Abstract
Mental retardation is the most common phenotypic abnormality seen in Down's syndrome (DS) patients, yet the underlying mechanism remains mysterious. DS critical region 1 (DSCR1), located on chromosome 21, is overexpressed in the brain of DS fetus and encodes an inhibitor of calcineurin, but its physiological significance is unknown. To study its functional importance and role in mental retardation in DS, we generated Drosophila mutants of nebula, an ortholog of human DSCR1. Here, we report that both nebula loss-of-function and overexpression mutants exhibit severe learning defects that are attributed by biochemical perturbations rather than maldevelopment of the brain. These results, combined with our data showing that the same biochemical signaling pathway is altered in human DS fetal brain tissue overexpressing DSCR1, suggest that alteration of DSCR1 expression could contribute to mental retardation in DS.
Down's syndrome (DS), a disorder that affects ≈1 in 700 live births across all ethnic groups, is the leading genetic cause of mental retardation (1). Other clinical abnormalities associated with DS include congenital heart disease, hypotonia, facial dismorphology, immune system defects, gastrointestinal anomalies, and the early development of the pathological and neurochemical changes of Alzheimer's disease (1, 2). Extensive studies on DS patients with partial trisomy 21 have allowed researchers to narrow the search for genes associated with the phenotypic features of DS to a segment of chromosome 21, the DS region (2). Among the genes located in the DS region is the DS critical region 1 (DSCR1) gene. DSCR1 is normally expressed in the central nervous system and the heart, but overexpressed in fetal DS brain (3, 4). DSCR1 belongs to a highly conserved calcineurin inhibitor family called calcipressin, which contains RCN1P in yeast (5), CBP1 in fungus (6), Nebula in Drosophila (GenBank accession no. AAD33987), and Dscr1 in mouse (7). Biochemical studies have shown that DSCR1 and other proteins in the calcipressin family can bind to and inhibit calcineurin, a serine/threonine protein phosphatase important for learning and memory (4-6, 8-12). The physiological significance of DSCR1 and its role in DS, however, are not well understood. Here, we generated Drosophila mutants of nebula, a homolog of human DSCR1, to address: (i) what is the role of nebula in learning and memory; and (ii) is mental retardation in DS related to DSCR1 overexpression?
Materials and Methods
Generation of Mutant Flies. nla1 was generated by using random hop with Drosophila lines carrying P{lacZ,w+}. The site of P element insertion was determined by plasmid rescue (13), and the presence of a single P element insertion was confirmed by genomic Southern blots. To excise the P element, we crossed nla1 with ry Ki P{ry+ Δ2-3}. The male jump-starters were then crossed to w;TM3/TM6 flies. Progeny with white eyes were made homozygous, and lines were established. Imprecise and precise P element excision lines were determined by PCR and confirmed by sequence analyses. All fly lines were outcrossed to Canton-S (CS) (Bloomington Stock Center, Bloomington, IN) background for several rounds. nlat1 and nlat2 transgenic flies were generated by a germ-line transformation method (14). The nebula transgene construct was generated by subcloning the coding region of nebula into the pINDY6 vector (L. Seroude and S. Benzer, personal communication). To drive overexpression of nebula, transgenic flies were crossed to either Act5C-GAL4/Tb (15), Elav-GAL4 3E (16), c739-GAL4 (17), Elav-GeneSwitch (18), or hsp70-GAL4 (19) fly lines.
Pavlovian Olfactory Learning and Memory Assays. Flies 2-7 days old were used in the Pavlovian olfactory conditioning assay (20). Before conditioning, the training apparatus was humidified for 2 h with a vacuum-connected air bubbler to allow saturation of humidity. Before training, flies were ensured to distribute equally between 3-octanol (OCT) and 4-methylcyclohexanol (MCH) used for training. Groups of ≈50-100 flies were trained with one odor (MCH) for 60 s paired with electric shock (at 60 V), and then exposed subsequently to the second odor (OCT) for 60 s without electric shock. Immediately after training, learning was measured by giving flies 120 s to choose between the two odors used during training in T maze arms. To examine long-term memory, flies were subjected to spaced training (10 training sessions with 15-min rest intervals between sessions) (21). Flies were then tested both immediately and 24 h after spaced training. All training was performed under dim red light, and performance tests were carried out in the absence of light. Performance index was calculated as the fraction of flies that avoided the conditioned stimulus (odor with electric shock) minus the fraction of flies that avoided the unconditioned stimulus (second odor) multiplied by 100. For odor acuity tests, flies were transported to the choice point of the T maze and allowed 120 s to choose between odor and air. For shock reactivity, flies were allowed 120 s to choose between the tube with electric shock pulses and the tube without shock. Individual performance index was calculated as reported (22).
