Abstract
With the development of many computational methods that predict the structural models of protein-protein complexes, there is a pressing need to benchmark their performance. As was the case for protein monomers, assessing the quality of models of protein complexes is not straightforward. An effective scoring scheme should be able to detect substructure similarity and estimate its statistical significance. Here, we focus on characterizing the similarity of the interfaces of the complex and introduce two scoring functions. The first, the interfacial Template Modeling score (iTM-score), measures the geometric distance between the interfaces, while the second, the Interface Similarity score (IS-score), evaluates their side chain contact similarity in addition to their geometric similarity. We first demonstrate that the IS-score is more suitable for assessing docking models than the iTM-score. The IS-score is then validated in a large-scale benchmark test on 1,562 dimeric complexes. Finally, the scoring function is applied to evaluate docking models submitted to the Critical Assessment of PRediction of Interactions (CAPRI) experiments. While the results according to the new scoring scheme are generally consistent with the original CAPRI assessment, the IS-score identifies models whose significance was previously underestimated.
Keywords: docking, protein-protein interaction, protein-protein interface, structure prediction, TM-score, IS-score, CAPRI
Introduction
In the quest to determine all protein-protein interactions in a given proteome, recent high-throughput technologies have enabled substantial progress1–3. Drafts for different model systems are emerging, though many details are still missing4–7. The mapping of protein-protein interactions, however, is just a starting point towards revealing their functional roles in living biosystems. In order to understand protein-protein interactions, it is necessary to structurally characterize all representative protein complexes at high resolution8.
Despite rapid growth in the number of structurally solved protein complexes9, the pace of structure determination lags far behind the pace of the detection of protein-protein interactions. To fill this gap, many computational approaches have been proposed for predicting the structures of protein complexes. They can be roughly categorized into two types: Template-Based (TB) and Template-Free (TF). In TB approaches10–15, one first builds a homology model based on a solved template structure, and then refines the model. In TF approaches16–23, also known as protein-protein docking methods, one docks unbound components that form the target complex. Both methods have advantages and disadvantages. TB approaches generally have higher accuracy, but suffer from low coverage because of their dependence on the availability of template structures. Although the issue of low coverage might be overcome by the recognition that the structural space of protein-protein interfaces is highly degenerate24, in practice identifying which protein pairs actually interact is very challenging. On the other hand, TF approaches can deal with a novel target whose quaternary structure does not match any solved template structure, but there is no guarantee of high quality docking models, particularly when bound structures undergo significant conformational changes from the unbound structures25. Furthermore, TF approaches require the information that two input proteins interact; that is, they are not reliable in predicting whether two proteins interact or not, largely due to the limitations of force fields used for evaluating interaction energy26. By comparison, TB methods usually contains (explicitly or implicitly) an evolutionary component, which prefers templates sharing conserved biological interactions with target proteins. Thus, in addition to predicting the structure of protein interactions, TB methods may be used to predict whether two proteins interact.
To benchmark the performance of docking methods, a community-wide experiment, known as CAPRI, has been carried out27–29. One central task is to measure the quality of a predicted docking model, using its target structure, usually a solved crystal structure, as the gold standard. Furthermore, in the case of template-based modeling, it is also critical to measure the quality of both the template and the final model. Thus, any improvement or deterioration resulting from the “refinement” procedure, designed to improve over the template alignment, can be evaluated. For these purposes, one needs to derive effective structure comparison metrics. The CAPRI assessors employed complex criteria based on the Root Mean Square Deviation (RMSD) and the fraction of conserved native contacts fnat29. While these criteria are convenient, they have three limitations: The first is that RMSD is often dominated by the largest deviations, and hence, may overlook substructure similarity. The second is that the statistical significance of a given RMSD value is length dependent30. The third is that the thresholds employed for model quality classification are often subjective, in the sense that an assessment of the statistical significance of the given structural comparison metric is lacking.
The problem of model quality assessment is not unique to protein docking experiments. An analogous situation was encountered in the evaluation of structural models predicted for monomeric proteins. In the recent Critical Assessment of Protein Structure Prediction (CASP), several commonly used scoring functions include the Global Distance Test (GDT) score31, the MaxSub score32, and the TM-score33. The statistical significance relative to random of both the GDT and the MaxSub scores are sensitive to the size of the target protein33. As a result, one often cannot tell whether a raw score indicates a significant prediction. By contrast, the TM-score corrects for length effects. Based on the statistics obtained from comparing random protein structures at various lengths, a TM-score of 0.4 or higher indicates a significant prediction33. Other statistically rigorous treatments also have been undertaken to calculate the significance (i.e., P-values) of protein models34,35.
Previously, we introduced the iTM-score and the IS-score in iAlign36, a program for the structural comparison of protein-protein interfaces based on interface structure alignments, where the equivalence of target and template residues is not a priori specified. It has been shown that the IS-score is an effective metric for evaluating structural alignments of protein-protein interfaces24,36. In this study, we examine both scoring functions for measuring the quality of docking models. The key difference between the previous study and the current one is that iAlign does not require any previously specified sequence correspondence, whereas in the current scenario, the mapping of equivalent target-template residues is specified in advance. As a result, one needs to adjust the random background and re-calibrate the statistical models, as detailed below. Furthermore, we performed large-scale benchmark tests to compare and validate our scoring schemes and applied the IS-score to docking models submitted to the CAPRI experiments.
Methods
A heavy-atom distance cutoff of 4.5 Å is employed to define an interfacial contact. A protein-protein interface is the collection of all residues with at least one interfacial contact between pairs of proteins.
Scoring function and search algorithm
Assuming that a native (target) structure has L interfacial residues, the iTM-score of a corresponding docking model is defined by comparing the geometric distances of the native interfacial residues of the model and the native structure33,36,
| (Eq. 1) |
where Na is the number of superimposed native interfacial residues, di is the Euclidean distance between the Cα atoms from the ith superimposed residue pair, and the empirical scaling factor is introduced to correct for length effects. Note that the definition of the iTM-score is exactly the same as used for assessing the model quality of the global structure alignment of monomeric proteins33. However, the TM-scores of interfaces and of individual proteins have a different level of statistical significance at the same numerical value (see below). To avoid confusion, we use the term iTM-score to denote the TM-score of interfaces and reserve the notation TM-score for the global comparison of a pair of structures.
