Abstract
Polyketides represent a significant fraction of all natural products. Many possess pharmacological activity which makes them attractive drug candidates. The production of the parent macrocyclic aglycones are catalyzed by multimodular polyketide synthases utilizing short-chain acyl-CoA monomers. When producing polyketides through heterologous hosts, one must not only functionally express the synthase itself, but activate the machinery used to generate the required substrate acyl-CoA’s. As a result, metabolic engineering of these pathways is necessary for high-level production of heterologous polyketides. In this study, we over-express three different pathways for provision of the two substrates (propionyl-CoA and (2S)-methylmalonyl-CoA) utilized for the biosynthesis of 6-deoxyerythronolide B (the macrolactone precursor to erythromycin): 1) a propionate → propionyl-CoA → (2S)-methylmalonyl-CoA pathway, 2) a methylmalonate → methylmalonyl-CoA → propionyl-CoA pathway, and 3) a succinate → succinyl-CoA → (2R)-methylmalonyl-CoA → (2S)-methylmalonyl-CoA → propionyl-CoA pathway. The current study revealed that propionate is a necessary component for > 5 mg l−1 titers. Deletion of the propionyl-CoA:succinate CoA transferase (ygfH) or over-expression of the transcriptional activator of short chain fatty acid uptake improved titer to over 100 mg l−1, while the combination of the two improved titer to over 130 mg l−1. The addition of exogenous methymalonate could also improve titer to over 100 mg l−1. Expression of a Streptomyces coelicolor A3(2) methylmalonyl-CoA epimerase, in conjunction with over-expression of E. coli’s native methylmalonyl-CoA mutase, allowed for the incorporation of exogenously fed succinate into 6-deoxyerythronolide B.
Keywords: heterologous, polyketide, propionate, methylmalonate, Escherichia coli, 6-deoxyerythronlide B
Introduction
Natural products have long served as sources of human therapeutics (Clardy et al. 2006; Newman 2008; Paterson and Anderson 2005). Of all small-molecule new chemical entities (NCE’s) between 1981 and 2006, 34% were natural products or semi-synthetic derivatives of such molecules. In fact, of the 109 antibacterial NCE’s and the 83 anticancer NCE’s, 74 and 45, respectively, were such molecules (Newman and Cragg 2007). Even with the growing contributions of medicinal chemistry and biopharmaceutical development programs, 13 natural product-derived drugs were approved in the United States between 2005 and 2007, with five of those compounds being the first members of new classes (Harvey 2008).
Polyketides represent a large family of natural product molecules with wide ranging pharmacological activities. Examples include erythromycin (an antibacterial), FK506 (an immunosuppressant), and lovastatin (anti-cholesterol) (Cane et al. 1998). Complex polyketides are constructed by modular polyketide synthases (PKSs) by successive rounds of decarboxylative Claisen condensation reactions between an acyl thioester and thioesterified malonate derivatives. Though many of the acyl-CoA starter units (such as acetyl-CoA and propionyl-CoA) for priming PKS’s are fairly common among prokaryotic and eukarotiyc systems, many of the thioesterified malonate derivates used as extender units are less common (such as (2S)-methylmalonyl-CoA, (2S)-ethylmalonyl-CoA, and chloroethylmalonyl-CoA) (Chan et al. 2009). Native hosts that produce such molecules naturally have the secondary metabolism to support the production of these starter and extender units.
Given the often fastidious nature of secondary metabolite producers, complex medium requirements, low specific growth rates, unclear metabolic network connectivity, and/or lack of established genetic engineering technologies, heterologous biosynthesis emerged as an option to produce these complex molecules in rapid-growing and well-characterized hosts (Pfeifer and Khosla 2001; Zhang et al. 2008). The ultimate goal of heterologous biosynthesis is to produce valuable metabolites in a cost-, time-, and personnel-efficient manner. With the advent of heterologous biosynthesis, one must not only express functional mega-synthases, but also engineer existing cellular metabolism to improve precursor supply, and/or introduce additional cellular metabolism to support initial precursor production. As such, metabolic engineering of substrate pathways is necessary for economically competitive production of such complex metabolites in heterologous hosts. Though established recombinant protocols exist for the inclusion of heterologous PKS systems, the tailoring of host cellular metabolism must be considered case-by-case in which heterologous biosynthesis is placed in the context of native metabolism.