RU486 Induction. RU486 (10 mg/ml, Mifepristone, Sigma) was dissolved in ethanol. RU486 was diluted and mixed with fly food to get a final concentration of 100 μg/ml for adult feeding and 20 μg/ml for larval feeding. The final ethanol concentration in both cases was 4%. To activate GeneSwitch transiently in adults, 1- to 5-day-old adult flies were transferred to fly food containing 100 μg/ml RU486, and Pavlovian olfactory learning tests were performed 24 h after feeding. To activate GeneSwitch during development, larvae were raised on fly food containing 20 μg/ml RU486; eclosed adults were promptly removed to normal food media. Adult flies that had been transferred to normal fly food media for 3-6 days were used in Pavlovian olfactory learning assays. For mock feeding, flies were raised as described above, except fly food contained 4% ethanol, but no RU486.
Materials. Human normal and trisomy 21 fetal brain tissues were obtained from the Brain and Tissue Bank at the University of Maryland, Baltimore.
Supporting Methods. For details on methods used, see Supporting Text, which is published as supporting information on the PNAS web site.
Results and Discussion
Generation of nebula Loss-of-Function Mutants. We created a Drosophila mutant with a P element insertion between the 5′ UTR and the translation start site of the nebula gene (nla1; Fig. 1A). Furthermore, by mobilizing the P element inserted in nla1, we isolated precise and imprecise P element excision lines, nlaPJ and nla2, respectively (Fig. 1 A). The nebula protein shares 43% identity and 64% similarity in amino acid sequence with human DSCR1 (Fig. 5, which is published as supporting information on the PNAS web site). Quantitative real-time RT-PCR analyses revealed reduced expression of nebula in the mutants (Fig. 1B), suggesting that both nla1 and nla2 are hypomorphic alleles. The nebula mutants are viable and fertile without visible abnormality. In situ hybridization using an antisense probe to the nebula gene showed that nebula is expressed in the cortex of the normal fly brain (Fig. 6, which is published as supporting information on the PNAS web site).
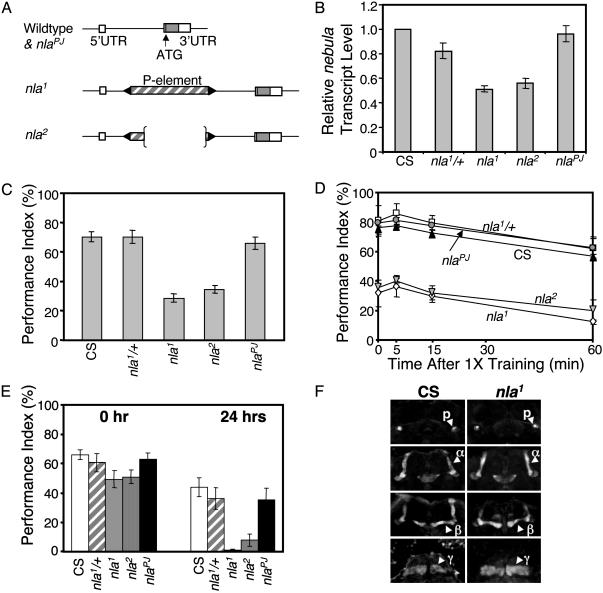
Fig. 1.
nebula mutants show defective learning and long-term memory, but normal short-term memory decay. (A) Schematics of different nebula alleles. Empty and filled boxes indicate untranslated and coding sequences, respectively. nlaPJ and nla2 show precise and imprecise P element excision, respectively, as determined by sequence analysis. (B) Quantitative real-time RT-PCR results showing relative fold change in nebula transcript level for each fly line. n = 4 independent experiments done in triplicate. Values represent mean ± SEM. (C) Performance of control (CS), nla1 heterozygotes (nla1/+), nla1 homozygotes (nla1), nla2 homozygotes (nla2), and nlaPJ homozygotes (nlaPJ) in Pavlovian olfactory conditioning test after training. nla1 and nla2 showed severe learning defects, whereas nlaPJ displayed normal learning. (D) Memory decay curves of flies after one training session. The same decay rate seen for all fly lines indicates that nebula is not required for short-term memory. (E) Long-term memory assayed immediately (0 h) or 24 h after spaced training. nla1 and nla2 showed a virtual absence of long-term memory, whereas CS, nla1/+, and nlaPJ showed normal memory retention. For C-E, n ≥ 5 for each fly strain and each time point. All values are expressed as mean ± SEM. (F) The nla1 mutant has normal mushroom bodies as revealed by Fasciclin II antibody staining. There is no difference in the structures of the α, β, γ lobes and peduncles (p) of nla1 flies as compared to the control (CS) flies.