In order to calculate the distance di, a subset of corresponding residues are superimposed using the Kabsch algorithm37, which minimizes their pairwise root-mean-square deviation, RMSD. Since there are many ways to select the subset, the notation max in Eq. 1 indicates that the iTM-score is the maximum out of all possible superimpositions. A heuristic iterative extension algorithm is employed to calculate the iTM-score33, similar to the one used for calculating the GDT-score31 and MaxSub32. Briefly, we select fragments of size Lsub = L, L/2, L/4, …, 4, respectively. When Lsub is less than L, initial fragments are selected by sliding continuously along the native interface. Starting from an initial fragment of size Lsub, the corresponding residues within Lsub in the model and native interfaces are superimposed. Then, all model/interface residue pairs within a distance less than d0 are collected and superimposed again. The process is iterated until the rigid-body transformation converges.
The second scoring function is the Interface Similarity score (IS-score), which measures not only geometric distances but also the conservation of interfacial contacts36. The IS-score is derived from the iTM-score as follows,
| (Eq. 2) |
| (Eq. 3) |
Here, the contact overlap factor fi ≡ (ci / ai + ci / bi) / 2, where ai is the number of interfacial contacts observed at the ith position of the native interface, bi is the number of interfacial contacts observed at the corresponding position in the model, and ci is the number of interfacial contacts conserved in both interfaces. If ci = 0, fi is 0, regardless of the value of bi. The scaling factor is introduced to make the means of the IS-scores length-independent among randomly selected interfaces (see below). Note that the scaling factor is slightly different from what was derived previously in iAlign36. The adjustment is introduced to correct for a small shift in the means of the IS-scores among random interfaces. The search algorithm for calculating the IS-score is essentially the same as describe above for the iTM-score.
Both the iTM/IS-score give a maximum score of one for a perfect model.
Statistical significance
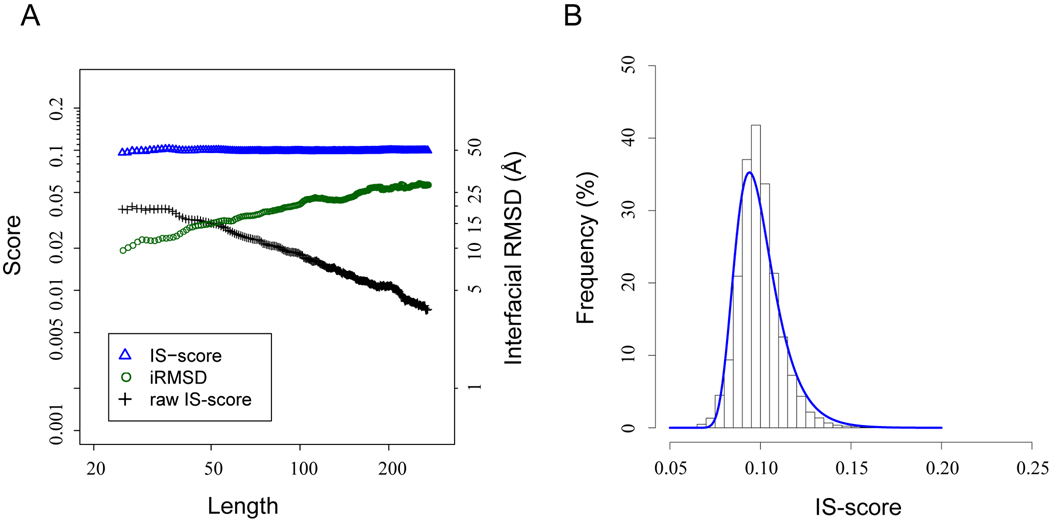
The statistical significance of the IS-score is estimated by comparing 24,120 randomly selected interface pairs of same lengths (see Data Set). In each pair, interfacial residues at the same positions in respective sequences are arbitrarily assigned as equivalent. Figure 1A shows the means of the IS-scores and iRMSD values of unrelated interfaces. Without applying the scaling factor, the raw IS-score calculated using Eq. 3 decreases exponentially as the length of the interface increases. Likewise, the mean random iRMSD value increases exponentially as the interface size increases. By comparison, the re-scaled IS-scores are approximately length-independent at a mean value of 0.10. It should be noted that the mean of random IS-scores calculated here is smaller than the mean of random IS-scores calculated previously with the program iAlign36. The reason is that iAlign does not a priori impose a one-to-one sequence correspondence. Therefore, iAlign usually finds a better correspondence (or alignment), which gives a higher IS-score even for randomly related interfaces.
Fig. 1.
Distributions of randomly selected protein-protein interface pairs. (A) Mean of IS-scores and interfacial RMSD values versus the size of protein interfaces. Horizontal dashed lines are located at 0.1. (B) Distribution of the IS-score among random interfaces. The histogram is the observed score distribution, and the solid line is the fit according to the Gumbel distribution (Eq. 5).
Since the IS-scores are maxima, the extreme value distribution is a suitable statistical model for describing their distribution. As shown in Figure 1B, the probability density function of the IS-scores calculated from the random background follows the extreme value distribution,
| (Eq. 4) |
where z denotes the Z-score given by z = (s − μ) / σ. The variable s denotes the IS-score; μ is the location parameter, and σ is the scale parameter. The corresponding P-value of the score can be calculated according to the formula
| (Eq. 5) |
The scores from random interfaces were fit to Eq. 4. The resulting P-values and their corresponding IS-scores are given in Table I. The calculated P-values according to the statistical model agree with the empirical values obtained by ranking the IS-scores of 24,120 random interface pairs. One may use these scores to quickly estimate statistical significance.
Table I.