The PKS responsible for synthesizing the erythromycin macrocycle, is called the deoxyerythronlide B synthase (DEBS), the product of which, has been the study of numerous seminal efforts in producing polyketides and understanding their biosynthesis including: cell-free synthesis (Pieper et al. 1995), heterologous biosynthesis (Kao et al. 1994; Pfeifer et al. 2001), the analysis of intermodular communication (Gokhale et al. 1999), combinatorial biosynthesis (Menzella et al. 2005), and structural analyses (Tang et al. 2006). The product of DEBS, 6-deoxyerythronolide B (6-dEB), has been produced in three different heterologous hosts, Streptomyces coelicolor (Kao et al. 1994), Streptomyces lividans (Xue et al. 1999), and Escherichia coli (Pfeifer et al. 2001), as well as a functional portion of the DEBS PKS in Saccharomyces cerevisiae (Mutka et al. 2006). E. coli strain BAP1 has been developed previously for the heterologous production of polyketide and nonribosomal peptide natural products (Pfeifer et al. 2001) and is used as the base production system in this study. To generate BAP1, the Bacillus subtilis surfactin phosphopantetheinyl transferase gene (sfp, required for posttranslational modification of the DEBS enzymes) (Quadri et al. 1998) was inserted into the prpRBCD location of the BL21(DE3) chromosome (removing E. coli’s primary propionate catabolic pathway, the methylcitrate pathway (Textor et al. 1997)), under the control of an inducible T7 promoter (Pfeifer et al. 2001). During this same genetic insertion, a T7 promoter was inserted before the native prpE gene (coding for a propionyl-CoA synthetase) to increase flux towards the production of propionyl-CoA, a direct precursor of 6-dEB. The ygf operon (also known as the scp operon) is responsible for the conversion of succinate to propionate in E. coli through succinyl-CoA, methylmalonyl-CoA, and propionyl-CoA intermediates (Haller et al. 2000).
There have been a number of studies focused on improving the stability of the large plasmids harboring the PKS genes (Murli et al. 2003), utilizing alternative substrate pathways for production (Dayem et al. 2002), and high-cell density bioprocess optimization (Lau et al. 2004) towards improving 6-dEB BAP1 production. Previously in our laboratory, we utilized metabolic modeling strategies for surveying heterologous hosts and medium compositions with respect to improving 6-dEB biosynthesis (Boghigian et al. 2010). Further, we analyzed the ygf operon by systematically deleting and over-expressing individual operon genes to understand their effect on 6-dEB biosynthesis (Zhang et al. 2010). While most of the individual deletions and over-expressions led to either the same or decreased 6-dEB production titers under the conditions tested, deletion of ygfH (propionyl-CoA:succinate CoA transferase), led to an approximately two-fold increase in production titer. In an effort to further understand the effect of these pathways on polyketide formation, and examine the interactions between these pathways, we applied a multi-scale engineering strategy to incorporate metabolic pathway engineering along with different bioprocess-related conditions (substrate feeding strategies). The results have implications for improving titers of both 6-dEB and other polyketides which utilize one or both of the acyl-CoA precursors examined here.
Materials & Methods
Background Strains & Plasmids
E. coli BAP1 was used as previously described (Pfeifer et al. 2001). TB3 is a derivative of BAP1 (Zhang et al. 2010) constructed by P1 transduction with a ygfH::kan (propionyl-CoA:succinate CoA transferase) mutant of BW25113 as a donor (Baba et al. 2006).
The genes required for the production of 6-dEB from propionate were cloned into plasmids pBP130 and pBP144, constructed previously (Pfeifer et al. 2001). Briefly, pBP130 (approximately 26kb) contains the eryA2 and eryA3 genes (coding for the DEBS2 and DEBS3 enzymes) under a single T7 promoter, on a pET21c background. Plasmid pBP144 (approximately 19kb) contains eryA1 under a T7 promoter and genes coding for the two subunits of the Streptomyces coelicolor propionyl-CoA carboxylase enzyme (accA1 and pccB) (Rodriguez and Gramajo 1999) under the control of second T7 promoter, on a pET28 background. All three eryA genes were cloned from the native erythromycin producer, Saccharopolyspora erythraea (Cortes et al. 1990; Donadio et al. 1991). pYW7317 is a derivative of pBP144 without the accA1 and pccB genes (Zhang et al. 2009).
