Abstract
We introduce cap analysis gene expression (CAGE), which is based on preparation and sequencing of concatamers of DNA tags deriving from the initial 20 nucleotides from 5′ end mRNAs. CAGE allows high-throughout gene expression analysis and the profiling of transcriptional start points (TSP), including promoter usage analysis. By analyzing four libraries (brain, cortex, hippocampus, and cerebellum), we redefined more accurately the TSPs of 11-27% of the analyzed transcriptional units that were hit. The frequency of CAGE tags correlates well with results from other analyses, such as serial analysis of gene expression, and furthermore maps the TSPs more accurately, including in tissue-specific cases. The high-throughput nature of this technology paves the way for understanding gene networks via correlation of promoter usage and gene transcriptional factor expression.
Keywords: full-length cDNA, transcriptome, sequencing, cap-trapping
Even the comparison of mammalian genome draft sequences (1) has left many unanswered questions with regard to the exact identification of expressed genes, their promoter elements, and the network of promoter/transcriptional factor usage that underlies gene expression. Partial identification of the promoter sites has been provided by gene discovery programs based on the sequencing of full-length cDNA libraries (2-4); these have been instrumental in identifying the sequence of promoter regions, including potentially different promoters (5). Several thousand promoters can be determined by sequencing 5′ ends from full-length cDNA libraries and mapping the sequences to the genome, thus determining which correspond to coding and regulatory regions, respectively. These analyses can produce statistics on transcriptional start sites derived from large numbers of 5′ end sequences. However, these methods lack the throughput to provide significantly abundant data for intermediately/lowly expressed genes, chiefly because the comprehensive sequencing of cDNA libraries is prohibitively expensive. On the other hand, microarrays for high-throughput tissue expression analysis do exist (6), but these cannot determine transcription starting points and therefore cannot be used to accurately identify the cis regulatory elements that will be essential for computing gene networks. Another limitation of microarrays is that the only genes/transcripts that can be studied are those that have already been identified by the sequencing, which is far from completion (2). Serial analysis of gene expression (SAGE) allows partial sequence information of short tags at the 3′ ends of mRNAs (7) to be obtained. Although the information is partial, it is amenable to relatively cheap high-throughput digital data collection, because it is based on the cloning and subsequent sequencing of concatamers of short DNA fragments derived from 3′ ends of multiple mRNAs (http://cgap.nci.nih.gov/SAGE). This method was further improved on by Long-SAGE, which allows for the cloning of 20-nt SAGE tags (8), which mainly identify single loci on the genome, highlighting the importance of new gene discovery based on sequencing mRNA-derived tags. Nevertheless, SAGE is suitable only for obtaining 3′ end sequencing information for counting transcriptional units (TUs) (9) but neither for the identification of promoters nor for full-length cDNA cloning. This is because SAGE cannot identify mRNA 5′ ends, which may lie tens or hundreds of kilobases upstream in the genomic sequence.
Promoter elements can be identified by comparing relatively evolutionarily distant genome sequences (1) by looking for regions of conservation upstream of annotated genes. However, this form of comparative genomics does not identify the conditions when certain promoters are activated. Furthermore, there is ongoing work that attempts to link the presence of genomic elements (10) of genes to expression. These are still preliminary approaches, because they do not describe the transcriptional starting point (TSP) or the precise activation point.
To solve these problems, here we introduce a previously undescribed method: cap analysis gene expression (CAGE), which allows high-throughput identification of sequence tags corresponding to 5′ ends of mRNA at the cap sites and the identification of the TSP. The method essentially uses cap-trapper full-length cDNAs (11), to the 5′ ends of which linkers are attached (12). This is followed by the cleavage of the first 20 base pairs by class II restriction enzymes (8), PCR, concatamerization, and cloning of the CAGE tags. CAGE tags derived by sequencing these libraries were mapped to the genome and used for TSP and expression analysis, as well as for the determination of the 5′ end borders of new transcriptional units. CAGE concatamer sequencing is more cost-effective than full-length cDNA library sequencing because of the much higher throughput of identified tags.