Effects of nebula Loss-of-Function Mutations on Learning and Memory. To investigate the role of nebula in learning and memory, we used a Pavlovian olfactory associative learning and memory test (20). Significant defect in learning was observed after a single trial of training for homozygous nla1 and nla2 mutants, as compared to nla1 heterozygotes, precise excision line (nlaPJ), and CS flies (Fig. 1C). The learning defects are not attributed by abnormal sensorimotor responses, because both odor and shock avoidances were indistinguishable among the fly lines tested (Fig. 7, which is published as supporting information on the PNAS web site). Nevertheless, we cannot completely rule out the possibility that some of the performance defects seen in the nebula mutants could be caused by subtle alteration in sensitivity to stimuli.
Short-term memory is thought to last 60 min after training in Drosophila (23). We thus investigated short-term memory in different nebula alleles by examining the performance at different time points after training. The same rate of normal memory decay seen for the different fly lines (Fig. 1D) suggests that short-term memory is likely intact and that the low performance value obtained right after training may be a reflection of defective learning, albeit we do not rule out the possibility that nebula is required for immediate short-term memory (during the 3 min in between training and the first possible test time point). To further examine the role of nebula in the memory pathway, we tested long-term memory in the nebula mutants. Long-term memory in Drosophila is associated with phosphorylated cAMP-responsive element binding protein (pCREB) and protein synthesis and is evident 1 day after 10 trials of spaced training with 15-min rest intervals between trainings (21, 23). Strikingly, we found that homozygous nebula mutants showed a virtual absence of long-term memory, whereas control flies displayed ≈40% memory retention 24 h after training (Fig. 1E). The observed defect in long-term memory is not caused by the initial deficiency in acquisition, because the performance of nla1 and nla2 homozygotes immediately after spaced training did not differ significantly from CS flies (P > 0.05 for CS vs. nla1 and CS vs. nla2 as determined by Student's t tests). Together, our results indicate that increasing the number of training sessions can improve the learning performance of nebula mutants, and more importantly, nebula is required for effective learning and long-term memory.
nebula Mutants Have Normal Mushroom Bodies but Altered Calcineurin-Mediated Signaling. Despite the overall normal brain structure (data not shown), it is possible that the learning and memory deficits seen for the nebula mutants are caused by structural defects specifically in the mushroom bodies, the center for learning and memory in Drosophila (24). To examine the integrity of the mushroom bodies, we visualized the α, β, and γ lobes with Fasciclin II antibody (25). Of all 60 nla1 homozygous mutant flies examined, no visible structural defect in the mushroom bodies was detected, and the α, β, and γ lobes were similar to the control flies (Fig. 1F). This finding implies that the nebula mutation does not cause maldevelopment of the mushroom bodies and that defects in learning and memory are not caused by gross structural defect.
To understand whether the mutation in nebula significantly alters the biochemical activity predicted for the gene, we examined calcineurin activity level in the nebula mutant flies. Calcineurin activity detected in the homogenate of nla1 homozygotes was ≈40% higher than CS control and nla1 heterozygotes, indicating that nebula functions as an endogeneous inhibitor of calcineurin as other members in the calcipressin family (4-6) (Fig. 2A). We further examined the signaling pathways downstream from calcineurin and found that homozygous nla1 mutants showed a substantial decrease in cAMP-dependent protein kinase (PKA) activity, the amount of pCREB, and the level of d-jun transcripts (Fig. 2). Given that PKA and pCREB are important for normal learning and long-term memory (26-28), the observed biochemical perturbations in the nebula mutant may contribute to the observed deficiency in learning and memory.