Plasmid pACYCDuet-matBC was kindly provided by Prof. Mattheos A.G. Koffas and contains matB (coding for a malonyl-CoA synthetase) and matC (coding for a dicarboxylate carrier protein) from the nitrogen fixing soil bacterium Rhizobium trifolii, under the control of two separate T7 promoters (An and Kim 1998; An et al. 1999; Leonard et al. 2008).
Plasmid Construction
Standard molecular biology protocols were conducted according to Sambrook (Sambrook and Russell 2001). GeneHogs (Invitrogen) or XL-1 Blue (Stratagene) strains were used depending on the resistance marker of the plasmid to be constructed. The endonucleases used in this study were all purchased from New England Biolabs (Ipswich, MA, USA). All genes native to E. coli were PCR amplified from the BL21(DE3) (Novagen) genome. All primers used for PCR amplification can be found in Table I. The propionyl-CoA synthetase (prpE) was amplified and cloned into MCS1 of pACYCDuet-1 utilizing BamHI and HindIII restriction sites, generating pACYCDuet-prpE. The transcriptional activator of the ATO system (atoC) was amplified and cloned into MCS2 of pACYCDuet-1 and pACYCDuet-prpE utilizing NdeI and XhoI restriction sites, generating pACYCDuet-atoC and pACYCDuet-prpE-atoC, respectively. An E. coli codon-optimized version of the Streptomyces coelicolor A3(2) methylmalonyl-CoA epimerase gene (mce) was synthesized by Operon (Huntsville, AL, USA) designed with flanking EcoRI and HindIII sites. This synthetic gene was inserted between the EcoRI and HindIII sites in MCS1 of pCDFDuet-1, generating pCDFDuet-mce. The methylmalonyl-CoA mutase gene (sbm, encoding a “sleeping beauty mutase”) was PCR amplified from E. coli BL21(DE3) and cloned into MCS2 of pCDFDuet-mce utilizing NdeI and XhoI restriction sites, generating pCDFDuet-mce-sbm. All plasmids were verified by Sanger sequencing at the Tufts University Core Facility. All the plasmids used in this study are listed in Table II.
Table I.
Oligonucleotide primers utilizing in this study. All sequences are 5’ → 3’ and restriction sites are denoted with an underline
| Name | Sequence (5’→3’) |
|---|---|
| BamHI_prpE_for | GGGGGATCCATGTCTTTTAGCGAATTTTATCAGCGTTC |
| HindIII_prpE_rev | GGGAAGCTTACCTACGGTTCAGGTCC |
| NdeI_atoC_for | GGGCATATGACTGCTATTAATCGCATCC |
| XhoI_atoC_rev | GGGCTCGAGTTATACATCCGCCGGATCG |
| NdeI_sbm_for | GGGCATATGTCTAACGTGCAGGAGTG |
| XhoI_sbm_rev | GGGGTCGAGTTAATCATGATGCTGGCTTATCAG |
| pKD13_operon_for | AATACCCTCATTTTGATTGCGTTTTACGGAGCAAATAATGATTCCGGGGATCCGTCGACC |
| pKD13_operon_rev | ATTGCTGAAGATCGTGACGGGACGAGTCATTAACCCAGCATGTAGGCTGGAGCTGCTTCG |
| k2 | CGGTGCCCTGAATGAACTGC |
| ver_operon_rev | CGCCCAGCCAGTTGAGTTCA |
Table II.
Plasmids and strains used in this study.