Statistical significance of the IS-scores derived from 24,120 pairs of random interfaces.
| P-value | IS-score | |
|---|---|---|
| Model | Empirical | |
| 0.05 | 0.125 | 0.120 |
| 0.01 | 0.142 | 0.134 |
| 0.005 | 0.149 | 0.141 |
| 0.001 | 0.166 | 0.160 |
| 1e-04 | 0.190 | 0.187 |
| 1e-05 | 0.214 | - |
| 1e-06 | 0.238 | - |
| 1e-08 | 0.286 | - |
| 1e-10 | 0.334 | - |
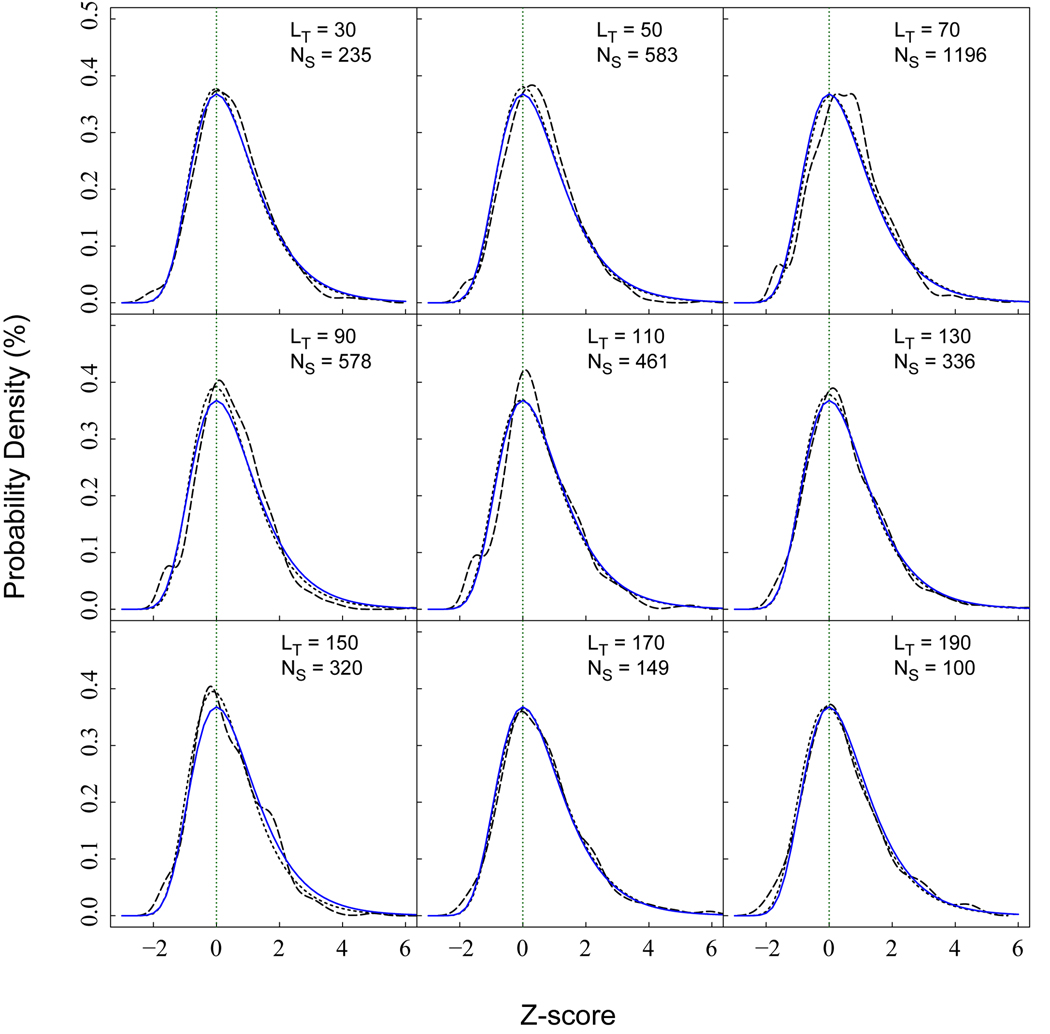
An improved estimation of statistical significance is obtained by modeling the distributions of scores at specific lengths. Fig. 2 shows the observed and modeled distributions at various lengths. Each distribution is modeled by the Gumbel distribution described in Eq. 4. The location and scale parameters can be estimated through linear regression fits,
| (Eq. 6) |
Fig. 2.
Distributions of the Z-score among random interfaces of various lengths. Long dashed lines are the observed probability density, and the short dashed lines are direct fits using the Gumbel distributions. Solid lines are probability densities calculated for the IS-scores with statistical models described by Eq. 4 and 6. LT represents the length of query, and NS is the number of samples.
The parameters a to d, given in Table II, were obtained by linear fitting to the location and scale parameters, which were obtained through maximum likelihood estimates with the EVD package in the statistical platform R (http://www.r-project.org/).
Table II.
Parameters for calculating the location and scale parameters in Eq. 6.
| Parameters | IS-score |
|
|---|---|---|
| LQ < 55 | LQ ≥ 55 | |
| a | 0.0806 | 0.0794 |
| b | 0.0034 | 0.0033 |
| c | 0.0277 | 0.0794 |
| d | −0.0040 | −0.0054 |
Analysis measures
In addition to the iTM/IS-score, we also define common metrics adopted for evaluating docking models29. The smaller/larger of the two monomers in a binary complex are termed as the ligand/receptor of the complex. Let Nc denote the number of interfacial contacts observed in the native complex structure, and n the number of native interfacial contacts preserved in the docking model. The fraction of native contacts is fnat ≡ n/Nc. The interfacial RMSD, iRMSD, is the RMSD of the Cα atoms of interfacial residues observed in a native structure with respect to their positions in a docking model, and the ligand RMSD, lRMSD, is the global RMSD of the Cα atoms of all ligand residues. The iRMSD is calculated after superimposing these native interfacial residues, whereas the lRMSD is calculated after superimposing the receptors.