Methods
CAGE Protocol. mRNA was prepared by standard methods (11). The cDNA synthesis was carried out by using a 25-μg mRNA and first-strand cDNA primer (oligo dT12-18) with SuperScript II RT in the presence of trehalose and sorbitol (13). Subsequently, full-length cDNA was selected with biotinylated cap-trapper (11). A specific linker, containing a recognition site for XhoI, I-CeuI, XmaJI, and the class II restriction enzyme MmeI (“upper oligonucleotide GN5” (sequence: biotin-agagagagacctcgagtaactataacggtcctaaggtagcgacctaggtccgacGNNNNN) and “upper oligonucleotide N6” (sequence: biotin-agagagagacctcgagtaactataacggtcctaaggtagcgacctaggtccgacNNN NNN) were mixed in a ratio of 4:1, and then this mixture in turn was mixed at 1:1 to the “lower oligonucleotide”: (sequence: phosphate group-gtcggacctaggtcgctaccttaggaccgttatagttactcgaggtctctctct-NH2), which was then ligated to the single-strand cDNA (12). To synthesize the second strand of the cDNA, we added to 10 μl of the cDNA sample: 6 μl of 100-ng/μl second-strand primer (bio-agagagagacctcgagtaactataacggtcctaaggtagcgacctaggtccgac), 7.2 μl of 5× A buffer (Invitrogen), 4.8 μl of 5× B buffer (Invitrogen), 6 μl of 2.5 mM dNTPs (Takara Bio, Shiga, Japan), and water up to 45 μl. The reaction mixture was heated to 65°C before 15 μl of 1 unit/μl Elongase polymerase (Invitrogen) was added. The reaction was performed in a thermocycler with the following settings: 5 min at 65°C, 30 min at 68°C, and 10 min at 72°C.
Preparation of the 5′ End Tags. The resulting double-stranded cDNA was cleaved with MmeI (3 units/μg cDNA), a class II restriction enzyme in 100 μl, and incubated at 37°C for 1 h. After Proteinase K treatment, samples were phenol/chloroform and chloroform extracted (from here, samples treated in this way are defined as being “purified”) and ethanol precipitated. Subsequently, the second linker (Upper-XbaI: Pi-tctagatcaggactcttctatagtgtcacctaaagtctctctctc-NH2 and Lower-XbaI: gagagagagactttaggtgacactatagaagagtcctgatctagaNN) was ligated to the 2-bp overhang at the cleavage site. Two microliters of cDNA solution was mixed to 4 μl of second linker DNA (0.4 μg/μl) and 8 μl of water. Before adding the ligase, the mixture was incubated at 65°C for 2 min followed by a brief incubation on ice. Then 2 μl of a 10× reaction buffer (NEB), 2 μl of T4 DNA ligase (NEB, 400 units/μl), and 2 μl of water were added, followed by incubation at 16°C for 16 h. Heating the reaction mixture at 65°C for 5 min terminated the ligation reaction.
Ligation products (with biotin at the 5′ ends) were separated from unmodified DNA with 200 μl of streptavidin magnetic beads (Dynabeads MP-280 Streptavidin, Dynal, Great Neck, NY). The beads were blocked by incubation with 100 μg of tRNA for ≈20 min at room temperature. Beads were then washed three times with 200 μl of 1 M NaCl/0.5 mM EDTA/5 mM Tris·HCl, pH 7.5 (1× B&W buffer) and resuspended in 200 μl of the same buffer.
The beads were combined with the samples and incubated with mild agitation at room temperature for 15 min to bind the modified 5′ cDNA tags to the beads. This was followed by the collection of the cap-tags/bead complex with a magnetic stand. The beads were rinsed twice with 200 μl of 1× B&W containing a BSA (200 μg/ml) buffer, twice with 200 μl of 1× B&W buffer, and finally twice with 200 μl of 0.1× TE buffer (1 mM Tris·HCl, pH 7.5/0.1 mM EDTA).
The 5′-end cDNA tags were released from the beads by treatment with excess free biotin (14). Biotin was solved at 1.5% (wt/vol) in 4 M guanidine thiocyanate/25 mM sodium citrate, pH 7.0/0.5% sodium N-lauroylsarcosinate. Incubation at 45°C for 30 min under occasional agitation with 50 μl of this solution allows the cDNA fragments to be released from the beads. This elution was repeated three times, and fractions were pooled. After the addition of 3.5 μg of glycogen, the sample was isopropanol precipitated and resuspended in a 50-μl 0.1× TE buffer. The cDNA tags were further purified by gel filtration on a G-50 spun column (Label IT Biotin Labeling Kit, Takara) followed by mixture treatment with RNaseI, purification, and isopropanol precipitation. The DNA was finally resuspended in 20 μl of 0.1× TE buffer.