| Name | Description | Source |
|---|---|---|
| pACYCDuet-1 | cat; P15A ori lacI T7lac | Novagen |
| pCDFDuet-1 | aadA; CloDF13 ori lacI T7lac | Novagen |
| pBP130 | bla; T7prom-eryA2-eryA3-T7term | (Pfeifer et al. 2001) |
| pBP144 | kan; T7prom-pccB-accA1-T7prom-eryA1-T7term | (Pfeifer et al. 2001) |
| pYW7317 | kan; T7prom-eryA1-T7term | (Zhang et al. 2009) |
| pACYCDuet-matBC | cat; T7prom-matB-T7term-T7prom-matC-T7term | (Leonard et al. 2008) |
| pACYCDuet-prpE | cat; T7prom-prpE-T7term | This study |
| pACYCDuet-atoC | cat; T7prom-atoC-T7term | This study |
| pACYCDuet-prpE-atoC | cat; T7prom-prpE-T7prom-atoC-T7term | This study |
| pCDFDuet-mce | aadA; T7prom-mce-T7term | This study |
| pCDFDuet-mce-sbm | aadA; T7prom-mce-T7prom-sbm-T7term | This study |
| pKD13 | bla, cat; template for chloramphenicol cassette amplification | (Datsenko and Wanner 2000) |
| pKD46 | bla; encodes γ, β, and exo under the control of a pBAD promoter | (Datsenko and Wanner 2000) |
Strain Construction
The entire ygf operon (sbm-ygfDGH) was deleted from the BAP1(araBAD:tet) chromosome by λ-Red recombination. First, BAP1(araBAD:tet) was transformed with plasmid pKD46 and expression of the γ, β, and exo genes was induced with 10 mM L-arabinose at 30°C. A kanamycin resistance gene (kan) flanked by two flipase recognition target (FRT) sites was PCR amplified from pKD13 (using the pKD13_operon_for and pKD13_operon_rev primer pair) containing 50bp of homology arms upstream of sbm and downstream of ygfH. This PCR reaction was digested with DpnI, gel purified, and approximately 100 ng of DNA was transformed into induced cells. Cells were then plated on LB-agar supplemented with 25 mg l−1 kanaymcin. Successful recombinants were verified by PCR (using the k2 and ver_operon_rev primer pair). This strain was stored as a glycerol stock, prepared electrocompetent, and transformed with pCP20 (Cherepanov and Wackernagel 1995) to excise the kan gene between the FRT sites, generating a kanamycin sensitive strain, BAB2. All strains used in this study are listed in Table III.
Table III.
Strains used in this study.
| Name | Description | Source |
|---|---|---|
| BL21(DE3) | F− ompT hsdSB (rB−, mB−) gal dcm (DE3) | Novagen |
| GeneHogs | F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (StrR) endA1 nupG fhuA::IS2 | Invitrogen |
| XL-1 Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F´ proAB lacIqZΔM15 Tn10 (TetR)] | Stratagene |
| BAP1 | BL21(DE3); ΔprpRBCD::T7prom-sfp-T7prom-prpE | (Pfeifer et al. 2001) |
| TB3 | BAP1; ΔygfH::FRT | (Zhang et al. 2010) |
| BAB2 | BAP1; Δsbm-ygfDGH::FRT | This study |
Initial Screening Cultures
All production cultures contained 5 g l−1 yeast extract, 10 g l−1 tryptone, 10 g l−1 sodium chloride, 15 g l−1 glycerol, 3 ml l−1 50% (v v−1) Antifoam B, 100 mM HEPES, and were adjusted to pH 7.60 with 5 M sodium hydroxide. Here, 3 ml cultures were conducted in 16 × 100 mm culture tubes.
For the initial screening study, the culture medium previously described was prepared supplemented with 60 mM sodium propionate, 60 mM disodium malonate, or 60 mM disodium methylmalonate. These were mixed with the production medium lacking the additional carbon source to create the various substrate concentrations desired. For precultures, a stab of glycerol stock was inoculated into 2 ml LB medium with appropriate antibiotics and grown overnight at 37°C and 250 rpm. For production cultures, 3 ml production medium was inoculated into 16 × 100 mm culture tubes with the precultures to an OD600nm = 0.1. These production cultures were grown for 72 hr at 22°C and 250 rpm. At the end of the culture period, cell-density was measured spectrophotometrically at 600 nm and a single. When needed, antibiotics were supplemented at concentrations of 100 mg l−1 for carbenicillin, 50 mg l−1 for kanamycin, and 34 mg l−1 for chloramphenicol. Induction of heterologous gene-expression was accomplished by supplementing 100 µM isopropyl β-D-1-thiogalactopyranoside (IPTG) to the culture medium.
Shake-Flask Production Cultures
Shake-flask cultures (15 ml in 125 ml Erlenmeyer flasks) containing production medium were used for subsequent production tests. Single colonies were picked from freshly streaked plates and inoculated into 1 ml production medium containing necessary antibiotics. These cultures were grown at 37°C and 250 rpm until OD600nm ≈ 0.6 and were used to inoculate 15 ml production medium with necessary antibiotics and 100 µM IPTG at a volumetric ratio of 5%. Cultures were then incubated at 22°C and 250 rpm for 120 hr. At the end of the culture period, cell-density was measured spectrophotometrically at 600 nm. Antibiotics and IPTG were added as described previously, when necessary. 6-dEB was analyzed, as described previously, by either RP-HPLC-ELSD (Wang et al. 2007) or MS (Zhang et al. 2010).