Data sets
(i) Random Background
The random background for statistical significance analysis was derived from the M-TASSER template library11. We first obtained all-against-all pairs of all dimeric complexes. A pair of dimers was then selected, if any two monomers, one from each dimer, have a global sequence identity < 30% and a global TM-score < 0.4. This selection led to a set of globally unrelated dimer pairs. Since IS-score requires that the two interfaces have the same length, we randomly removed interfacial residues of the longer interface, if the two interfaces are of different size. The removal was carefully done by requiring that all remaining interfacial residues maintain at least one interfacial contact. To prevent possible over-representation of any given dimer, we further required that no dimer appears more than 20 times in the final selections. The procedure yielded 24,120 pairs of interfaces, which were used for estimating the statistical significance of the IS-score. In each pair, two interfacial residues were assigned as equivalent if they appear at the same positions in respective sequences after removing all non-interfacial residues.
(ii) Decoy Set
For the comparison between the iTM-score and the IS-score, we used a decoy set from the Dockgound38. The decoy set was curated from docking models generated with unbound protein structures for 61 target complexes. We further define a near native docking model if it has lRMSD ≤ 5 Å and fnat > 30%, and define an incorrect model if it has lRMSD > 5 Å and fnat = 0%. The procedure produced 425 near native models and 5,232 incorrect models.
(iii) Docking Set
From the M-TASSER template library11, we selected 1,526 complexes whose individual proteins are less than 500 amino acids in length. Rigid-body docking using the bound structures from the complexes were subsequently carried out with the program FT-Dock23 using default parameters. The top 100 docking models, ranked by shape complementarity, were retained for validating the statistical significance of the IS-scores. In total, we collected 152,600 models by pooling together the top 100 docking models from all complexes.
(iv) CAPRI Models
The docking models for recent CAPRI targets were downloaded from the official web site (http://www.ebi.ac.uk/msd-srv/capri/). We selected ten recent protein-protein targets (T24 – T36, except for cancelled T26, and RNA/protein targets T33 and T34), for which the docking models were available to the public. The criteria adopted by the CAPRI assessors for model quality evaluation are the following29:
High – fnat ≥ 0.5 & (lRMSD ≤ 1 Å ‖ iRMSD ≤ 1 Å),
Medium – (fnat ≥ 0.5 & lRMSD > 1 Å & iRMSD > 1 Å) ‖ (fnat ≥ 0.3 & fnat < 0.5 & lRMSD ≤ 5 Å & iRMSD ≤ 2 Å),
Acceptable – (fnat ≥ 0.3 & lRMSD > 5 Å & iRMSD > 2 Å) ‖ (fnat ≥ 0.1 & fnat < 0.3 & lRMSD ≤ 10 Å & iRMSD ≤ 4 Å),
Incorrect – fnat < 0.1 ‖ (lRMSD > 10 Å & iRMSD > 4 Å).
The notions & and ‖ denote logical conjunction and disjunction, respectively. It should be noted that the CAPRI assessors employed distance cutoffs of 5 and 10 Å to define interfacial residues separately for calculating fnat and iRMSD. In this study, we only used the final assessments (i.e., High, Medium, Acceptable, and Incorrect) provided by the CAPRI assessors.
Availability
The data sets and IS-score software package including the source code are freely available at http://cssb.biology.gatech.edu/isscore.
Results
IS-score versus iTM-score
We first compare the performance of the IS-score and iTM-score on evaluating the quality of docking models. For this comparison, we selected 425 near native and 5,232 incorrect docking models from a Dockgound decoy set generated with unbound protein structures (see Methods). As shown in Fig. 3A, the distributions of the IS-scores for near native and incorrect docking models are well separated. Near native docking models all have an IS-score above 0.17, and 97% of the IS-scores > 0.25, whereas incorrect models all have the scores < 0.12. By comparison, an overlapping regime in the iTM-scores is observed between near native and incorrect models. Incorrect docking models have their iTM-scores ranging from 0.33 to 0.68; and a peak is observed at 0.5. The peak is due to the superimposition of one side of the protein interface. Most unbound protein structures used for docking are structurally very close to their bound structural forms. In these cases, at least half of a native interface can be superimposed to its counterpart in a docking model, despite the fact that the other side of the interface is far away from it native position in an incorrect model. Such superimposition gives a significant iTM-score > 0.4, as overlapping the score regime of the near native models from 0.4 to 0.9.
Fig. 3.
Comparison of the iTM/IS-scores for assessing the quality of protein docking models. (A) Score distributions of incorrect docking models and of near native docking models. iTMS and ISS denote iTM-score and ISS score, respectively. (B) ROC curves of sensitivity versus false positive rate.
The performance of IS-score and iTM-score is further displayed in the Receiver Operating Characteristic (ROC) curves (Fig. 3B), where the sensitivity is the fraction of near-native models, and the false positive rate is the fraction of incorrect models. The ROC curves were obtained by varying the thresholds of the iTM/IS-score. The IS-score has a perfect ROC curve with the value of AUC0.2 (Area Under Curve up to a 20% false positive rate) of 1, whereas the iTM-score has an AUC0.2 value of 0.76. Overall, the analysis demonstrates that a similarity metric based purely on geometric distances has an intrinsic flaw for evaluating docking models and that the IS-score yields a much more accurate assessment by taking interfacial contacts explicitly into account.