Amplification of CAGE Tags. The DNA fragments were amplified in a PCR step by using the following two linker-specific primers: Primer 1 (uni-PCR): 5′-biotin-GAGAGAGAGACTTTAGGTGACACTA-3′; Primer 2 (MmeI-PCR): 5′-biotin-AGAGAGAGACCTCGAGTAACTATAA-3′. Twenty parallel PCRs were performed in a total volume of 50 μl by using 1 μl of cDNA-tags/5 μl of 10× buffer/3 μl of DMSO/12.5 μl of 2.5 mM dNTPs/0.5 μl of Primer 1 (350 ng/μl)/0.5 μl of Primer 2 (350 ng/μl)/27.5 μl of ddH2O/0.5 μl of ExTaq (5 units/μl, Takara). After incubating at 94°C for 1 min, 15 cycles were performed for 30 sec at 94°C, 1 min at 55°C, 2 min at 70°C followed by 5 min at 70°C. The resulting PCR products were pooled, purified, ethanol precipitated, and finally resuspended in 50 μlof0.1× TE buffer. The PCR products were purified on a 12% polyacrylamide gel. The appropriate 119-bp band was cut out of the gel, crushed, and twice extracted with 150 μl of elution buffer (0.5 M ammonium acetate/10 mM magnesium acetate/1 mM EDTA, pH 8.0/0.1% SDS) for 1 h at 65°C. Tags were filtrated with MicroSpin Empty Columns (Amersham Biosciences) by centrifugation at 600 × g for 2 min. The centrifugation was repeated after applying a further 50 μlof0.1× TE. The resulting extract was then purified, the DNA was ethanol precipitated, and finally resuspended in 20 μl of 0.1× TE buffer.
Large-Scale Tag Production. Purified bands were PCR-amplified once more in a similar way to that described above. Twenty tubes were heated: step1, 94°C for 1 min; step 2, 94°C for 30 sec; step 3, 55°C for 1 min; and step 4, 70°C for 2 min. This was repeated for seven cycles followed by a final elongation at 70°C for 5 min. The PCR products were pooled, purified, isopropanol precipitated, and finally redissolved in 50 μl of 0.1× TE buffer. The sample was next purified with G-50 purification, isopropanol precipitated, and resuspended in 30 μl of 0.1× TE buffer. The purified PCR products were digested with XmaJI (MBI Fermentas, Vilnius, Lithuania) and XbaI (NEB, Beverly, MA), followed by sample purification, ethanol precipitation, and resuspension in 10 μl of 0.1× TE buffer.
Separation of 32-nt Tag. The desired 32-bp DNA tags were separated from the free DNA ends cut off during restriction by incubation with streptavidin-coated magnetic beads, which retain the biotin-labeled DNA ends (15). The cleaved tags were mixed with the beads (300 μl) and incubated at room temperature for 15 min with mild agitation. Then the supernatant was collected after removal of the magnetic beads. The beads were rinsed with 50 μl of 1× B&W buffer, and pooled 32-nt tags from both supernatants were isopropanol precipitated and resuspended in 10 μl of 0.1× TE buffer. The sample was supplemented with 5 μl of 10× RNase I Buffer (Promega)/2 μl of 5 units/μl RNase I in 50 μl; incubated for 15 min at 37°C; treated with 1 μl of 10 μg/μl Proteinase K/1 μl of 0.5 M EDTA/1 μl of 10% SDS followed by further incubation at 15 min at 45°C. The sample was purified and isopropanol precipitated, followed by resuspension in 40 μl of 0.1× TE buffer.
The tags were further purified on a 12% polyacrylamid gel. The desired 32-nt band was cut out of the gel, crushed, and eluted with the previously used elution buffer for 1 h at 37°C, followed by purification, addition of 3.5 μl of glycogen, and ethanol precipitation. The DNA was finally resuspended in 4 μl of water.
Formation of Concatemers and Cloning. The tags were ligated to form concatemers by adding them to 2.4 μl of the tags, 0.3 μl of 10× T4 DNA ligase buffer (NEB), and 0.3 μl of T4 DNA Ligase (NEB). After an incubation of 10 min at 16°C, a 1/60 (molar ration) of XbaI linearized pZErO-1 cloning vector and 0.2 μl of 10× T4 DNA ligase buffer (NEB)/0.3 μl of T4 DNA Ligase (NEB) were added to a volume of 5 μl. The reaction was allowed to proceed overnight at 16°C to ligate concatemers to the vector. Subsequently, the reaction mixture was purified, and the DNA was precipitated with isopropanol. The DNA was finally resuspended in 6 μl of ddH2O.
The obtained ligations were finally electroporated into DH10b cells (Invitrogen) at 2.5 kv/cm, plated on zeocin plates (50 μg/ml), and the obtained plasmid was sequenced with “forward” primer as described (16).
