by a luciferase reporter. Luciferase expression, which is dependent on viral RNA replication and RNA-dependent mRNA synthesis, was measured in intact yeast cells. Approximately 4,500 yeast deletion strains (≈80% of yeast genes) were screened in duplicate and selected strains analyzed further. This functional genomics approach revealed nearly 100 genes whose absence inhibited or stimulated BMV RNA replication and/or gene expression by 3- to >25-fold. Several of these genes were shown previously to function in BMV replication, validating the approach. Newly identified genes include some in RNA, protein, or membrane modification pathways and genes of unknown function. The results further illuminate virus and cell pathways. Further refinement of virus screening likely will reveal contributions from additional host genes.
Many or most steps in virus infections involve interactions of viral and host factors. Such virus–host interactions thus are crucial determinants of virus host range, replication, and pathology. Studies of virus–host interactions have advanced understanding of viral and cellular function and can provide targets for antiviral development. Positive-strand RNA [(+)RNA] viruses are the largest genetic class of viruses and include significant human pathogens such as hepatitis C virus, West Nile virus, and the coronavirus causing severe acute respiratory syndrome (SARS). Whereas recent studies show that host factors are critical for (+)RNA virus genome replication, mRNA synthesis, and other steps (1, 2), identifying such factors remains difficult.
Brome mosaic virus (BMV), a member of the alphavirus-like superfamily of human-, animal-, and plant-infecting (+)RNA viruses, has been studied as a model for viral RNA replication, encapsidation, recombination, and other processes (3). BMV has three genomic RNAs. RNAs 1 and 2 encode the interacting, multifunctional 1a helicase-like and 2a polymerase RNA replication factors (4, 5), which form endoplasmic reticulum (ER) membrane-associated RNA replication complexes with functional similarities to the replicative cores of retrovirus and double-strand (ds)RNA virus virions (6). RNA3 encodes protein 3a that enables infection spread between cells in natural hosts. The negative-strand [(-) RNA]3 replication intermediate also serves as a template for synthesis of a subgenomic (sg) mRNA, RNA4, which encodes the viral coat protein (Fig. 1A).
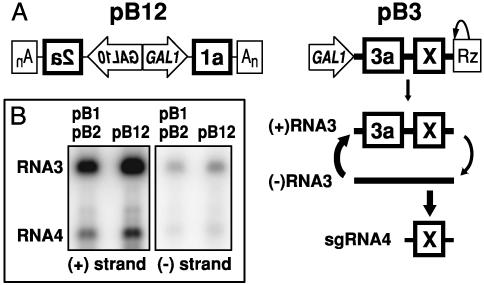
Fig. 1.
(A) Viral cDNAs (bold lines) and flanking expression elements in pB12, expressing 1a and 2a, and pB3, expressing RNA3. X, the BMV coat protein gene or any gene replacing it, such as Rluc. (Right) pB3 transcription produces an initial (+)RNA3 inoculum, used by 1a and 2a to direct synthesis of (-)RNA3, which then is used as a template to greatly amplify (+)RNA3 and produce sgRNA4. An, poly(A) signal; GAL1/GAL10, yeast promoters; Rz, self-cleaving ribozyme. (B) pB12 supports WT RNA3 replication at a level similar to that of independent plasmids expressing 1a and 2a.
The yeast Saccharomyces cerevisiae has proven a valuable model for normal and disease processes in human and other cells. The unusual ability of BMV to direct its genomic RNA replication, gene expression, encapsidation, and other processes in this yeast (7, 8) has allowed traditional yeast mutagenic analyses that have identified host genes involved in multiple steps of BMV RNA replication and gene expression. Such host genes encode a wide variety of functions and contribute to diverse replication steps, including supporting and regulating viral translation, selecting and recruiting viral RNAs as replication templates, activating the RNA replication complex through chaperones, and providing a lipid profile compatible with membrane-associated viral RNA replication (9–14; reviewed in refs. 2 and 15).
Here, we sought to develop a more rapid, global method to systematically identify yeast host factors with effects on BMV RNA replication by using an ordered array of yeast deletion strains (16) to assay virus replication in the absence of each of ≈4,500 yeast factors, which is ≈80% of the yeast genome. We describe screening this deletion array by using a whole-cell assay based on BMV-directed Renilla luciferase (Rluc) expression by pathways dependent on viral RNA replication and viral RNA-directed sg mRNA synthesis. The assay identified nearly 100 host genes whose absence repressed or enhanced BMV-directed Rluc expression by 3- to 25-fold. The results provide a significantly expanded view of virus–host interactions and should advance understanding of virus and cell pathways.
Materials and Methods
Yeast. YMI04 and ded1i yeast were described (11). Strains BY4743 (WT; ref. 17) and the homozygous diploid deletion series (BY4743 strain background; ref. 16) were from Research Genetics (Huntsville, AL). Standard yeast techniques were used (18), except for 96-well transformations, which were based on a one-step procedure (19). Briefly, yeast were grown to saturation overnight at 30°C in 96-well plates (1.2 ml per well), pelleted, suspended in 100 μl of transformation mix (0.18 M LiAc, pH 5.5, 36% polyethylene glycol-3350, 90 mM DTT, 0.5 mg/ml sheared salmon sperm DNA, and 20 μg/ml of each plasmid), incubated at 45°C for 60 min, and plated on solid minimal media.