Discriminating docking models
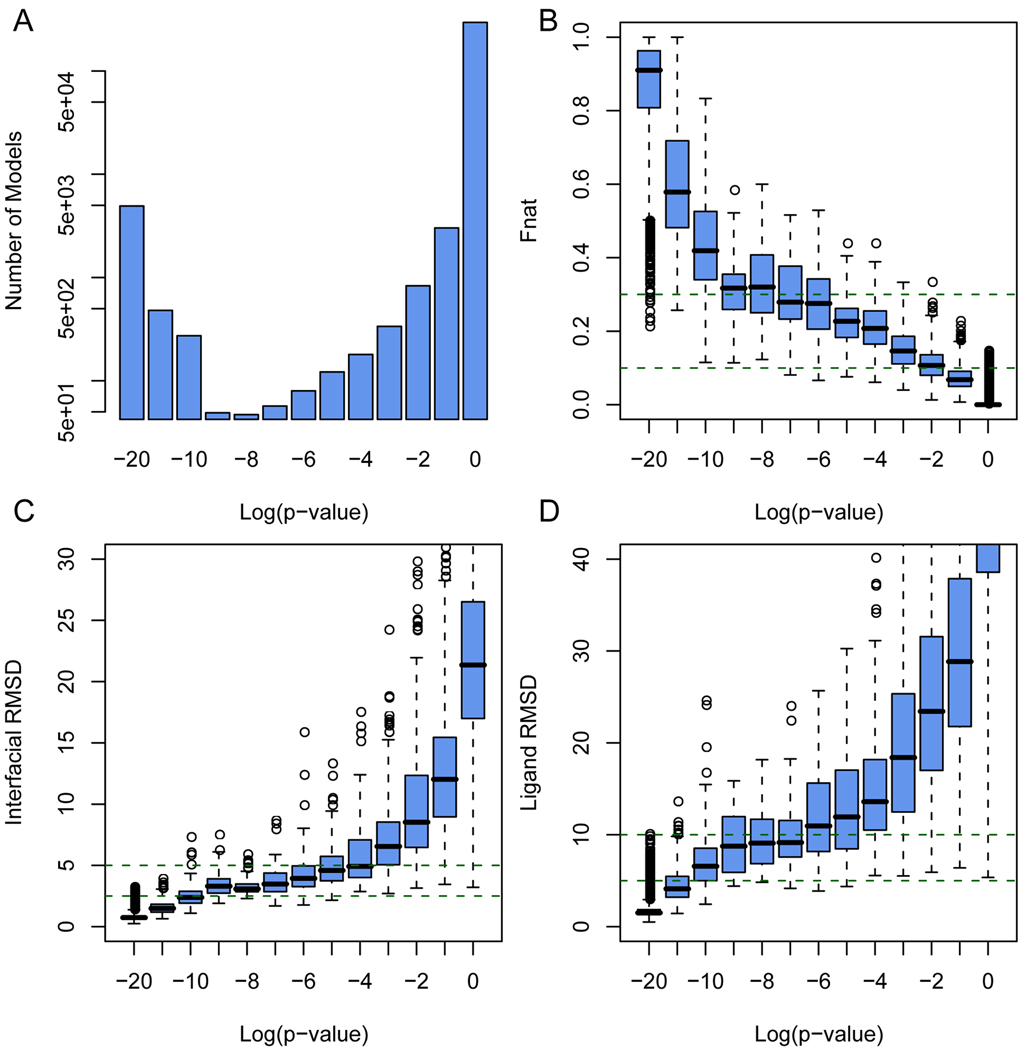
To further examine whether the IS-score returns a reasonable estimate of statistical significance, we further performed large scale tests on a total of 152,600 docking models for the 1,526 target complexes. Each model was assessed according to the IS-score with respect to the native structure. As expected, the vast majority (96%) of these models have an insignificant IS-score with P > 0.01, while a small fraction (3.2%) of docking models resemble the native structure at a high level of similarity with P < 1×10−10 (Fig. 4A).
Fig. 4.
Quality assessments of 152,600 docking models generated for 1,526 protein complexes. (A) Number of docking models according to the IS-score P-values. Box plots of docking models according to (B) fraction of native contacts preserved in models, (C) interfacial and (D) ligand RMSDs. The lower, middle and upper quartiles of each box are the 25th, 50th, and 75th percentile; whiskers extend to a distance of up to 1.98 times the interquartile range. Outliers and means are represented by circles.
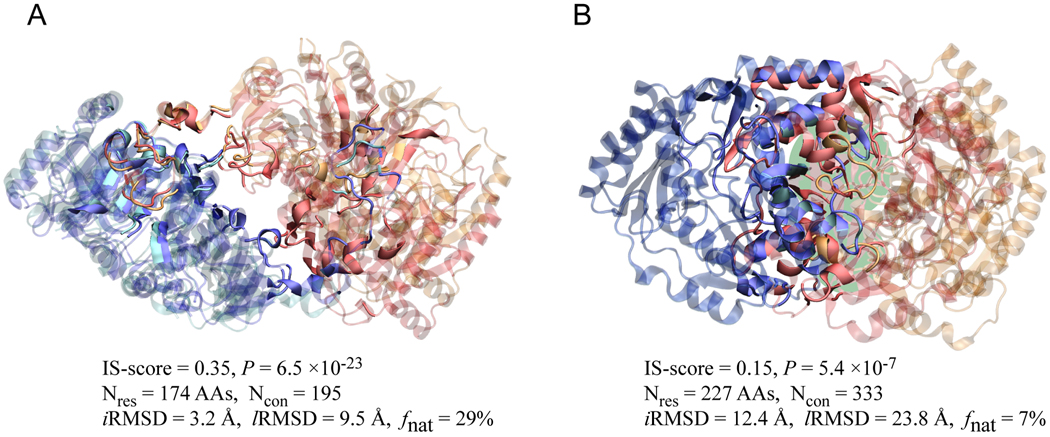
As shown in Fig. 4B and C, all docking models within 2.5 Å iRMSD from native structures or with a fnat value > 30% have a significant P better than 1×10−6, mostly, better than 1×10−10. Conversely, almost all interfaces with P < 1×10−6 have an iRMSD of less than 2.5 Å and a fnat of more than 30%. In rare exceptions, a docking model has a significant P < 1×10−6, while exhibiting a relatively high iRMSD/lRMSD > 3/8 Å and low native contacts < 30%. These cases are from docking very large complexes with usually more than 150 interfacial amino acids. Two cases are shown in Fig. 5. Despite a high lRMSD of 9.5 Å, visual inspection suggests that the docking model shown in Fig. 5A resembles very well the native structure, validating the estimated high P-value of 6.5×10−23. In Fig. 5B, the docking model has a P of 5.4×10−7, due to the maintenance of 22 native contacts, despite a different orientation from the native docking pose.
Fig. 5.
Two docking models for (A) a putative citrate lyase (PDB code: 1xr4, chain A and B) and (B) an aminotransferase (PDB code: 1dty, chain A and B). In each snapshot, the two chains from docking model are colored in cyan/orange, and the corresponding chains in the native structures are colored in blue/red. For clarity, interface/non-interface regions are shown in solid/transparent colors, respectively. Overlapped interface regions are indicated by a green background in (B). Molecular images were created with VMD39. The length of the interface and the number of interfacial contacts are denoted as Nres and Ncon, respectively.