Sequencing Operation and Insert Masking. CAGE libraries were sequenced with forward primers essentially as described (16) with minor modifications to use zeocin for selection of recombinants. We used in-house developed algorithms for the extraction of tags and for masking the vectors. CAGE tags were extracted with the following parameters: vector masking, minimum 12-bp recognition allowed; linker (13 bp) masking: maximum mismatch, 2 bp allowed; XbaI site maximum mismatch, 2 bp allowed; tag length, 17-24 bp.
Mapping of CAGE-Tag Sequences to Mouse Genome. A preliminary mapping of the 20 mers derived from 1,000 full-length cDNAs (9) to the genome assembly at http://genome.cse.ucsc.edu/goldenPath/mmFeb2002/chromosomes by using BLAST (17) showed that sites mapped by a continuous stretch of at least 18 bases has enough specificity (89%) to hit the true loci. CAGE tags were compared with a Fantom-2 TU set (9) mapped to mouse genome version UCSCmm3. Also, LocusLink mRNAs were taken from the National Center for Biotechnology Information (2003/06/30), mapped on UCSCmm3 with default BLAST, and repeat unmasked; “representative” means 5′ slippage in each locus. Tags are matched with the nearest sequence 5′ end (negative distance means tag is upstream).
Results and Discussion
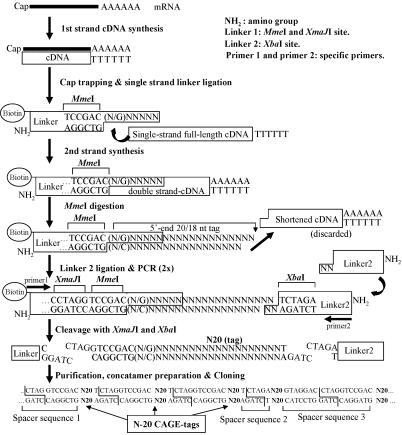
Rationale of Method and Design. DNA sequences upstream of TSPs usually encompass most of the regulatory elements that control gene expression (3). To map the TSPs and their usage, including those rarely expressed mRNAs, we combined the cap-trapper full-length cDNA selection (11) with the production of sequencing tags of sufficient length for unique mapping to mammalian genomes (Fig. 1). After cap trapping, we added a primeable sequence at the 5′ ends of the cDNAs (12), modified by the addition of MmeI, a class II restriction enzyme, which cleaves 20/18 bp outside the recognition sequence, generating 20-nt 5′ end CAGE tags. Assuming complete randomness of the genome sequence, an arbitrary sequence of 20 nucleotides will appear on average once in ≈1.1 × 1012 nucleotides; this seemed a better choice than other available class II restriction enzymes (GsuI) that would produce shorter16-nt tags, appearing on average once every ≈4 × 109 nucleotides.
Fig. 1.
Schematic procedure of the CAGE protocol as detailed in Methods.
We have produced and initially sequenced four CAGE libraries (Table 1), for which we present the analysis.
Table 1. Sequencing and mapping of CAGE tags to the mouse genome.
| Total tags | Tags mapped | Mapping rate, % | Sites mapped* | Sites/tags mapped | Unique tags† | Unique tag rate, % | |
|---|---|---|---|---|---|---|---|
| Whole brain | 21,070 | 11,102 | 52.7 | 235,400 | 21.2 | 7,899 | 71.1 |
| Cortex | 18,264 | 11,106 | 60.8 | 196,811 | 17.7 | 7,183 | 64.7 |
| Hippocampus | 9,172 | 5,997 | 65.4 | 153,687 | 25.6 | 4,009 | 66.9 |
| Cerebellum | 12,416 | 7,426 | 59.8 | 114,455 | 15.4 | 6,251 | 84.2 |
| Total/average | 60,922 | 35,631 | 58.5 | 700,353 | 19.7 | 25,342 | 71.1 |
Redundant number of tag sites mapped, including repeats
Tags that are mapped in a single site
Effectiveness of Mapping. Table 1 shows the efficiency of mapping the 20 nucleotides of the CAGE tags. Between 52.7% and 65.8% of all tags could be uniquely mapped to the mouse genome, whereas the remainder either mapped to multiple sites on the genome or could not be mapped due to stringent criteria allowed for mapping (see Methods), to keep the false-positive matches below a theoretical value of 3.65%. The average 60-70% GC content of CAGE tags, which decreases sequencing quality and length, was in part a cause of the mapping failure; protocols are being developed to maximize sequencing performance. Quite a large part of the unique tags hit single loci of the genome (64.7%∼84.2%). A few tags matched many loci; these generally consisted of retrotransposon elements or pseudogenes. The most highly expressed redundant in brain hit a repeat element mapped to >400 loci. Further investigations will be required to verify whether it is related to a repeat tag that is overexpressed in rat hippocampus SAGE libraries (18). Another multiple mapping CAGE tag (76 loci) is the GAPD, which has a large number of pseudogenes. Assuming complete randomness, a random sequence of 20 nt has an ≈1/400th chance to appear in the genome. However, the observed number of CAGE tags that map ≥2 loci (Table 1; and 9.5% of all nonredundant tags) is far greater than would be expected by chance. Other CAGE tags mapping to multiple genomic loci may identify either repeats in the TSP, motifs that are preferred in TSP, or potential conserved regulatory elements in the 5′ UTR. Further analysis will be based on correlating sequence and transcript features. Although these tags are ambiguous in terms of gene expression, their identification may shed light on mRNA function by associating the appearance of tags to the biological phenomena under investigation.