Plasmids. pB12 (laboratory designation pB12VG1) expresses BMV 1a and 2a from the GAL1 and GAL10 promoters, respectively (Fig. 1 A). To construct pB12, the PacI–BamHI 1a-containing fragment of pB1YT3H (4) replaced the PacI–BamHI 2a-containing fragment of pB2YT5 (4) to make pB1VG17, then the 2a ORF and ADH1 poly(A) site (T4 DNA polymerase-blunted PstI–SphI fragment from pB2YT5) were inserted into EcoRI-linearized/blunted pB1VG17. pB3Rluc, based on pB3RQ39 (20), uses a truncated GAL1 promoter [GALL (21)] to express RNA3 with the coat protein ORF replaced by the Rluc ORF (from pRL-null; Promega). pB3VG1 is pB3RQ39 with the GAL1 promoter replaced by the CUP1 promoter, which was PCR-amplified from pSal1 (22) and contained a 5′ EcoRI site used for subcloning into EcoRI- and SnaBI-opened pB3RQ39. To make pCUP1B3 (laboratory designation pB3VG1ura), the EcoRI–SalI fragment from pB3VG1, containing the CUP1 promoter, WT RNA3, and a hepatitis δ ribozyme, was subcloned into EcoRI- and SalI-opened pRS316 (23). pB3GUSura was constructed by subcloning the EcoRI–SalI fragment from pB3MI20 (M. Ishikawa and P.A., unpublished results), containing the GAL1 promoter and RNA3 with the β-glucuronidase (GUS) gene replacing the coat protein sequence, into pRS316.
RNA and Protein Analyses. Transformed yeast were inoculated into liquid medium containing galactose, were grown overnight, subcultured, and grown overnight to mid-exponential phase (approximate OD600 = 0.6). Yeast containing pCUP1B3 were grown without added copper to use basal CUP1 promoter activity. For Rluc assays, yeast were pelleted and washed twice with assay buffer (10 mM NaPi, pH 7/20 mM EDTA, pH 8). For single-tube assays, cell lysates or a fixed number of cells based on OD600 reading were assayed in assay buffer plus 1 μM coelenterazine in 0.1% methanol by using a Pharmingen Monolight 3010 luminometer. For 96-well assays, fixed volumes of cells were similarly assayed by automated coelenterazine injection with a MicroLumat Plus (Perkin–Elmer). The OD600 of a second fixed volume of cells was measured to determine relative light units (RLU) per cell. For each pass through the ≈4,500 deletion strains, BMV-directed Rluc expression in each strain was calculated as the ratio of the strain RLU per 5 × 105 cells to the pass average RLU per 5 × 105 cells, and converted to a percentage. Total yeast RNA isolation, protein extractions, Northern and Western blots, and GUS assays were as described (11).
Results
Initiating and Assaying BMV RNA Replication. We previously used three plasmids to express BMV RNA replication factors 1a and 2a and RNA3 in yeast (20). To facilitate high-throughput transformation, we developed a two-plasmid BMV system. pB12 (Fig. 1 A) inducibly expresses 1a and 2a from the bidirectional, galactose-induced GAL1-GAL10 promoters (24). Yeast transformed with pB12 and a plasmid expressing WT RNA3 supported RNA3 replication and sgRNA4 synthesis to levels similar to those when 1a and 2a were expressed from individual plasmids (Fig. 1B).
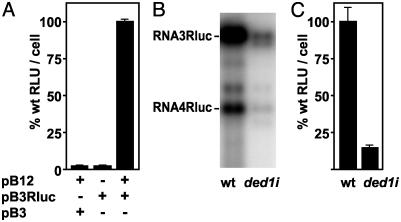
As sgRNA4 is templated by the (-)RNA3 replication intermediate (Fig. 1 A), its synthesis depends on, and can serve as, a reporter for BMV RNA replication (20). To assay sgRNA4 more simply and rapidly than by Northern blotting, we used a whole-cell reporter gene assay based on pB3Rluc, a plasmid expressing RNA3 with Rluc replacing the capsid gene. In the presence of 1a and 2a, such RNA3 derivatives are replicated and express the substituted foreign gene (Fig. 1 A and refs. 7 and 25). To avoid the need to lyse yeast to measure BMV-directed Rluc activity, we used a whole-cell assay (based on refs. 26 and 27). In this whole-cell assay, the Rluc signal from galactose-induced yeast with pB12 and pB3Rluc was 50-fold higher than the background in yeast with pB3Rluc alone or with pB12 and a plasmid expressing WT RNA3 (Fig. 2A). We also compared WT and isogenic ded1i yeast, in which BMV 2a polymerase translation and thus BMV replication are selectively inhibited by mutation of an RNA helicase encoded by DED1, a yeast gene essential for cell viability (11). Northern blots showed that WT RNA3 and RNA3-Rluc replication in ded1i yeast were 4% (11) and 10% (Fig. 2B) of that in WT yeast. In the whole-cell assay, Rluc activity for ded1i was 15% of WT, confirming the ability to detect defects in BMV replication (Fig. 2C).
Fig. 2.
(A) Rluc activity assayed on intact WT BY4743 yeast transformed with the indicated combinations of pB12, pB3Rluc, and/or pB3, a plasmid expressing WT RNA3 (20). (B) Northern blot analysis of BMV RNA replication products, RNA3Rluc and sg mRNA RNA4Rluc, in WT YPH500 or isogenic ded1i yeast cells transformed with pB12 and pB3Rluc. (C) Rluc activity assayed on intact WT YPH500 or isogenic ded1i yeast transformed with pB12 and pB3Rluc.
Identification of Genes Facilitating BMV-Directed Replication and Expression of Rluc. The BY4743 homozygous diploid yeast deletion strain array is composed of 4,792 yeast strains, each with both alleles of a specific, nonessential gene deleted, arrayed in 96-well plates (16). The diploid nature of the strains protects against phenotypic effects from any spontaneous mutations in untargeted loci. Two hundred thirty five strains annotated as “slow/petite” or “quality control failures” were not assayed. Another group of 66 strains with similar notated growth anomalies (plate 0372) transformed inefficiently and was only tested once. The remaining 4,491 strains were tested twice in a wholly independent manner. In each of these passes, pB12 and pB3Rluc were cotransformed into the deletion strains, and the resulting strains were grown and assayed for Rluc activity as described in Materials and Methods. Complete RLU data for both passes through the deletion strain array plus notes on occasional strain-specific variations in transformation efficiency or post-transformation growth rate are in Table 3, which is published as supporting information on the PNAS web site. Strains with an average change in BMV-directed Rluc expression of at least 3-fold, and at least a 2.5-fold change in each pass, are listed in Tables 1 and 2. Identity of these strains was verified by strain-specific PCR (16).