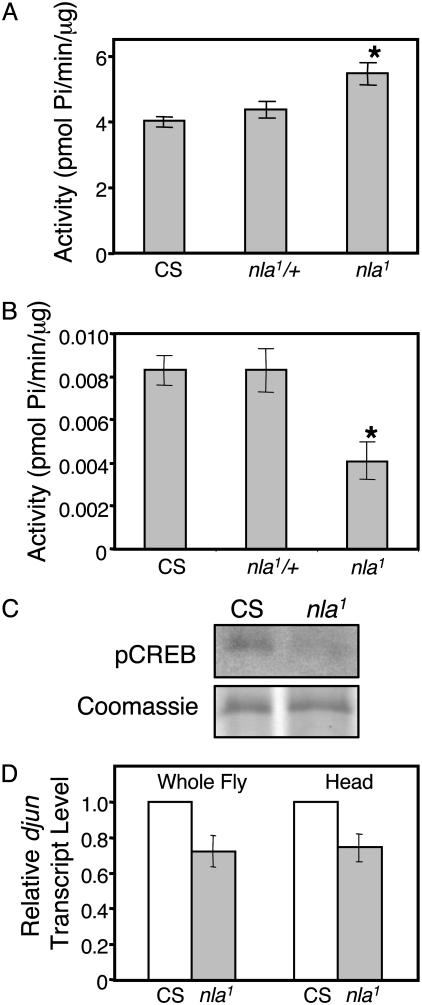
Fig. 2.
nebula mutation causes perturbations in biochemical signaling. (A) Calcineurin activity determined from extracts of CS, nla1/+, and nla1. Calcineurin activity in nla1 increased by 40%. (B) PKA activity in nla1 is ≈50% of the CS flies. (A and B) n = 3 experiments done in triplicate (*, P < 0.05). (C) Western blot analysis with anti-pCREB antibody shows that the level of pCREB in nla1 is significantly reduced. Equal intensity bands seen by Coomassie blue staining of the SDS protein gel confirm equal loading. (D) Quantitative real-time RT-PCR results. d-jun transcript level in nla1 is ≈30% lower than in the CS flies whether total RNA from whole flies or adult heads was used. n = 4 independent experiments in triplicate. Values represent mean ± SEM.
Transgenic Flies Overexpressing nebula Exhibit Severe Defects in Learning. Our findings that nebula, the Drosophila homolog of human DSCR1, can regulate learning and long-term memory led us to hypothesize that overexpression of DSCR1 in DS may be responsible for mental retardation. To create a model for studying the role of DSCR1 in mental retardation in DS, we generated transgenic flies overexpressing nebula by using the UAS/GAL4 system (29). Flies containing the nebula transgene on the third chromosome (nlat1) were crossed with both the Act5C-GAL4 (Act5C) and Elav-GAL4 (Elav) driver lines to yield nebula overexpression throughout the fly or specifically in the neurons, respectively (15, 16). Fig. 3A shows that the level of nebula transcripts was elevated ≈1.5-fold in transgenic flies containing the indicated driver. Remarkably, Pavlovian olfactory learning tests revealed that both Act5C/nlat1 and Elav/nlat1 had virtually no learning (Fig. 3B), whereas the flies showed no visible defect and responded normally to odor and electric shock (Fig. 7). To further restrict spatial expression of the nebula transgene, we used the c739-GAL4 driver, which drives UAS-transgene expression preferentially in the α and β lobes of the mushroom bodies (17, 30). Fig. 3B illustrates that overexpression of nlat1 or nlat2 (UAS-nebula transgene on the second chromosome) with the c739-GAL4 driver (c739;nlat1 and c739/nlat2) also caused severe learning defects, implying that specific over-expression of nebula in the mushroom bodies is sufficient to disrupt learning. Furthermore, overexpression of nebula in the mushroom bodies on the nebula mutant background completely restored learning to normal, confirming that nebula is the gene responsible for the learning defects seen in the nebula mutants (Fig. 3B). Next, we examined the level of pCREB in nebula transgenic flies and found that Act5C/nlat1 showed an increase in pCREB level as compared to the control lines (Fig. 3C), indicating the existence of altered biochemical signaling. The result that defective learning in the transgenic flies is accompanied by an increase in pCREB is consistent with recent findings by Yuan et al. (31) that transient overexpression of CREB and the subsequent elevation in the level of pCREB before odor preference training interfered with learning in rats. This finding suggests that the global increase in the level of pCREB may saturate the signaling pathway and thereby make the signaling pathway less receptive to further regulation by kinases after training.