Virtually all insignificant models at P > 0.01 has an iRMSD > 3 Å and fnat < 10%. About 1% of docking models exhibit an interface that bears a significant similarity to the native interface with a P between 0.01 and 1×10−6. These model interfaces typically have a iRMSD between 5 to 10 Å and preserve 10 to 30% of native contacts. They usually overlap a part of the native interface.
Assessing CAPRI models
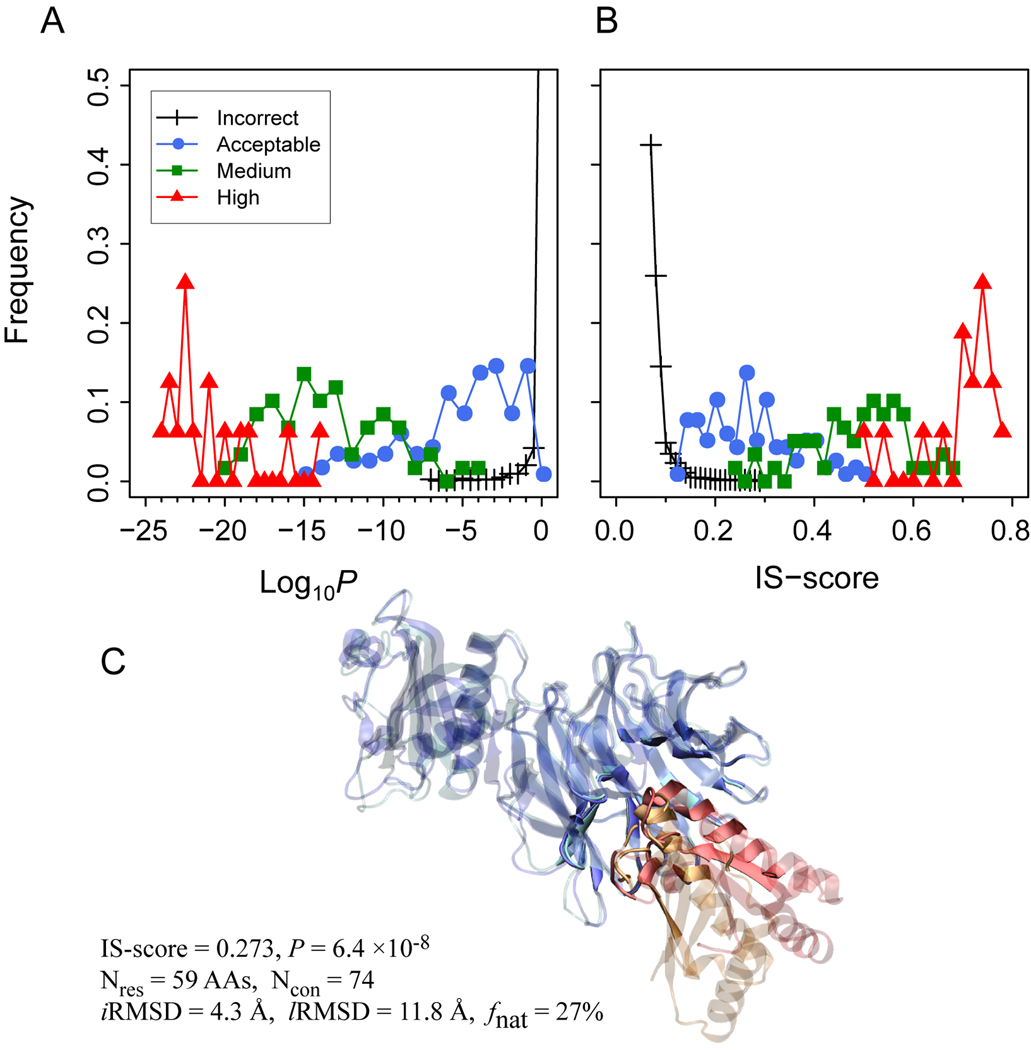
Finally, we applied the IS-score to assess the quality of docking models submitted by various research groups for ten recent CAPRI targets. The results of the IS-score evaluations are compared to the official assessments provided by the CAPRI organizers, who categorized each model into one of four groups: Incorrect, Acceptable, Medium, and High, according to iRMSD, lRMSD, and fnat (see Methods). A total of 2,874 Incorrect, 117 Acceptable, 59 Medium, and 16 High quality models for these ten targets were evaluated. Consistent with the CAPRI assessments, the overall distributions of the four groups of docking models are clearly separated according to either the IS-scores or their P-values (Fig. 6). The means of the IS-scores/Log10P are 0.08/−0.21 (I), 0.26/−5.7 (A), 0.48/−14.0 (M), and 0.69/−21.2 (H), respectively.
Fig. 6.
Distribution of CAPRI models according to (A) the IS-score P-values and (B) the IS-score. Legends indicate model quality provided by the CAPRI assessors. (C) One example (model ID: T26_P41.M02) of CAPRI docking models for target T26. The model was categorized as Incorrect according to the CAPRI assessors, but shows significant interface similarity. The coloring scheme is the same as that employed in Fig. 5.
Out of 192 models with Acceptable or better quality, 174 (91%) and 185 (96%) have a significant P < 0.01 and 0.05, respectively. Only seven Acceptable models have a P > 0.05. These models, from targets T24, T25, T27, and T29, have about 10% to 15% native contacts correctly modeled. However, the numbers of preserved native contacts are small, considering that their native interfaces consist of about 40 native contacts or less. On the other hand, a total of 263 models have a significant similarity to their target interface at a P < 0.01 or better. Among these significant models, 89 were assigned as Incorrect. Most of these significant Incorrect models are from targets T26 and T32, which have relatively large interfaces with more than 60 native contacts. The difference between the CAPRI and IS-score assessments can be attributed to two main reasons. First, the CAPRI assessment uses fnat and RMSDs, with a size-dependence issue, whereas the IS-score takes the length effect into account. Second, the IS-score only considers interface similarity but ignores global orientation. A slight rotation could lead to a large lRMSD, despite the fact that the iRMSD is relatively small. An example of an Incorrect model with significant interface similarity is shown in Fig. 6C. Visual inspection suggests that the model has good interface similarity at iRMSD of 4.3 Å and 27% fnat. However, a slight tilt around the interface leads to a large lRMSD of 12 Å.