Table 1. Yeast deletion strains in which BMV-directed Rluc expression was impaired at least 3-fold.
| Yeast ORF | Gene | Average percent Rluc expression* | Function/phenotype |
|---|---|---|---|
| YGR063c | SPT4 | 1 ± 0 | Transcription elongation |
| YBR173c | UMP1 | 3 ± 0 | Proteasome activator |
| YDL160c | DHH1 | 4 ± 1 | mRNA decapping, translation |
| YDR156w | RPA14 | 5 ± 1 | RNA polymerase I subunit |
| YDR069c | DOA4 | 8 ± 5 | Ubiquitin-specific protease |
| YGL025c | PGD1 | 8 ± 1 | RNA polymerase II regulation |
| YPL178w | CBC2 | 8 ± 1 | RNA cap-binding protein |
| YGR037c | ACB1 | 9 ± 2 | Fatty acid transport, metabolism |
| YJL101c† | GSH1 | 10 ± 2 | Glutathione synthesis |
| YGR188c | BUB1 | 11 ± 4 | Protein tyrosine kinase activity |
| YJL101c† | GSH1 | 11 ± 3 | Glutathione synthesis |
| YDR363w-a | SEM1 | 11 ± 3 | Exocytosis/cell cycle regulation |
| YHR167w | THP2 | 13 ± 1 | Transcription elongation |
| YNL229c | URE2 | 13 ± 4 | Nitrogen utilization |
| YKL213c | DOA1 | 14 ± 1 | Ubiquitin metabolism |
| YML062c | MFT1 | 15 ± 1 | Transcription elongation |
| YJL148w | RPA34 | 16 ± 9 | RNA polymerase I subunit |
| YMR125w | STO1 | 16 ± 4 | RNA cap-binding protein |
| YPL084w | BRO1 | 19 ± 3 | Protein ubiquitination |
| YHL011c | PRS3 | 19 ± 1 | Ribose phosphate diphosphokinase |
| YGR184c | UBR1 | 19 ± 1 | Ubiquitin ligase |
| YGR135w | PRE9 | 19 ± 2 | 205 proteasome core α subunit |
| YNL032w | SIW14 | 20 ± 5 | Protein tyrosine phosphatase |
| YGL125w | MET13 | 20 ± 4 | Methionine biosynthesis |
| YNL056w | 20 ± 4 | Hypothetical ORF/unknown function | |
| YPL055c | LGE1 | 20 ± 2 | Protein ubiquitination/cell size control |
| YML013w | SEL1 | 21 ± 10 | Secretion regulation |
| YGL042c | 21 ± 0 | Hypothetical ORF/unknown function | |
| YHL029c | 22 ± 5 | Hypothetical ORF/unknown function | |
| YFR010w | UBP6 | 22 ± 6 | Protein deubiquitination |
| YDL020c | RPN4 | 23 ± 2 | Proteasome and cell cycle regulation |
| YPL226w | NEW1 | 23 ± 7 | ATP-binding cassette transporter |
| YOR246c | 23 ± 1 | Oxidoreductase activity | |
| YGL194c | HOS2 | 23 ± 4 | Histone deacetylase |
| YER119c-a | 24 ± 1 | Hypothetical ORF/unknown function | |
| YCR077c | PAT1 | 24 ± 9 | mRNA catabolism, translation |
| YDR241w | BUD26 | 24 ± 9 | Bud site selection |
| YML010c-b | 24 ± 9 | Hypothetical ORF/unknown function | |
| YKL160w | ELF1 | 24 ± 5 | Unknown function |
| YGL043w | DST1 | 25 ± 6 | Transcription elongation |
| YDR067c | 26 ± 5 | Hypothetical ORF/unknown function | |
| YJL124c | LSM1 | 26 ± 13 | mRNA catabolism/RNA cap-binding |
| YGR001c | 27 ± 2 | Hypothetical ORF/unknown function | |
| YCR095c | 28 ± 7 | Hypothetical ORF/unknown function | |
| YCR033w | SNT1 | 28 ± 6 | Histone deacetylase |
| YGL127c | SOH1 | 28 ± 8 | DNA repair |
| YPR046w | MCM16 | 28 ± 7 | Chromosome segregation |
| YER120w | SCS2 | 28 ± 1 | Myoinositol metabolism |
| YML010w-a | 29 ± 4 | Hypothetical ORF/unknown function | |
| YDR378c | LSM6 | 29 ± 0 | mRNA catabolism/splicing |
| YLR055c | SPT8 | 30 ± 1 | Histone acetylation |
| YBL027w | RPL19B | 30 ± 1 | Ribosome component |
| YDR477w | SNF1 | 30 ± 9 | Protein kinase |
| YHR179w | OYE2 | 31 ± 4 | NADPH dehydrogenase |
| YNL099c | OCA1 | 31 ± 5 | Protein tyrosine phosphatase activity |
| YKL010c | UFD4 | 31 ± 1 | Protein ubiquitination |
| YMR022w | QRI8 | 31 ± 6 | Ubiquitin conjugating enzyme |
| YIL107c | PFK26 | 31 ± 4 | Phosphofructokinase |
| YBR103w | SIF2 | 32 ± 5 | Histone acetylase |
Average Rluc expression of both passes
A few deletions, such as YJL101c, are present twice in the library
Table 2. Yeast deletion strains in which BMV-directed Rluc expression was enhanced at least 3-fold.