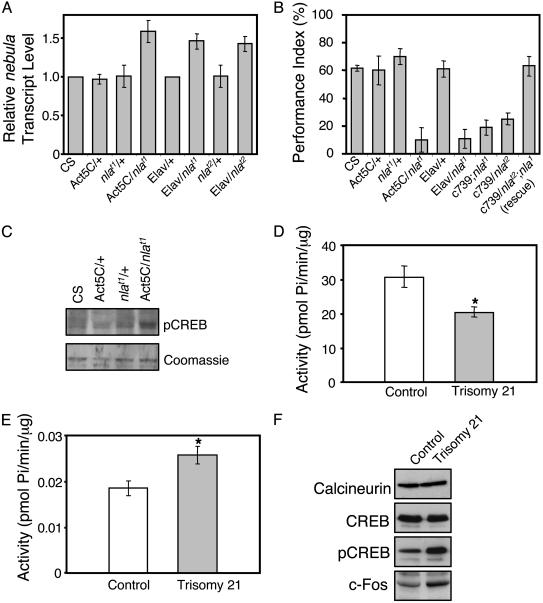
Fig. 3.
Transgenic flies overexpressing nebula show defective learning, and human trisomy 21 fetal brain tissues overexpressing DSCR1 exhibit altered calcineurin-mediated signaling. (A) Quantitative real-time RT-PCR results. n = 3 independent experiments done in triplicate. Note that nlat1 and nlat2 transgenic flies show the same level of nebula overexpression in the presence of Elav-GAL4 driver. (B) Performance index values for Pavlovian olfactory learning. Flies overexpressing nebula by using the Act5C driver (Act5c/nlat1), Elav driver (Elav/nlat1), and mushroom bodies-specific driver (c739/nlat1 and c739/nlat2) show severely impaired learning. Overexpression of nebula in the mushroom bodies of the homozygous nebula loss-of-function mutant (c739/nlat2;nla1) shows complete rescue of the learning defect. (C) Western blot analysis shows that the level of pCREB in Act5C/nlat1 is higher than that of the control lines (CS, Act5C/+, nlat1/+). Coomassie staining confirms the amount loaded. (D) Calcineurin activity in control and trisomy 21 fetal brain tissues. (E) PKA activity in control and trisomy 21 fetal brain tissues. (F) Western blots showing calcineurin, CREB, pCREB, and c-Fos protein levels in control and trisomy 21 fetal brain tissues. All values represent mean ± SEM. For A, n = 4 experiments done in triplicate. n = 4 for each fly strain in B. For D and E, n = 3 experiments done in triplicate.*, P < 0.05 as determined by Student's t test.
Overexpression of DSCR1 in Human Trisomy 21 Fetal Brain Tissue also Leads to Disturbed Calcineurin-Mediated Signaling. To directly correlate the findings obtained by using our Drosophila model with DSCR1 overexpression in DS, we examined the level of DSCR1 transcripts and DSCR1-mediated signaling in human trisomy 21 fetal brain tissues. Using quantitative real-time RT-PCR, we detected a 1.55 ± 0.14 (n = 3 experiments done in triplicate; values represent mean ± SEM)-fold increase in the level of DSCR1 transcripts in human DS fetal brain. Compared to the control, trisomy 21 fetal brain tissue also demonstrated altered calcineurin-mediated signaling. Trisomy 21 fetal brain showed a 40% reduction in calcineurin activity (Fig. 3D), but 40% increase in PKA activity level (Fig. 3E). In addition, overexpression of DSCR1 led to a substantial increase in the amount of pCREB and c-Fos, whereas the overall level of CREB remained constant (Fig. 3F), consistent with altered biochemical signaling seen in the nebula transgenic flies.