Distribution of CAGE Tags. We next verified whether CAGE tags could be used to identify mouse TSP and promoters. We compared the tags mapping to single sequences in the genome with known annotated mouse cDNAs (Table 2) by using the Fantom-2 set, which is the largest collection of mouse full-length cDNA (9), and the LocusLink representative mRNA sequences (19). With respect to the Fantom-2 clone set, the CAGE tags show a good overall correspondence in mapping with the 5′ ends of described TUs, but with some significant differences. Forty-four percent of the tags map at least 100 nt upstream of the Fantom-2 sequence (Table 2). Because 67% of the Fantom-2 sequences represent perfect full-length cDNAs without 3′/5′ truncations, and about half of the artifacts were 5′-end truncated, these data suggest that between 11% and 27% of the CAGE tags added a >100-nt extension to what was previously thought to be the TSP, describing with better accuracy the 5′ end of mRNAs. More than 2,630 tags mapped at least 10 kb upstream of either the Fantom-2 or the Reference Sequence database (RefSeq) mRNA, identifying new TSPs that are invaluable in identifying new promoter elements (3). The relatively large number of such upstream CAGE tags suggests also that traditional cloning and sequencing of full-length cDNAs are suboptimal because of difficulties in cloning and sequencing long cDNA (2), whereas cloning short CAGE tags is very promising for gene discovery. Only 10.8% of the CAGE tags mapped within 100 nt upstream of a known Fantom-2 TSP, and among them only 4% within 10 nt, thus suggesting large variability of the TSP. These await production of more CAGE tags for further fine TSP tuning and accurate searching for upstream elements. Accordingly, 29.9% of the CAGE tags mapped downstream of the described TSP but before the beginning of the second exon, suggesting the existence of a shorter TSP. Although most of the tags mapped within the first exon, 2.6% of the tags mapped in the annotated first intron, suggesting that new promoters with probable alternative splicing may be located within currently annotated introns.
Table 2. Mapping of CAGE tags relative to existing cDNA/mRNA sequences.
| Upstream
|
Internal
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Mapping of CAGE tags relative to existing cDNA/mRNA sequence | Total unique tags | Far upstream | Within 100 nt | Within 10 nt | In exon | In first exon | Intron | Far downstream |
| Comparison with Fantom-2 clones | ||||||||
| Whole brain | 7,899 | 3,907 | 780 | 287 | 2,151 | 1,977 | 177 | 884 |
| Cortex | 7,183 | 3,452 | 663 | 251 | 1,973 | 1,858 | 157 | 938 |
| Hippocampus | 4,009 | 2,063 | 355 | 134 | 1,062 | 977 | 79 | 450 |
| Cerebellum | 6,251 | 1,726 | 934 | 356 | 2,404 | 2,161 | 234 | 953 |
| Total | 25,342 | 11,148 | 2,732 | 1,028 | 7,590 | 6,973 | 647 | 3,225 |
| Comparison with LocusLink representative mRNA sequence (RefSeq) | ||||||||
| Whole brain | 7,899 | 1,644 | 1,695 | 380 | 986 | 705 | 177 | 3,397 |
| Cortex | 7,183 | 1,690 | 1,587 | 386 | 748 | 589 | 175 | 2,983 |
| Hippocampus | 4,009 | 780 | 862 | 177 | 469 | 364 | 93 | 1,805 |
| Cerebellum | 6,251 | 1,942 | 1,594 | 370 | 1,310 | 933 | 263 | 1,142 |
| Total | 25,342 | 6,056 | 5,738 | 1,313 | 3,513 | 2,591 | 708 | 9,327 |
About 12.7% of the CAGE tags mapped far downstream in the annotated cDNAs. Besides the CAGE tags may represent real TSPs far downstream, there may be a certain background number of false ones. Statistical analysis of more massive datasets including clusters of CAGE tags is necessary to distinguish the two cases.