Fig. 7 shows the quality of individual docking models for each target. For all targets with the exception of T25, unbound or homology structures were provided as the starting structures for docking experiments. Overall, it is clear that higher ranked models have better quality, consistent with the official assessments. In particular, for targets T35 and T36, where only one Acceptable model was found, these two Acceptable models were the best as assessed by the P-value of the IS-score. Additionally, corresponding to the assessment that no Acceptable model was found for T28, the top ranked docking model of the same target has a marginal P value of 0.047.
Fig. 7.
The quality of individual docking models submitted by different research groups for ten CAPRI targets. The target ID is shown in the lower left corner of each plot. The horizontal dashed lines are located at P = 0.05 according to the IS-score.
Discussion and Conclusion
Currently, iRMSD and lRMSD are metrics commonly employed in docking studies. The major advantages of the RMSD metrics are two-fold: First, the overall quality of a docking model is guaranteed if one uses a very conservative RMSD criterion; second, the calculation of RMSD is very straightforward. However, RMSD metrics also have two significant disadvantages. First, it is well-known that the statistical significance of a given RMSD value is length dependent (e.g., Fig. 1A). As a result, there is no straightforward relationship between RMSD values and the statistical significance of docking models. This is reflected in a simple fact that, at the same iRMSD value (e.g., 3 Å), to build a docking model for a 100 residue interface is more difficult than for a 30 residue interface. In addition, RMSD metrics are global metrics, meaning that local similarity may not be properly characterized by RMSDs. One extreme example is shown in Fig. 8, where the docking model has a highly significant IS-score of 0.43 (P = 2×10−18), despite a large iRMSD value of 13.2 Å, caused by the rotations of two helical segments. Other than the two helical segments, the remainder (60%) of the interface superimposes with an RMSD of less than 2 Å between the model and the native structure. Obviously, the model in this example is not a random prediction. For the purpose of assessing a docking method, it is important to differentiate such a case from a random model prediction. Overall, one should be cautious in using RMSD metrics to assess the quality of a docking model.
Fig. 8.
An example illustrates that local structural similarity is captured by the IS-score but not by iRMSD. The model structure is superimposed onto the native structure in (A) top view and (B) side view. The model structure overlaps the native structure (PDB code 2cwq) in the interface region, except for two helical segments (labeled as H1 and H2) exhibiting almost 180 degree of rotations, one of which is indicated by a grey arrow. Interfacial regions of the native structure and the corresponding residues in the model structure are shown in solid colors, and other regions are transparent.
We have introduced and examined the performance of two scoring schemes, the iTM-score and the IS-score, for use in assessing the quality of protein-protein docking models. Both scores are able to detect significant substructure similarity if it exists. While the iTM-score is based on geometric distances, the IS-score combines both interfacial contacts and geometric distances. In benchmark tests of 425 near native models and 5,232 randomly related, incorrect models, generated from rigid-body docking of unbound protein structures, the IS-score achieves a perfect classification at an AUC0.2 value of 1, whereas the iTM-score gives an inferior performance at an AUC0.2 value of 0.76. The main issue with the iTM-score is that the interaction pose is not explicitly taken into account. As a result, an artificially high iTM-score may be obtained through the superimposition of one side of the interface, while the other side of the interface may be far away from its native position. The issue is intrinsic to all scoring functions based solely on geometric distances. By comparison, the introduction of the contact overlap factor in the IS-score scheme eliminates this issue.
For a proper model quality assessment, it is important to assess statistical significance of predicted models. Using random interfaces as the background, we have derived statistical models for estimating the significance of the IS-score. The estimation is validated on 156,200 randomly selected docking models. Virtually all highly significant interfaces with P > 10−6 are native-like, and conversely, all native-like docking models display a highly significant P > 10−6, mostly > 10−10. By contrast, insignificant models with P > 10−6 have a iRMSD > 3 Å and a fnat < 10%. Models with P between 0.01 and 1×10−6 have some interfacial similarity, but may exhibit a rotation that gives a relatively large lRMSD.
The IS-score is further applied to evaluate the docking models for ten recent CAPRI targets. Overall, the evaluation of the IS-score is consistent with the official CAPRI assessment. On average, the mean of the IS-scores are 0.26, 0.48, and 0.69, for Acceptable, Medium, and High resolution models, respectively. However, it appears that the official assessment is somewhat conservative. According to the P-values of the IS-scores, we identified quite a few models whose significance is under-estimated. The IS-score scheme is conceptually simple and statistically sound. One further application of the scheme is to use it as the objective function for method optimization.
Acknowledgments
This work is supported by the National Institutes of Health Grant No. GM-48835.