GeneSwitch Expression System to Determine the Cause of Deficient Learning. To better understand whether the learning deficits in nebula-overexpressing flies are caused by excessive expression of nebula at the time of the learning test or by developmental defects, we used the inducible UAS/GeneSwitch expression system (18, 32). First, we overexpressed nebula transiently and specifically in the neurons of adult flies by feeding Elav-GeneSwitch/nlat1 flies (Elav-GS/nlat1) RU486. In the presence of RU486, Elav-GS, a conditional GAL4-progesterone-receptor fusion protein controlled by the neuron-specific promoter Elav, could bind to the UAS and activate transcription of nebula. Second, we overexpressed nebula throughout development by feeding Elav-GS/nlat1 larvae with RU486 and examined the learning performance of eclosed adults 3-6 days after removal to normal food media. Fig. 4A shows adult Elav-GS/nlat1 flies that had been fed for 1 day with media containing RU486 showed a robust increase in nebula transcript level; Elav-GS/nlat1 larvae fed with RU486 showed elevated nebula transcript only during development (larvae), but not in adulthood (Fig. 4B). Pavlovian olfactory learning tests demonstrated that transient elevation of nebula transcripts in adult flies by RU486 feeding is sufficient to cause defective learning, similar to flies that constantly overexpress nebula (Figs. 3B and 4C). This result was further confirmed by transient overexpression of nebula by using the heat-shock driver. Note that the learning impairment is not caused by abnormal sensorimotor responses because both RU486-fed flies and heat-shocked flies responded normally to odor and shock avoidances (Fig. 7). In contrast, Elav-GS/nlat1 flies that overexpress nebula during development, but not at the time of olfactory learning tests, showed normal learning in comparison to control flies that had been raised the same way (Fig. 4C). Together with data that nebula is a regulator of calcineurin-mediated signaling, our data strongly support that biochemical defects, rather than maldevelopment of the brain, are the cause of learning impairment in the transgenic flies.
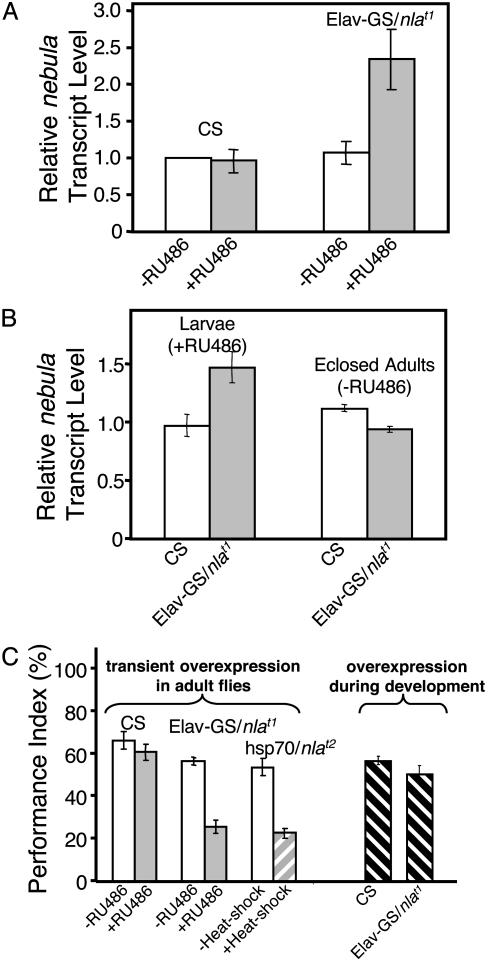
Fig. 4.
Deficient learning in nebula-overexpressing flies is not caused by maldevelopment of the brain. (A) Quantitative real-time RT-PCR detects an increase in the level of nebula transcripts in adult heads 24 h after feeding Elav-GS/nlat1 flies with 100 μg/ml RU486 (+RU486), but not if flies were mock fed (-RU486). (B) Feeding Elav-GS/nlat1 larvae with 20 μg/ml RU486 during development induced nebula overexpression. Adults that had been transferred to normal food media for 3-6 days show normal nebula transcript levels. (C) Performance index values for Pavlovian olfactory learning tests. Transient overexpression of nebula in adult flies (Elav-GS/nlat1 and hsp70/nlat2) impaired learning. However, learning performance of adult Elav-GS/nlat1 flies that had been fed with RU486 during development as described in B was not altered as compared to CS flies that had been raised the same way. For A and B, n = 4 experiments done in triplicate; for C, n = 4 for each strain. All values represent mean ± SEM.
Concluding Remarks. By using Drosophila as a simple model organism, we demonstrated that nebula mediates learning and long-term memory. Our observations that both nebula loss-of-function and overexpression mutants displayed learning defects suggest that precise regulation of nebula-mediated calcineurin signaling is necessary to maintain optimum learning. Furthermore, these results, along with data that the same biochemical pathway is disrupted in human trisomy 21 fetal brain tissue, strongly implicate the involvement of DSCR1 in mental retardation in DS.