| Yeast ORF | Gene | Average percent Rluc expression* | Function/phenotype |
|---|---|---|---|
| YCR063w | BUD31 | 1377 ± 385 | Bud site selection |
| YLR373c | VID22 | 856 ± 168 | Vacuolar protein catabolism |
| YGL214w† | 769 ± 46 | Hypothetical ORF/unknown function | |
| YPR189w | SKI3 | 606 ± 35 | mRNA catabolism/antiviral protein |
| YEL003w | GIM4 | 529 ± 98 | Tubulin binding/folding |
| YCR094w | CDC50 | 522 ± 124 | Transcription regulation |
| YKL149c | DBR1 | 493 ± 10 | RNA catabolism |
| YOR076c | SKI7 | 454 ± 34 | mRNA catabolism/antiviral protein |
| YGR200c | ELP2 | 444 ± 112 | Transcription elongation |
| YGL213c | SKI8 | 440 ± 81 | mRNA catabolism/antiviral protein |
| YNL119w | 428 ± 15 | Hypothetical ORF/unknown function | |
| YJR074w | MOG1 | 422 ± 27 | Nuclear protein import |
| YJL183w | MNN11 | 414 ± 144 | Mannosyltransferase/glycosylation |
| YPL086c | ELP3 | 409 ± 16 | Transcription elongation |
| YHR004c | NEM1 | 407 ± 18 | Nuclear membrane/ER morphology |
| YOL064w | MET22 | 399 ± 65 | Methionine biosynthesis |
| YAL026c | DRS2 | 395 ± 21 | Phospholipid translocating ATPase |
| YPL102c | 381 ± 28 | Hypothetical ORF/unknown function | |
| YHR111w | UBA4 | 374 ± 53 | Ubiquitin-activating enzyme |
| YJL204c | RCY1 | 373 ± 2 | Endocytosis, membrane recycling |
| YHR207c | SET5 | 367 ± 93 | Unknown function |
| YKL110c | KTI12 | 367 ± 47 | Carbon utilization/toxin resistance |
| YBL079w | NUP170 | 366 ± 51 | Nucleocytoplasmic transport |
| YLR398c | SKI2 | 359 ± 61 | mRNA catabolism/antiviral protein |
| YLR357w | RSC2 | 355 ± 2 | Chromatin modeling |
| YOR051c | 354 ± 2 | Hypothetical ORF/unknown function | |
| YPL101w | ELP4 | 348 ± 52 | Transcription elongation |
| YCR098c | GIT1 | 346 ± 90 | Glycerophosphoinositol uptake |
| YPR061c | 343 ± 22 | Hypothetical ORF/unknown function | |
| YGL214w† | 342 ± 35 | Hypothetical ORF/unknown function | |
| YNL148c | ALF1 | 340 ± 28 | Tubulin folding |
| YLR384c | ELP1 | 337 ± 39 | Transcription elongation |
| YOR014w | RTS1 | 337 ± 47 | Protein phosphatase |
| YGL211w | 336 ± 69 | Hypothetical ORF/unknown function | |
| YML017w | PSP2 | 333 ± 42 | Unknown function |
| YNL120c | 333 ± 21 | Hypothetical ORF/unknown function | |
| YJL128c | PBS2 | 328 ± 61 | Osmoregulatory MAP kinase |
| YMR312w | ELP6 | 324 ± 53 | Pol II transcription elongation |
| YAL009w | SPO7 | 323 ± 71 | Nuclear membrane/ER morphology |
| YLR320w | MMS22 | 306 ± 36 | Ubiquitination, DNA repair |
Average Rluc expression of both passes
A few deletions, such as YGL214w, are present twice in the library
Table 1 lists genes whose deletion inhibited BMV-directed Rluc expression. The 58 ORFs in Table 1 include 48 for which at least suggested functions were previously annotated in the Saccharomyces Genome Database (www.yeastgenome.org). A number of these genes are implicated in regulating varied aspects of RNA function or turnover (CBC2, STO1, DHH1, LSM1, LSM6, and PAT1). Prior work from our group showed that LSM1 is required for efficient template selection for BMV RNA replication (10). Other genes (ACB1 and SCS2) are involved in membrane lipid synthesis or metabolism, and may modulate activity of the membrane-associated viral RNA replication complex by affecting membrane lipid composition, as previously found for fatty acid desaturase OLE1 (12), which is essential for cell viability and therefore not tested in the deletion array. For some Table 1 genes, the link(s) to BMV RNA replication were less obvious. Examples include genes involved in protein turnover (e.g., DOA1, PRE9, and QRI8), glutathione synthesis (GSH1), nitrogen utilization (URE2), etc. Table 1 also includes 10 hypothetical ORFs of >100 amino acids for which functions have not yet been determined (www.yeastgenome.org). The varied possible contributions to BMV replication by the Table 1 genes are considered further in Discussion.
BMV-directed Rluc-expression was inhibited to varying degrees by deleting a number of genes that facilitate transcription elongation, including SPT4, THP2, MFT1, and DST1 (Table 1). Several of these are involved in modifying chromatin structure and have effects on chromosome segregation. Deleting these or other genes with effects on transcription (PGD1, SNT1, and SNF1) might affect DNA-directed transcription of BMV 1a or 2a mRNAs, BMV RNA3, or yeast genes required for BMV replication. THP2 and DST1 deletion decreased 2a and 1a plus 2a protein levels, respectively (see below). The possibility that such deletions also might act by modulating expression of other yeast genes was emphasized by our finding that deleting ELP1, -2, -3, -4, or -6, the five nonessential components of another transcription elongation complex, increased BMV-directed Rluc expression (see Table 2 below) and that deleting the transcription-suppressing histone deacetylase HOS2 or its cofactor SNT1, inhibited Rluc expression (Table 1).