Comparing the same dataset to LocusLink representative mRNA sequences (RefSeq) suggests a quite different figure (Table 2). The number of CAGE tags mapping far upstream is decreased to 23%, which is still considerably high despite >550,000 5′ ESTs from full-length cDNA libraries (2) available to build the RefSeq sequences. These are the longest possible in silico assemblies of ungapped ESTs and may therefore include nonrepresentative long assemblies. Despite this, a large amount of CAGE tags (17.4%) extended the RefSeq from 10 to 100 nt, allowing further accurate mapping of TSPs. A striking difference, if compared with the Fantom-2 set, is the larger number of CAGE tags (36.7%) mapping internally “far downstream” from the annotated first exon. Further analysis will be necessary to compare the difference in the Fantom-2 set, which is based on the annotation of an actual collection of cDNA clones, to the current release of RefSeq (19), which is based on computer annotation and selection of the longest sequence assembly. Besides the possibility that many CAGE tags do not represent starting points, it is also possible that naturally occurring internal starting points may not have been included in the final RefSeq mRNAs sequence.
Interestingly, the ratio of “far upstream” to far downstream is inverted in the two datasets depending on what we assume to be the complexity of the tissue (Table 2): the supposed complexity is, in order, (i) whole brain, which includes the whole brain transcripts; (ii) cortex and hippocampus; and (iii) cerebellum, which is composed by ≈10 main cell types. The number of far downstream tags is similar in the Fantom-2 dataset for all tissues, whereas the number of Fantom-2 far upstream is largest for the brain and lowest for the cerebellum, suggesting there are many upstream variants in complex tissues compared with annotated cDNAs. Tissue complexity might be reflected in the complexity of promoter usage.
In contrast, in the RefSeq dataset, the cerebellum shows the largest far upstream and the lowest far downstream tags counts, whereas the largest number of far downstream is derived from the most complex tissues. In most complex tissues (as above), there may be wide use of promoters to increase diversity, and the build of the RefSeq may not take into account internal tissue-specific promoter usage complexity. A detailed comparison of the two datasets is beyond the scope of this report.
We verified that the CAGE tags map real TSP by RT-PCR of 32 tags mapping 150-300 nt upstream (the far-upstream category) with respect to the Fantom-2 dataset (not shown). We could amplify 19 of them, of which seven gave the exact product and the remainder produced apparently spliced forms. The lack of amplification of the remaining primers may be due to poor RT-PCR condition, artifacts of CAGE tags, or the fact that the exons represented in the Fantom-2 clone may not be transcribed in the nervous tissues we used.
We verified that minor TSPs were detected by CAGE tags. For instance, the ATPase Na+/K+ transporting β-1 polypeptide is transcribed from two different TSP ≈350 bp apart. Three CAGE tags perfectly overlapped, at the upstream TSP, with the 5′ end of two full-length cDNA clones (clone IDs = 1200016M21 from lung and 2410046B18 from embryonic stem cells) and 17 tags mapped the downstream TSP, matching the 5′ end of four additional overlapping independent full-length cDNAs (clone IDs = 1500005L15 and 150000A19, both from cerebellum; C130050K08 from embryo 16 head; and 6330539J10 from medulla oblungata). These TSPs were confirmed by an additional 23 ESTs from our full-length libraries and the shorter TSP also by a Mammalian Gene Collection full-length cDNA. As a second example, tags mapping on the cystatin C gene revealed that CAGE tags correlated well over multiple TSPs, verified by nine full-length cDNAs and 5′ end clusters from 223 ESTs from full-length libraries.
Our data highlight the importance of aligning comprehensive CAGE tag collections to the genome, full-length cDNAs, ESTs, and predicted exons to shed light on the complexity of alternative promoter usage and TSPs. Subsequently, regulatory sequences and promoters can be better analyzed by searching for transcription factor-binding sites (20) and their location and correlation with CAGE tags.