Here, we focused our study on the function of DSCR1 in learning and memory to address its role in mental retardation associated with DS. However, DSCR1 is also expressed in other tissues such as the heart and skeletal muscle (3) and may thus regulate diverse calcineurin-dependent processes (33-35). Furthermore, calcineurin has been shown to regulate the transcription of DSCR1, suggesting the existence of a complex negative feedback regulation circuit (36). It will be interesting to investigate whether such regulatory mechanism also controls nebula expression in flies. Combined with its amenability to genetic manipulations and behavioral assays, Drosophila can be used to identify some of the genotype-phenotype relations in DS. In addition, our finding that defective learning in nebula overexpressing flies is likely caused by functional defects rather than developmental defects raises the exciting possibility of pharmacological intervention to ameliorate at least some of the cognitive deficits in DS patients. Drosophila may be used as a tool to rapidly screen for drugs that treat learning and memory deficits by restoring the balance of kinases and phosphatases. Genetic screens that identify suppressors of nebula in the learning and memory pathway will also provide insights into the underlying mechanism of mental retardation in DS.
Supplementary Material
Acknowledgments
We thank A. DiAntonio (Washington University School of Medicine, St. Louis) for providing the Elav-GAL4 line and B. White (National Institutes of Health) for providing the Elav-GeneSwitch line. This research was supported by an intramural grant from the National Institute of Neurological Disorders and Stroke, the National Institutes of Health, and the Ellison Medical Foundation (to K.-T.M.).
Abbreviations: DS, Down's syndrome; DSCR1, DS critical region 1; PKA, cAMP-dependent protein kinase; CREB, cAMP-responsive element binding protein; pCREB, phosphorylated CREB; CS, Canton-S.
References
- 1.Reeves, R. H., Baxter, L. L. & Richtsmeier, J. T. (2001) Trends Genet. 17, 83-88. [DOI] [PubMed] [Google Scholar]
- 2.Epstein, C. J. (1995) in The Metabolic and Molecular Bases of Inherited Disease, eds. Scriver, C. R., Beaudet, A. L., Sly, W. S. & Vaile, D. (McGraw-Hill, New York), pp. 749-794.
- 3.Fuentes, J. J., Pritchard, M. A., Plana, A. M., Bosch, A., Ferrer, I. & Estivill, X. (1995) Hum. Mol. Genet. 4, 1935-1944. [DOI] [PubMed] [Google Scholar]
- 4.Fuentes, J. J., Genescà, L., Kingsbury, T. J., Cunningham, K. W., Pérez-Riba, M., Estivill, X. & de la Luna, S. (2000) Hum. Mol. Genet. 9, 1681-1690. [DOI] [PubMed] [Google Scholar]
- 5.Kingsbury, T. J. & Cunningham, K. W. (2000) Genes Dev. 14, 1595-1604. [PMC free article] [PubMed] [Google Scholar]
- 6.Görlach, J., Fox, D. S., Cutler, N. S., Cox, G. M., Perfect, J. R. & Heitman, J. (2000) EMBO J. 19, 3618-3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casas, C., Martínez, S., Pritchard, M. A., Fuentes, J. J., Nadal, M., Guimerà, J., Arbonés, M., Flórez, J., Soriano, E., Estivill, X., et al. (2000) Mech. Dev. 101, 289-292. [DOI] [PubMed] [Google Scholar]
- 8.Klee, C. B., Ren, H. & Wang, X. (1998) J. Biol. Chem. 273, 13367-13370. [DOI] [PubMed] [Google Scholar]
- 9.Mansuy, I. M., Mayford, M., Jacob, B., Kandel, E. R. & Bach, M. E. (1998) Cell 92, 39-49. [DOI] [PubMed] [Google Scholar]
- 10.Rothermel, B., Vega, R. B., Yang, J., Wu, H., Bassel-Duby, R. & Williams, R. S. (2000) J. Biol. Chem. 275, 8719-8725. [DOI] [PubMed] [Google Scholar]