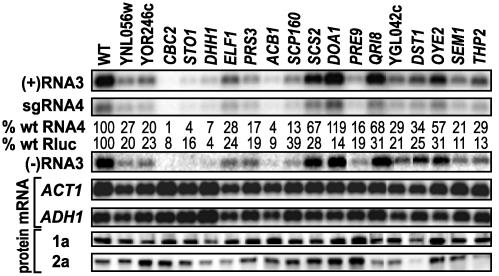
Eighteen deletion strains from Table 1 were selected for more detailed analysis (Fig. 3). These strains were selected from those for which BMV-directed Rluc expression was decreased by at least 3-fold on each of the two independent passes through the deletion strain array. In addition, the genes deleted were chosen to represent a range of different functions and pathways, and varied levels of virus inhibition to test the linearity of the Rluc results. WT BY4743 and each selected deletion strain were transformed with pB12 and pCUP1B3, a plasmid expressing WT RNA3 from the CUP1 promoter. WT RNA3 crosschecks for possible defects specific to the Rluc reporter and the CUP1 promoter crosschecks for possible defects specific to the GAL1-promoted B3Rluc transcription used in the Rluc assays. Moreover, for these and the prior Rluc assays, yeast were grown on galactose, requiring galactose induction of chromosomal GAL1 and GAL10, whose promoters were used to express BMV 1a and 2a (Fig. 1 A), and 1a and 2a expression were verified directly (Fig. 3). The yeast were grown to midexponential phase, and total RNA and proteins extracted for Northern and Western analyses. Whereas yeast ACT1 and ADH1 mRNA levels in the deletion strains were similar to WT, RNA4 levels generally were reduced to a level comparable to the reduction in Rluc expression. Thus, reduced Rluc activity in these strains was not due to defects in RNA4 translation, but in RNA4 synthesis and/or accumulation. Yeast lacking ACB1 or CBC2, e.g., had 8–9% of WT Rluc activity and ≈1–4% of WT RNA4 levels. Other deletions with similar behavior included YNL056w, YOR246c, STO1, DHH1, ELF1, PRS3, and PRE9. Yeast lacking DOA1, QRI8, THP2, or SCS2 had lesser defects in WT RNA3 replication and WT RNA4 production than in BMV-directed Rluc expression, suggesting defects exacerbated by B3Rluc.
Fig. 3.
Northern and Western analyses of BMV RNAs and proteins from WT BY4743 yeast and selected deletion strains with reduced BMV-directed Rluc expression. Each strain was transformed with pB12 to express 1a and 2a and with pCUP1B3 to express WT RNA3. Percent of WT sgRNA4 accumulation and Rluc expression are indicated. Northern blots of yeast ACT1 and ADH1 mRNAs are shown for comparison. Although not listed in Table 1, SCP160 was included due to its membrane association.
Consistent with inhibition of viral RNA replication, levels of (-)RNA3, the replication intermediate that serves as a template for sgRNA4, generally were reduced in parallel with RNA4, as were the levels of the other (-)RNA3-templated product, (+)RNA3. For most strains in Fig. 3, 1a and 2a proteins accumulated to normal or somewhat increased levels. Reduced 1a and 2a protein levels were seen on deleting DHH1, which we recently found is required for efficient translation of BMV genomic RNAs and their mRNA derivatives (ref. 13 and A.O.N. and P.A., unpublished results), or on deleting DST1 (see above). Reduced 2a protein levels were seen on deleting ELF1, QRI8, THP2, or YGL042 (which overlaps DST1).
Yeast Genes Inhibiting BMV-Directed Replication and Expression of Rluc. Table 2 lists genes whose deletion stimulated BMV-directed Rluc expression. The 39 ORFs in Table 2 include 32 ORFs for which proposed functions were previously annotated, and 7 hypothetical ORFs. They include four SKI genes (SKI2, -3, -7, and -8) that are involved in degrading nonpolyadenylated RNAs and inhibit the yeast L-A dsRNA replicon (28). Other genes listed are implicated in lipid or membrane synthesis, transport or remodeling (NEM1, DRS2, RCY1, GIT1, and SPO7), tubulin folding (GIM4 and ALF1), and other processes.
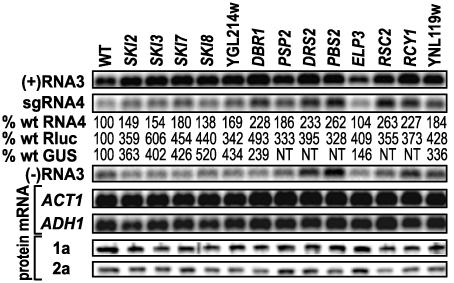
Thirteen deletion strains with increased BMV-directed Rluc expression were selected for more detailed analysis. In these strains (Fig. 4), yeast ACT1 and ADH1 mRNAs accumulated to WT levels but, in keeping with the increased Rluc expression, BMV RNA3 and sgRNA4 accumulated to increased levels, except in yeast lacking ELP3. Intriguingly, 1a and 2a protein levels generally were slightly reduced from WT.
Fig. 4.
Northern and Western analyses of BMV RNAs and proteins from WT BY4743 yeast and selected deletion strains with enhanced BMV-directed Rluc expression. As in Fig. 3, WT RNA3 from pCUP1B3 was used as the replication template. Percent of WT sgRNA4 accumulation and Rluc and GUS expression are indicated. NT, not tested.
Many tested deletions, including RNA catabolism genes SKI2, -3, -7, -8, and DBR1, left (-)RNA3 levels close to WT (Fig. 4), suggesting that the increased (+)RNA3 and sgRNA4 levels might primarily be due to effects on RNA stability rather than synthesis. By contrast, (-)RNA3 levels were increased by deleting DRS2, PBS2, or RCY1, encoding factors related to membrane composition, trafficking and function (Table 2 and Discussion).