Metabolite Quantification by CEX-HPLC-RID
Precursor and byproduct metabolites were quantified by the previously mentioned HPLC system coupled to a Refractive Index Detector (RID). Clarified culture supernatant (20 µl) was applied to a Bio-Rad Aminex® HPX-87H Ion Exchange (300 mm × 7.8 mm, 9 µm) column, preceded by a 30 mm guard column of the same resin. The isocratic analysis used a 9.5 mM H2SO4 solvent held at a flow rate of 0.3 ml min−1. These conditions were identified by using an iterative stochastic search HPLC optimization program based on the compounds anticipated to be present in the culture medium (Dharmadi and Gonzalez 2005). The elution order was as follows: pyruvate (16.7 min), malonate (18.9 min), methylmalonate (20.9 min), succinate (22.7 min), lactate (24.2 min), glycerol (25.1 min), formate (26.8 min), acetate (29.1 min), propionate (34.5 min), and ethanol (41.3 min).
Results
Initial Screening Study
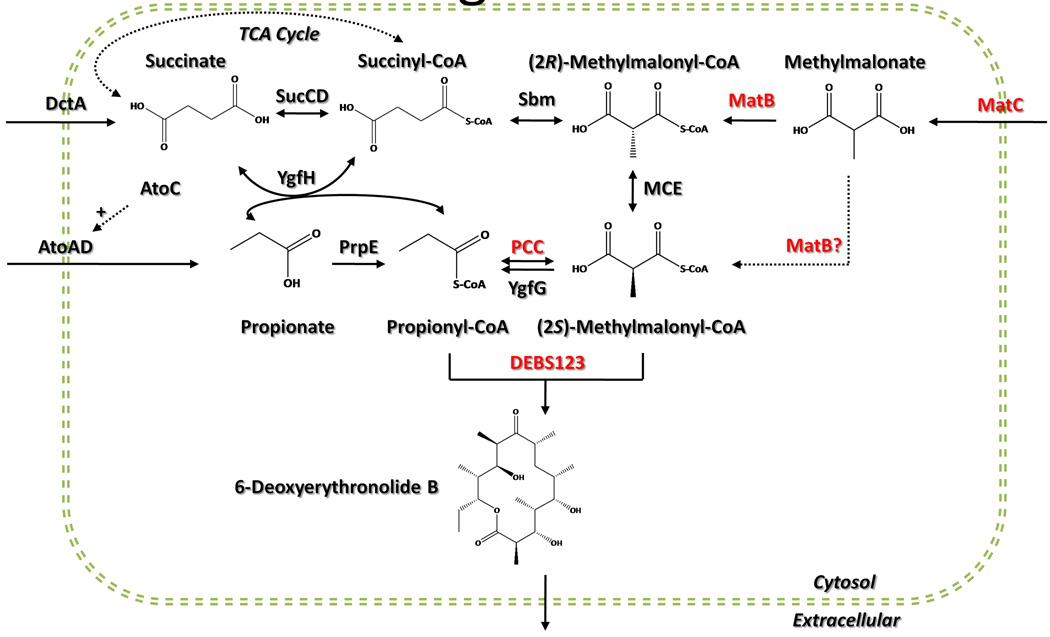
Upon inspection of generalized metabolic maps, and given the DEBS (2S)-methymalonyl-CoA requirement, we attempted to construct a metabolic pathway capable of converting exogenous methylmalonate to (2S)-methymalonyl-CoA (Figure 1). This pathway would include E. coli’s native YgfG or the heterologous reversible PCC to provide propionyl-CoA. The MatBC pathway responsible for methylmalonate-methylmalonyl-CoA conversion from R. trifolii was then reconstructed in E. coli. While these MatB and MatC synthases have a preference for malonate as a substrate, it has been shown that they can also activate methylmalonate at 20.4% the in vitro rate of malonate (An and Kim 1998).
Figure 1.