References
- 1.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422(6928):198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 2.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17(10):1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 3.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403(6770):623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 4.Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440(7084):631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 5.Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302(5651):1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 6.Krogan NJ, Cagney G, Yu HY, Zhong GQ, Guo XH, Ignatchenko A, Li J, Pu SY, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440(7084):637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 7.Li SM, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JDJ, Chesneau A, Hao T, et al. A map of the interactome network of the metazoan C-elegans. Science. 2004;303(5657):540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russell RB, Alber F, Aloy P, Davis FP, Korkin D, Pichaud M, Topf M, Sali A. A structural perspective on protein-protein interactions. Curr Opin Struct Biol. 2004;14(3):313–324. doi: 10.1016/j.sbi.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 9.Tuncbag N, Gursoy A, Guney E, Nussinov R, Keskin O. Architectures and functional coverage of protein-protein interfaces. J Mol Biol. 2008;381(3):785–802. doi: 10.1016/j.jmb.2008.04.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aloy P, Russell RB. Interrogating protein interaction networks through structural biology. Proc Natl Acad Sci USA. 2002;99(9):5896–5901. doi: 10.1073/pnas.092147999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen HL, Skolnick J. M-TASSER: An algorithm for protein quaternary structure prediction. Biophys J. 2008;94(3):918–928. doi: 10.1529/biophysj.107.114280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu L, Lu H, Skolnick J. MULTIPROSPECTOR: An algorithm for the prediction of protein-protein interactions by multimeric threading. Proteins: Struct Funct Genet. 2002;49(3):350–364. doi: 10.1002/prot.10222. [DOI] [PubMed] [Google Scholar]
- 13.Gunther S, May P, Hoppe A, Frommel C, Preissner R. Docking without docking: ISEARCH--prediction of interactions using known interfaces. Proteins. 2007;69(4):839–844. doi: 10.1002/prot.21746. [DOI] [PubMed] [Google Scholar]
- 14.Keskin O, Nussinov R, Gursoy A. PRISM: protein-protein interaction prediction by structural matching. Methods Mol Biol. 2008;484:505–521. doi: 10.1007/978-1-59745-398-1_30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sinha R, Kundrotas PJ, Vakser IA. Docking by structural similarity at protein-protein interfaces. Proteins. 2010;78(15):3235–3241. doi: 10.1002/prot.22812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sandak B, Wolfson HJ, Nussinov R. Flexible docking allowing induced fit in proteins: Insights from an open to closed conformational isomers. Proteins: Struct Funct Genet. 1998;32(2):159–174. [PubMed] [Google Scholar]
- 17.Chen R, Li L, Weng ZP. ZDOCK: An initial-stage protein-docking algorithm. Proteins: Struct Funct Genet. 2003;52(1):80–87. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- 18.Dominguez C, Boelens R, Bonvin A. HADDOCK: A protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125(7):1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 19.Fernandez-Recio J, Totrov M, Abagyan R. Soft protein-protein docking in internal coordinates. Protein Sci. 2002;11(2):280–291. doi: 10.1110/ps.19202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozakov D, Brenke R, Comeau SR, Vajda S. PIPER: An FFT-based protein docking program with pairwise potentials. Proteins: Struct Funct Bioinform. 2006;65(2):392–406. doi: 10.1002/prot.21117. [DOI] [PubMed] [Google Scholar]
- 21.Vakser IA. Protein docking for low-resolution structures. Protein Eng. 1995;8(4):371–377. doi: 10.1093/protein/8.4.371. [DOI] [PubMed] [Google Scholar]
- 22.Wang C, Bradley P, Baker D. Protein-protein docking with backbone flexibility. J Mol Biol. 2007;373(2):503–519. doi: 10.1016/j.jmb.2007.07.050. [DOI] [PubMed] [Google Scholar]
- 23.Gabb HA, Jackson RM, Sternberg MJE. Modelling protein docking using shape complementarity, electrostatics and biochemical information. J Mol Biol. 1997;272(1):106–120. doi: 10.1006/jmbi.1997.1203. [DOI] [PubMed] [Google Scholar]
- 24.Gao M, Skolnick J. Structural space of protein-protein interfaces is degenerate, close to complete, and highly connected. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1012820107. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonvin AM. Flexible protein-protein docking. Curr Opin Struct Biol. 2006;16(2):194–200. doi: 10.1016/j.sbi.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 26.Kastritis PL, Bonvin AM. Are scoring functions in protein-protein docking ready to predict interactomes? Clues from a novel binding affinity benchmark. J Proteome Res. 2010;9(5):2216–2225. doi: 10.1021/pr9009854. [DOI] [PubMed] [Google Scholar]
- 27.Janin J. Assessing predictions of protein-protein interaction: The CAPRI experiment. Protein Sci. 2005;14(2):278–283. doi: 10.1110/ps.041081905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lensink MF, Mendez R, Wodak SJ. Docking and scoring protein complexes: CAPRI 3rd edition. Proteins: Struct Funct Bioinform. 2007;69(4):704–718. doi: 10.1002/prot.21804. [DOI] [PubMed] [Google Scholar]
- 29.Mendez R, Leplae R, De Maria L, Wodak SJ. Assessment of blind predictions of protein-protein interactions: Current status of docking methods. Proteins: Struct Funct Bioinform. 2003;52(1):51–67. doi: 10.1002/prot.10393. [DOI] [PubMed] [Google Scholar]
- 30.Reva BA, Finkelstein AV, Skolnick J. What is the probability of a chance prediction of a protein structure with an rmsd of 6 angstrom? Fold Des. 1998;3(2):141–147. doi: 10.1016/s1359-0278(98)00019-4. [DOI] [PubMed] [Google Scholar]
- 31.Zemla A. LGA: a method for finding 3D similarities in protein structures. Nucleic Acids Res. 2003;31(13):3370–3374. doi: 10.1093/nar/gkg571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siew N, Elofsson A, Rychiewski L, Fischer D. MaxSub: an automated measure for the assessment of protein structure prediction quality. Bioinformatics. 2000;16(9):776–785. doi: 10.1093/bioinformatics/16.9.776. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins: Struct Funct Bioinform. 2004;57(4):702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]
- 34.Levitt M, Gerstein M. A unified statistical framework for sequence comparison and structure comparison. Proc Natl Acad Sci USA. 1998;95(11):5913–5920. doi: 10.1073/pnas.95.11.5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ortiz AR, Strauss CEM, Olmea O. MAMMOTH (Matching molecular models obtained from theory): An automated method for model comparison. Protein Sci. 2002;11(11):2606–2621. doi: 10.1110/ps.0215902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao M, Skolnick J. iAlign: a method for the structural comparison of protein-protein interfaces. Bioinformatics. 2010;26(18):2259–2265. doi: 10.1093/bioinformatics/btq404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kabsch W. Solution for best rotation to relate two sets of vectors. Acta Crystallogr Sect A. 1976;32(SEP1):922–923. [Google Scholar]
- 38.Liu S, Gao Y, Vakser IA. DOCKGROUND protein-protein docking decoy set. Bioinformatics. 2008;24(22):2634–2635. doi: 10.1093/bioinformatics/btn497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graphics. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]