Expression Analysis and Different Promoter Usage. The CAGE analysis is designed to detect the expression level and difference in promoter usage underlying gene expression. Although detailed analysis of expression will require more tags and well deserves future analysis, we verify here the expression (or lack) of known genes/TUs in specific libraries. Several genes/TUs were represented at similar levels, such as a tag from ferritin heavy chain (chromosome 19, + strand, position 8986422∼), which was the second or third highly expressed gene in all of the libraries (not shown). Similarly, several tags from genes highly expressed in brain were found among the top 90 genes, such as calmodulin, prostaglandin D synthase, myelin basic protein, hemoglobin β minor, cystatin C, tubulin α 1, cytochrome C oxydase subunits, and several more. Despite the preliminary annotation, we were reassured to find that six of them were present at high frequency in a hippocampus SAGE library (18). There were other genes that were apparently expressed more specifically. In particular, the lower complexity and different specialization of cerebellum were noteable. For instance, among the other top 90 expressed tags, we did not find genes such as β-3 (a basic helix-loop-helix protein), the protein A930039A15, a tag mapping close to the unclassifiable RIKEN F630050H04 gene, and seven more unannotated tags. In contrast, in cerebellum, the Purkinje cell protein 2(L7) and zebrin 2 (a cerebellum-specific protein) were overrepresented.
Although among the 30 most expressed mRNAs there are many housekeeping genes, which are not expected to be extensively transcribed from multiple promoters, we have investigated promoter usage differences among these top mRNAs. Note that clustering of CAGE-tags was not performed at this time, and therefore this analysis is biased for mRNAs that start exactly at the same point and usually have a defined TATA-box (3). Consequently, genes for which the TSP is more widespread are under-represented in this analysis; here we have analyzed only cases for which we have found at least three exactly overlapping tags.
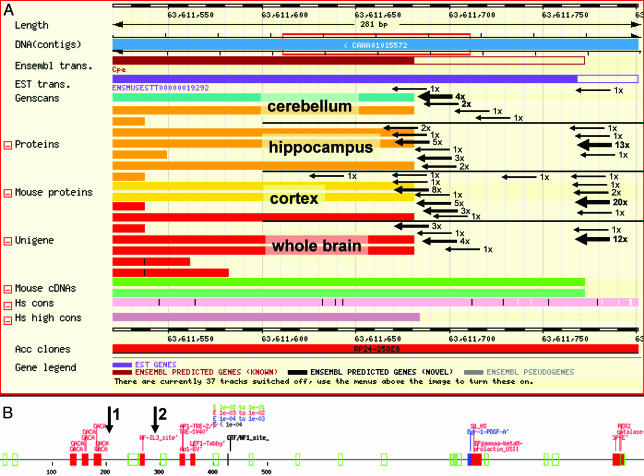
Among the three genes that clearly showed an alternative transcription starting point was carboxypeptidase E (chromosome 8, strand-, tags positions 63611683-63611784). Six of 10 tags for cerebellum were centered on position 63611701, the most frequently used in cerebellum. The cerebellum library produced only one tag around 6361176 +/- 3 nt, which was the major TSP for the other three tissues with 50 tags in total (Fig. 2).
Fig. 2.
Starting-point identification for the Carboxypeptidase E gene. (A) In-field arrows within an ensembl screenshot show TSPs detected by CAGE tags from the four libraries. The thick arrows show the main TSP for all tissues. TSP shows specificity only for cerebellum. (B) Alignment of promoter-predicted elements for 1,200 nt upstream to 63,611,500. Arrows 1 and 2 indicate the main two TSPs from A. Significance increases from green (lowest), red, and blue, to black (highest).
Also, Purkinje cell protein 2 showed a unique starting point for the cerebellum (chromosome 11, strand +, 79108451 +/-1; 13 tags), whereas the brain library produced three tags 54 bp apart (chromosome 11, strand +, 79108397), which are likely to represent tags from different TSPs under the control of a different promoter element. These data show that CAGE tags are instrumental in detecting alternative TSPs, which allows finding/mapping promoter/alternative promoter elements (Fig. 2B).
Perspectives
CAGE allowed the identification of 2,630 tags that mapped to regions in the genome that are >10 kbp from known transcripts and also the identification of a version further upstream of between 11% and 27% of known genes. CAGE tags can be used to synthesize primers to clone very rarely expressed mRNAs by long RT-PCR. So far this has been hampered by unreliable TSP predictions and ignorance of the tissue in which the desired rare mRNA is expressed. Furthermore, differential TSP usage can be correlated to the presence/absence of transregulatory factors (transcriptional factors, repressors, etc.). This information can be used to construct a matrix encompassing various biological phenomena and perturbations, correlating promoter status activity to the presence of transcription factors. This matrix will pave the way for a large-scale understanding of gene networks. Additionally, such technology will be a tool for SNP analysis in promoter regions and for selective collection of the promoters with PCR genomic regions adjacent to the transcriptional start site.
Acknowledgments
We thank Y. Mitsuiki, H. Isaka, and H. Nishibe for secretarial assistance; S. Kanagawa, H. Nishiyori, N. Sakazume, D. Koma, and K. Yoshida for technical assistance; T. Hayashi for support; and M. Yamamoto for technical advice. This study was supported by a Research Grant for the RIKEN Genome Exploration Research Project from the Ministry of Education, Culture, Sports, Science, and Technology of the Japanese Government (to Y.H.).