- 11.Malleret, G., Haditsch, U., Genoux, D., Jones, M. W., Bliss, T. V. P., Vanhoose, A. M., Weitlaug, C., Kandel, E. R., Winder, D. G. & Mansuy, I. M. (2001) Cell 104, 675-686. [DOI] [PubMed] [Google Scholar]
- 12.Zeng, H., Chattarji, S., Barbarosie, M., Rondi-Reig, L., Philpot, B. D., Miyakawa, T., Bear, M. F. & Tonegawa, S. (2001) Cell 107, 617-629. [DOI] [PubMed] [Google Scholar]
- 13.Hamilton, B. A., Palazzolo, M. T., Chang, J. H., VijayRaghavan, K., Mayeda, C. A., Whitney, M. A. & Meyerowitz, E. M. (1991) Proc. Natl. Acad. Sci. USA 88, 2731-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Montell, C., Jones, K., Hafen, E. & Rubin, G. (1985) Science 230, 1040-1043. [DOI] [PubMed] [Google Scholar]
- 15.Ito, K., Awano, W., Suzuki, K., Hirom, Y. & Yamamoto, D. (1997) Development (Cambridge, U.K.) 124, 761-771. [DOI] [PubMed] [Google Scholar]
- 16.Simpson, J. H., Kidd, T., Bland, K. S. & Goodman, C. S. (2000) Neuron 28, 753-766. [DOI] [PubMed] [Google Scholar]
- 17.Armstrong, J. D., de Belle, J. S., Wang, Z. & Kaiser, K. (1998) Learn. Mem. 5, 102-114. [PMC free article] [PubMed] [Google Scholar]
- 18.Osterwalder, T., Yoon, K. S., White, B. H. & Keshishian, H. (2001) Proc. Natl. Acad. Sci. USA 98, 12596-12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brand, A. H., Manoukian, A. S. & Perrimon, N. (1994) Methods Cell Biol. 44, 535-654. [DOI] [PubMed] [Google Scholar]
- 20.Tully, T. & Quinn, W. G. (1985) J. Comp. Physiol. A 157, 263-277. [DOI] [PubMed] [Google Scholar]
- 21.Tully, T., Préat, T., Boynton, S. C. & Del Vecchio, M. (1994) Cell 79, 35-47. [DOI] [PubMed] [Google Scholar]
- 22.Dura, J. M., Préat, T. & Tully, T. (1993) J. Neurogenet. 9, 1-14. [DOI] [PubMed] [Google Scholar]
- 23.Dubnau, J. & Tully, T. (1998) Annu. Rev. Neurosci. 21, 407-444. [DOI] [PubMed] [Google Scholar]
- 24.de Belle, J. S. & Heisenberg, M. (1994) Science 263, 692-695. [DOI] [PubMed] [Google Scholar]
- 25.Crittenden, J. R., Skoulakis, E. M., Han, K. A., Kalderon, D. & Davis, R. L. (1998) Learn. Mem. 5, 38-51. [PMC free article] [PubMed] [Google Scholar]
- 26.Yin, J. C., Wallach, J. S., Del Vecchio, M., Wilder, E. L., Zhou, H., Quinn, W. G. & Tully, T. (1994) Cell 79, 49-58. [DOI] [PubMed] [Google Scholar]
- 27.Drain, P., Folkers, E. & Quinn, W. G. (1991) Neuron 6, 71-82. [DOI] [PubMed] [Google Scholar]
- 28.Goodwin, S. F., Del Vecchio, M., Velinzon, K., Hogel, C., Rossell, S. R. H., Rully, T. & Kaiser, K. (1997) J. Neurosci. 17, 8817-8827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brand, A. H. & Perrimon, N. (1993) Development (Cambridge, U.K.) 118, 401-415. [DOI] [PubMed] [Google Scholar]
- 30.McGuire, S. E., Le, P. T. & Davis, R. L. (2001) Science 293, 1271-1272. [DOI] [PubMed] [Google Scholar]
- 31.Yuan, Q., Harley, C. W., Darby-King, A., Neve, R. L. & McLean, J. H. (2003) J. Neurosci. 23, 4670-4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roman, G., Endo, K., Zong, L. & Davis, R. L. (2001) Proc. Natl. Acad. Sci. USA 98, 12602-12607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crabtree, G. R. & Olson, E. N. (2002) Cell 109, S67-79. [DOI] [PubMed] [Google Scholar]
- 34.Ermak, G., Harris, C. D. & Davies, K. J. (2002) FASEB J. 16, 814-824. [DOI] [PubMed] [Google Scholar]
- 35.Vega, R. B., Rothermel, B. A., Weinheimer, C. J. Kovacs, A., Naseem, R. H., Bassel-Duby, R., Williams, R. S. & Olson, E. N. (2003) Proc. Natl. Acad. Sci. USA 100, 669-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang, J., Rothermel, B., Vega, R. B., Frey, N., McKinsey, T. A., Olson, E. N., Bassel-Duby, R. & Williams, R. S. (2000) Circ. Res. 87, E61-E68. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.