The increases in WT sgRNA4 levels for the SKI2, -3, -7, -8, YGL214w, DBR1, ELP3, and YNL119w deletion strains relative to WT yeast were ≥2-fold lower than the increase in Rluc activity (Fig. 4). This result may be because WT RNA3 replicates to ≈6-fold higher levels than B3Rluc even in WT yeast, leaving less potential to increase RNA yield after removing the antagonistic influence of these host genes. To test this hypothesis, we assayed the same strains for GUS expression from B3GUS, an RNA3 derivative that has the coat gene replaced by GUS and replicates to levels similar to B3Rluc (9). As predicted, GUS activity in the SKI2, -3, -7, -8, YGL214w, and YNL119w deletion strains increased relative to WT yeast in parallel with Rluc (Fig. 4). In yeast lacking DBR1 and ELP3, GUS activity was higher than in WT yeast but lower than corresponding Rluc levels, suggesting some Rluc-specific effects in these strains.
Discussion
The continuing emergence of pathogenic viruses like the SARS coronavirus, another (+)RNA virus, underscores the importance of advancing understanding of virus–host interactions. Here, we used a yeast gene deletion library (16) and the unusual ability of BMV to replicate in yeast to conduct a systematic search for host genes affecting viral replication. The assay was validated by finding several genes previously implicated in BMV replication. Overall, we identified nearly 100 yeast genes that, when absent, altered viral-directed Rluc expression by 3-fold to over 25-fold. The assays likely underestimated the contribution of many of these functions to viral replication because many nonessential yeast gene deletions are partially compensated by other genes (29). Moreover, just as high multiplicity of infection overcomes antiviral resistance in some cell lines (e.g., ref. 30), the high level, continuous expression of BMV replication proteins and RNA replication templates used in this initial screen likely reduced virus dependence on some host functions, relative to natural infections initiated by a single copy of viral RNA. Below, we discuss selected genes and pathways and their potentially diverse roles in virus replication.
Regulation of Viral RNA Translation, Replication, and Degradation. Among the genes identified, we previously found three, LSM1, LSM6, and PAT1, to be involved in translating BMV RNAs and recruiting them from translation to RNA replication, and to link these steps to a cellular mRNA turnover pathway (10, 13). Here, we also identified additional host factors involved in RNA regulation, including CBC2, STO1, and DHH1. CBC2 and STO1 encode a cap-binding complex (31), which was implicated in initial rounds of translation (32). CBC2 or STO1 deletion did not impair translation of the capped and polyadenylated 1a and 2a mRNAs (Fig. 3), but may have affected entry of the capped but nonpolyadenylated WT RNA3 templates into translation, from which they must be recruited for replication (33). The role of DHH1 in BMV RNA replication may be interrelated, because the encoded Dhh1p promotes mRNA decapping (34) and complexes with Lsm1p (35). On DHH1 deletion, 1a and 2a protein levels were slightly reduced (Fig. 3), which was consistent with our recent finding of a role for DHH1 in translating some viral mRNAs and their derivatives (ref. 13 and A.O.N. and P.A., unpublished results). However, the modest reduction in 1a and 2a appears insufficient to explain the 12- to 20-fold reduction in Rluc expression and RNA4 levels, implying that cells lacking DHH1 are defective in supporting RNA replication steps besides 1a and 2a accumulation.
Further links to translation and ribosome biogenesis were found for RPA14 and RPA34, whose deletion decreased Rluc expression 20- and 6-fold (Table 1). RPA14/34 are RNA polymerase I subunits that are dispensable for cell growth individually, but not simultaneously, suggesting related functions. Rpa14p forms an ssRNA-binding heterodimer with Rpa43p, an RNA polymerase I subunit that binds the polymerase I transcription factor encoded by RRN3. RRN3 is essential for cell viability, but we recently identified rrn3 mutations that preserve yeast growth while inhibiting BMV RNA replication (A.O.N. and P.A., unpublished results). Moreover, BMV-directed Rluc expression was inhibited >3-fold by deleting RPL19B, one of two genes encoding identical ribosomal protein L19 sequences. Mutations in other 60S ribosomal proteins or other mutations reducing free 60S ribosome levels inhibit replication of the M1 satellite RNA of yeast dsRNA replicon L-A, apparently by preferentially inhibiting translation of nonpolyadenylated mRNAs such as L-A mRNAs (28) and BMV RNA replication products, (+)RNA3 and sgRNA4.
BMV-directed Rluc or GUS expression was increased 4- to 6-fold by deleting SKI2, -3, -7, or -8, which encode factors for exosome-mediated 3′ to 5′ mRNA decay and inhibit accumulation of nonpolyadenylated mRNAs like those of L-A (28, 36, 37). Consistent with a similar effect on BMV RNA stability, all four SKI deletions increased WT (+)RNA3 and sgRNA4 accumulation with little change in (-)RNA3 (Fig. 4).
Regulated Levels and Interaction of Viral Replication Factors. Deletion of PRE9, the only nonessential 20S proteasome core component, reduced BMV-directed Rluc expression and WT RNA3 replication by 5- to 6-fold (Table 1 and Fig. 3). PRE9-containing proteasomes localize primarily to the nuclear envelope–ER network (38), the site of BMV RNA replication (6, 39), and BMV 2a polymerase accumulation increased significantly in yeast lacking PRE9 (Fig. 3). Like retroviruses, many (+)RNA viruses down-regulate expression of their polymerases, and this down-regulation is linked to higher replicative fitness (ref. 6 and references therein). The 2a-homologous nsP4 polymerase of Sindbis virus, another member of the alphavirus superfamily, is regulated by the ubiquitin/proteasome-dependent N-end rule degradation pathway (40). Absence of several other yeast genes involved in ubiquitination or protein turnover also dramatically reduced BMV-directed Rluc expression (Table 1). However, some of these genes (e.g., DOA1, UBP6, UBR1, and UMP1) had little or no effect on accumulation of WT BMV sgRNA4 (Fig. 3 and data not shown). Whether the more selective effects of these genes were due to the higher replicative fitness of WT RNA3 compared to the RNA3-Rluc derivative (see Results) or to other effects specific to Rluc requires further study. BMV 2a also is regulated at the translational level, in part through specific dependence on host RNA helicase DED1 (11).