Abbreviations: CAGE, cap analysis gene expression; TSP, transcriptional starting point; SAGE, serial analysis of gene expression; TU, transcriptional unit; RefSeq, Reference Sequence database.
References
- 1.Mouse Genome Sequencing Consortium (2002) Nature 420, 520-562. [DOI] [PubMed] [Google Scholar]
- 2.Carninci, P., Waki, K., Shiraki, T., Konno, H., Shibata, K., Itoh, M., Aizawa, K., Arakawa, T., Ishii, Y., Sasaki, D., et al. (2003) Genome Res. 13, 1273-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suzuki, Y., Tsunoda, T., Sese, J., Taira, H., Mizushima-Sugano, J., Hata, H., Ota, T., Isogai, T., Tanaka, T., Nakamura, Y., et al. (2001) Genome Res. 11, 677-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ueda, H. R., Chen, W., Adachi, A., Wakamatsu, H., Hayashi, S., Takasugi, T., Nagano, M., Nakahama, K., Suzuki, Y., Sugano, S., et al. (2002) Nature 418, 534-539. [DOI] [PubMed] [Google Scholar]
- 5.Zavolan, M., Kondo, S., Schonbach, C., Adachi, J., Hume, D. A., RIKEN GER Group, GSL Members, Hayashizaki, Y. & Gaasterland, T. (2003) Genome Res. 13, 1290-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miki, R., Kadota, K., Bono, H., Mizuno, Y., Tomaru, Y., Carninci, P., Itoh, M., Shibata, K., Kawai, J., Konno, H., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 2199-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Velculescu, V. E., Zhang, L., Vogelstein, B. & Kinzler, K. W. (1995) Science 270, 484-487. [DOI] [PubMed] [Google Scholar]
- 8.Saha, S., Sparks, A. B., Rago, C., Akmaev, V., Wang, C. J., Vogelstein, B., Kinzler, K. W. & Velculescu, V. E. (2002) Nat. Biotechnol. 20, 508-512. [DOI] [PubMed] [Google Scholar]
- 9.Okazaki, Y., Furuno, M., Kasukawa, T., Adachi, J., Bono, H., Kondo, S., Nikaido, I., Osato, N., Saito, R., Suzuki, H., et al. (2002) Nature 420, 563-573.12466851 [Google Scholar]
- 10.Jegga, A. G., Sherwood, S. P., Carman, J. W., Pinski, A. T., Phillips, J. L., Pestian, J. P. & Aronow, B. J. (2002) Genome Res. 12, 1408-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carninci, P. & Hayashizaki, Y. (1999) Methods Enzymol. 303, 19-44. [DOI] [PubMed] [Google Scholar]
- 12.Shibata, Y., Carninci, P., Watahiki, A., Shiraki, T., Konno, H., Muramatsu, M. & Hayashizaki, Y. (2001) BioTechniques 30, 1250-1254. [DOI] [PubMed] [Google Scholar]
- 13.Carninci, P., Shiraki, T., Mizuno, Y., Muramatsu, M. & Hayashizaki, Y. (2002) BioTechniques 32, 984-985. [DOI] [PubMed] [Google Scholar]
- 14.Mizuno, Y., Carninci, P., Okazaki, Y., Tateno, M., Kawai, J., Amanuma, H., Muramatsu, M. & Hayashizaki, Y. (1999) Nucleic Acids Res. 27, 1345-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Powell, J. (1998) Nucleic Acids Res. 26, 3445-3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shibata, K., Itoh, M., Aizawa, K., Nagaoka, S., Sasaki, N., Carninci, P., Konno, H., Akiyama, J., Nishi, K., Kitsunai, T., et al. (2000) Genome Res. 10, 1757-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Datson, N. A., van der Perk, J., de Kloet, E. R. & Vreugdenhil, E. (2001) Hippocampus 11, 430-444. [DOI] [PubMed] [Google Scholar]
- 19.Pruitt, K. D. & Maglott, D. R. (2001) Nucleic Acids Res. 29, 137-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heinemeyer, T., Wingender, E., Reuter, I., Hermjakob, H., Kel, A. E., Kel, O. V., Ignatieva, E. V., Ananko, E. A., Podkolodnaya, O. A., Kolpakov, F. A., et al. (1998) Nucleic Acids Res. 26, 362-367. [DOI] [PMC free article] [PubMed] [Google Scholar]