An overview of propionate, methylmalonate, and succinate metabolism and their relation to 6-dEB production in E. coli. Heterologous enzymes are shown in red text. Abbreviations: SucCD = succinyl-CoA synthetase; Sbm = sleeping beauty mutase = methylmalonyl-CoA mutase; MatB = malonyl-CoA synthetase; MatC = R. trifolii dicarboxylate carrier protein; MCE = methymalonyl-CoA epimerase; PCC = propionyl-CoA carboxylase; YgfG = methylmalonyl-CoA decarboxylase; PrpE = propionyl-CoA synthetase; YgfH = propionyl-CoA:succinate CoA transferase; DEBS123 = deoxyerythronolide B synthase; AtoC = transcriptional activator of the ATO system; AtoAD = acetyl-CoA:acetoacetyl-CoA transferase; DctA E. coli dicarboxylate carrier protein.
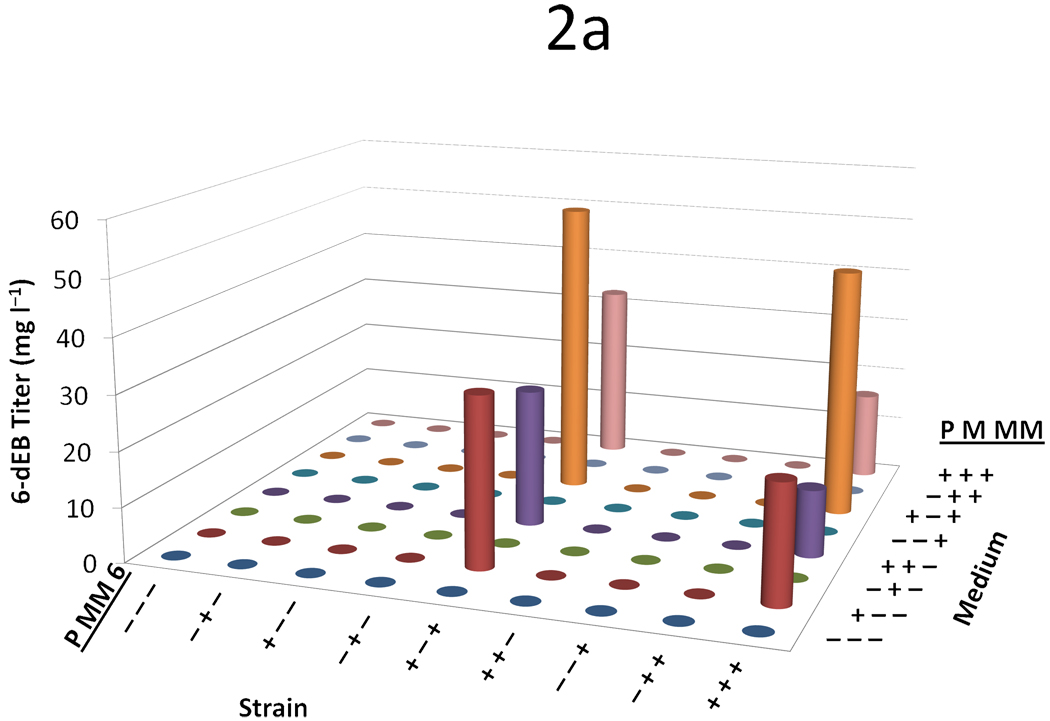
We devised a three-variable (propionate, malonate, and methylmalonate), two-level (0 mM or 20 mM) full-factorial supplementation experiment across nine strains with a variety of plasmid combinations (containing the propionate pathway, the malonate pathway, and the 6-dEB biosynthetic pathway). No 6-dEB (<5 mg l−1) was made in the absence of propionate supplementation (Figure 2a). The addition of malonate did not improve 6-dEB titers under any conditions tested. When propionate and methylmalonate were both supplied, 6-dEB production increased approximately two-fold compared to propionate supplementation alone. However, the overall 6-dEB titers were lower in the presence of the MatBC pathway.