Deletion of protein tyrosine phosphatases SIW14 or OCA1, inhibited BMV-directed Rluc expression by 5- and 3-fold (Table 1). These phosphatases might contribute to BMV replication by jointly counteracting N-terminal phosphorylation of 2a polymerase, which, for a closely related bromovirus, has been shown to occur in vivo and to inhibit 2a interaction with 1a to promote replication complex assembly (41). Thus, replication complex assembly may be regulated by cell phosphatases as well as kinases.
Host Membrane Involvement. All (+)RNA viruses replicate their RNA genomes on host cell membranes. BMV RNA replication occurs in virus-induced invaginations of the outer perinuclear ER membrane (6) with locally altered lipid composition (42). Mutations in the essential Δ9 fatty acid desaturase OLE1 cause a reduction in unsaturated fatty acids that blocks BMV RNA synthesis after viral RNA template recruitment (12). Here, we found that replication of WT BMV RNA3 was inhibited 25-fold (Fig. 3) by deleting ACB1, encoding an acyl-CoA-binding factor involved in membrane sphingolipid biosynthesis (43). This finding underscores the importance of membrane properties to BMV RNA replication and implies that multiple classes of membrane lipids are required for a functional RNA replication complex.
Deleting SEL1, encoding a transmembrane protein that inhibits secretion, suppressed BMV-directed Rluc expression 5-fold (Table 1). Lesser defects in Rluc expression and WT RNA3 replication were caused by deleting SCS2 (Table 1 and Fig. 3), encoding an ER protein involved in ER targeting of other proteins and in inositol metabolism, with localized homology to a human VAMP-associated protein that interacts with hepatitis C virus RNA replication factor, NS5A (44). Conversely, Rluc expression, WT BMV RNA replication, and (-)RNA3 levels were increased by deleting DRS2, RCY1, or PBS2, encoding a phospholipid-translocating ATPase, a membrane recycling factor, and a MAP kinase required for resistance to osmotic stress, respectively, any of which might affect replication complex-associated membranes (Table 2 and Fig. 4). BMV-directed Rluc expression similarly increased on deleting NEM1 or SPO7, encoding interacting factors controlling nuclear membrane expansion and morphology, or GIT1, encoding a factor involved in glycerophosphoinositol uptake (Table 2).
As in prior virus–host interaction studies, further studies on the genes discussed above may provide important insights to host cell function. Moreover, Tables 1 and 2 list 17 hypothetical ORFs of unknown function whose deletion affected BMV-directed Rluc expression. Six of these ORFs (YER119c-a, YGL042c, YGL214w, YML010w-a, YNL120c, and YPL102c) reside on the opposite strand from another ORF (SCS2, DST1, SKI8, YML010c-b, YNL119w, and ELP4, respectively) whose deletion had parallel effects on BMV. Thus, the former deletions might affect BMV because they inactivate the latter ORFs on the opposite strand. Consistent with a possible lack of biological function, the former ORFs are not conserved in closely related yeasts (45). Conversely, we found 11 uncharacterized ORFs (YCR095c, YDR067c, YGL211w, YGR001c, YHL029c, YML010c-b, YNL056w, YNL119w, YOR051c, YOR246c, and YPR061c) that do not overlap other ORFs and are conserved in other yeasts (45). The effects of deleting these ORFs on BMV further supports their biological relevance and should shed light on their functions.
The high-throughput, functional genomics approach described here identified many host factors that affect BMV replication and implicated previously unconsidered pathways in the virus life cycle. Further studies will determine more directly how implicated host factors affect the virus and how such effects illuminate cellular functions and pathways. Additional screens more closely duplicating natural infection likely will reveal further relevant host factors. Finally, additional studies should further illuminate the roles of host genes essential for cell growth, which necessarily were not present in the deletion library, but which we have already found to make major contributions to BMV replication (refs. 11 and 12 and unpublished results).
Supplementary Material
Acknowledgments
We thank Chris Bradfield and Guang Yao for valuable advice on the yeast deletion strain array and data analysis, April Wicks and Kimberly Luke for technical assistance, other members of our laboratory for valuable discussions, and Paul Lambert for comments on the manuscript. This work was supported by National Institutes of Health Grant GM35072. P.A. is an Investigator of the Howard Hughes Medical Institute. D.B.K. was partially supported by National Institutes of Health Training Grant T32 CA09075.
Abbreviations: BMV, brome mosaic virus; (+)RNA, positive-strand RNA; (-)RNA, negative-strand RNA; ER, endoplasmic reticulum; sg, subgenomic; Rluc, Renilla luciferase; GUS, β-glucuronidase; RLU, relative light unit.
References
- 1.Lai, M. M. C. (1998) Virology 244, 1-12. [DOI] [PubMed] [Google Scholar]
- 2.Ahlquist, P., Noueiry, A. O., Lee, W. M., Kushner, D. B. & Dye, B. T. (2003) J. Virol. 77, 8181-8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kao, C. C. & Sivakumaran, K. (2000) Mol. Plant Pathol. 1, 91-98. [DOI] [PubMed] [Google Scholar]
- 4.Ahola, T., den Boon, J. A. & Ahlquist, P. (2000) J. Virol. 74, 8803-8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, J. & Ahlquist, P. (2000) J. Virol. 74, 4310-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwartz, M., Chen, J., Janda, M., Sullivan, M., den Boon, J. & Ahlquist, P. (2002) Mol. Cell 9, 505-514. [DOI] [PubMed] [Google Scholar]
- 7.Janda, M. & Ahlquist, P. (1993) Cell 72, 961-970. [DOI] [PubMed] [Google Scholar]
- 8.Krol, M. A., Olson, N. H., Tate, J., Johnson, J. E., Baker, T. S. & Ahlquist, P. (1999) Proc. Natl. Acad. Sci. USA 96, 13650-13655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishikawa, M., Diez, J., Restrepo-Hartwig, M. & Ahlquist, P. (1997) Proc. Natl. Acad. Sci. USA 94, 13810-13815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diez, J., Ishikawa, M., Kaido, M. & Ahlquist, P. (2000) Proc. Natl. Acad. Sci. USA 97, 3913-3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noueiry, A. O., Chen, J. & Ahlquist, P. (2000) Proc. Natl. Acad. Sci. USA 97, 12985-12990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee, W. M., Ishikawa, M. & Ahlquist, P. (2001) J. Virol. 75, 2097-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noueiry, A. O., Diez, J., Falk, S. P., Chen, J. & Ahlquist, P. (2003) Mol. Cell. Biol. 23, 4094-4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomita, Y., Mizuno, T., Diez, J., Naito, S., Ahlquist, P. & Ishikawa, M. (2003) J. Virol. 77, 2990-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noueiry, A. O. & Ahlquist, P. (2003) Annu. Rev. Phytopathol. 41, 77-98. [DOI] [PubMed] [Google Scholar]
- 16.Winzeler, E. A., Shoemaker, D. D., Astromoff, A., Liang, H., Anderson, K., Andre, B., Bangham, R., Benito, R., Boeke, J. D., Bussey, H., et al. (1999) Science 285, 901-906. [DOI] [PubMed] [Google Scholar]
- 17.Brachmann, C. B., Davies, A., Cost, G. J., Caputo, E., Li, J., Hieter, P. & Boeke, J. D. (1998) Yeast 14, 115-132. [DOI] [PubMed] [Google Scholar]
- 18.Guthrie, C. & Fink, G. R. (1991) Methods Enzymol. (Academic, San Diego), Vol. 194.
- 19.Chen, D. C., Yang, B. C. & Kuo, T. T. (1992) Curr. Genet. 21, 83-84. [DOI] [PubMed] [Google Scholar]
- 20.Ishikawa, M., Janda, M., Krol, M. A. & Ahlquist, P. (1997) J. Virol. 71, 7781-7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mumberg, D., Muller, R. & Funk, M. (1994) Nucleic Acids Res. 22, 5767-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mascorro-Gallardo, J. O., Covarrubias, A. A. & Gaxiola, R. (1996) Gene 172, 169-170. [DOI] [PubMed] [Google Scholar]
- 23.Sikorski, R. S. & Hieter, P. (1989) Genetics 122, 19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnston, M. & Davis, R. W. (1984) Mol. Cell. Biol. 4, 1440-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.French, R., Janda, M. & Ahlquist, P. (1986) Science 231, 1294-1297. [DOI] [PubMed] [Google Scholar]
- 26.Vieites, J. M., Navarro-Garcia, F., Perez-Diaz, R., Pla, J. & Nombela, C. (1994) Yeast 10, 1321-1327. [DOI] [PubMed] [Google Scholar]
- 27.Srikantha, T., Klapach, A., Lorenz, W. W., Tsai, L. K., Laughlin, L. A., Gorman, J. A. & Soll, D. R. (1996) J. Bacteriol. 178, 121-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wickner, R. B. (1996) Microbiol. Rev. 60, 250-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu, Z., Steinmetz, L. M., Gu, X., Scharfe, C., Davis, R. W. & Li, W. H. (2003) Nature 421, 63-66. [DOI] [PubMed] [Google Scholar]
- 30.Gao, G., Guo, X. & Goff, S. P. (2002) Science 297, 1703-1706. [DOI] [PubMed] [Google Scholar]
- 31.Izaurralde, E., Lewis, J., McGuigan, C., Jankowska, M., Darzynkiewicz, E. & Mattaj, I. W. (1994) Cell 78, 657-668. [DOI] [PubMed] [Google Scholar]
- 32.Ishigaki, Y., Li, X., Serin, G. & Maquat, L. E. (2001) Cell 106, 607-617. [DOI] [PubMed] [Google Scholar]
- 33.Sullivan, M. & Ahlquist, P. (1999) J. Virol. 73, 2622-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fischer, N. & Weis, K. (2002) EMBO J. 21, 2788-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coller, J. M., Tucker, M., Sheth, U., Valencia-Sanchez, M. A. & Parker, R. (2001) RNA 7, 1717-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benard, L., Carroll, K., Valle, R. C., Masison, D. C. & Wickner, R. B. (1999) J. Virol. 73, 2893-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Araki, Y., Takahashi, S., Kobayashi, T., Kajiho, H., Hoshino, S. & Katada, T. (2001) EMBO J. 20, 4684-4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Enenkel, C., Lehmann, A. & Kloetzel, P. M. (1998) EMBO J. 17, 6144-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Restrepo-Hartwig, M. & Ahlquist, P. (1996) J. Virol. 70, 8908-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.deGroot, R. J., Rümenapf, T., Kuhn, R. J., Strauss, E. G. & Strauss, J. H. (1991) Biochemistry 88, 8967-8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim, S. H., Palukaitis, P. & Park, Y. I. (2002) EMBO J. 21, 2292-2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee, W. M. & Ahlquist, P. (2003) J. Virol. 77, 12819-12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gaigg, B., Neergaard, T. B., Schneiter, R., Hansen, J. K., Faergeman, N. J., Jensen, N. A., Andersen, J. R., Friis, J., Sandhoff, R., Schroder, H. D. & Knudsen, J. (2001) Mol. Biol. Cell 12, 1147-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tu, H., Gao, L., Shi, S. T., Taylor, D. R., Yang, T., Mircheff, A. K., Wen, Y., Gorbalenya, A. E., Hwang, S. B. & Lai, M. M. (1999) Virology 263, 30-41. [DOI] [PubMed] [Google Scholar]
- 45.Kellis, M., Patterson, N., Endrizzi, M., Birren, B. & Lander, E. S. (2003) Nature 423, 241-254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.