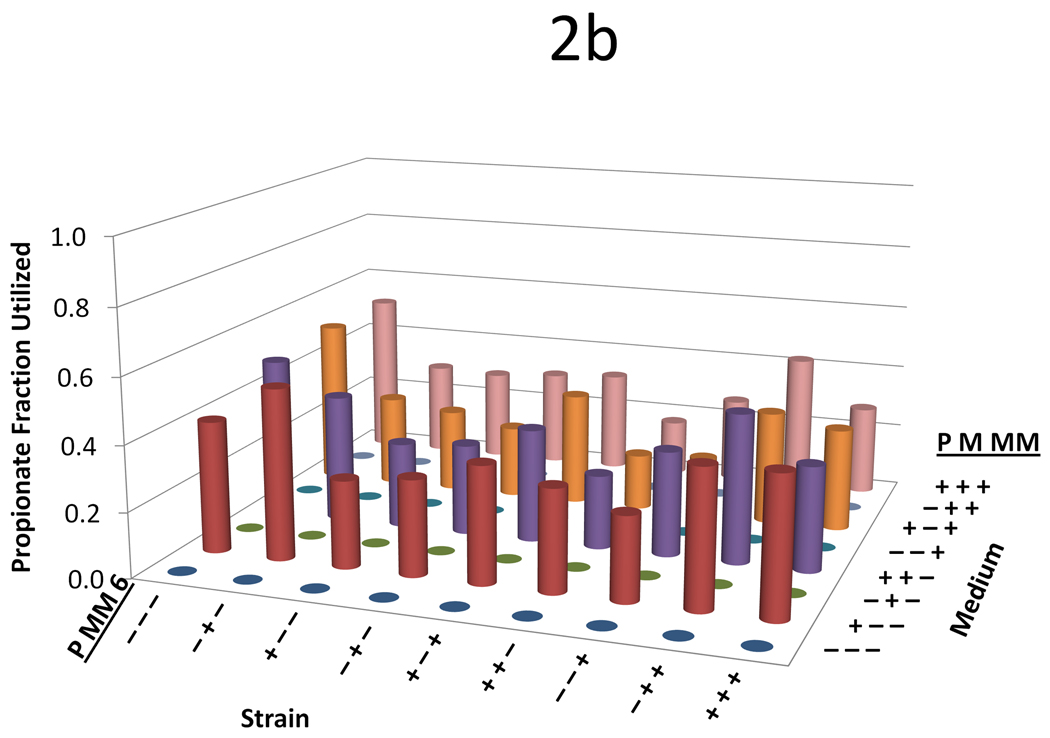
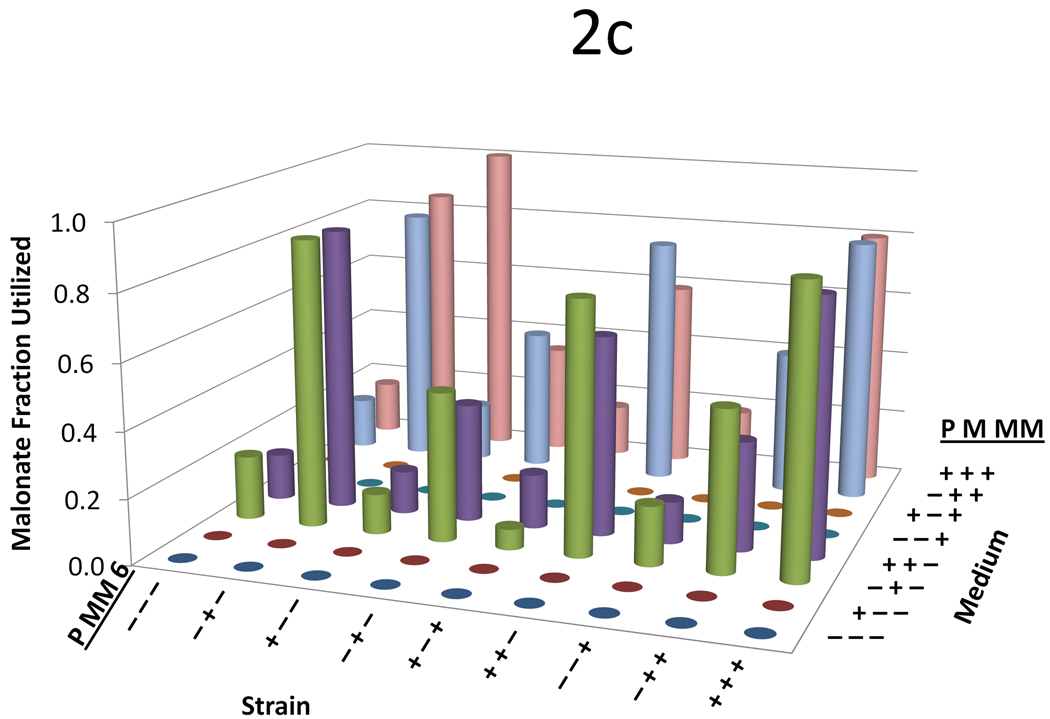
Figure 2.
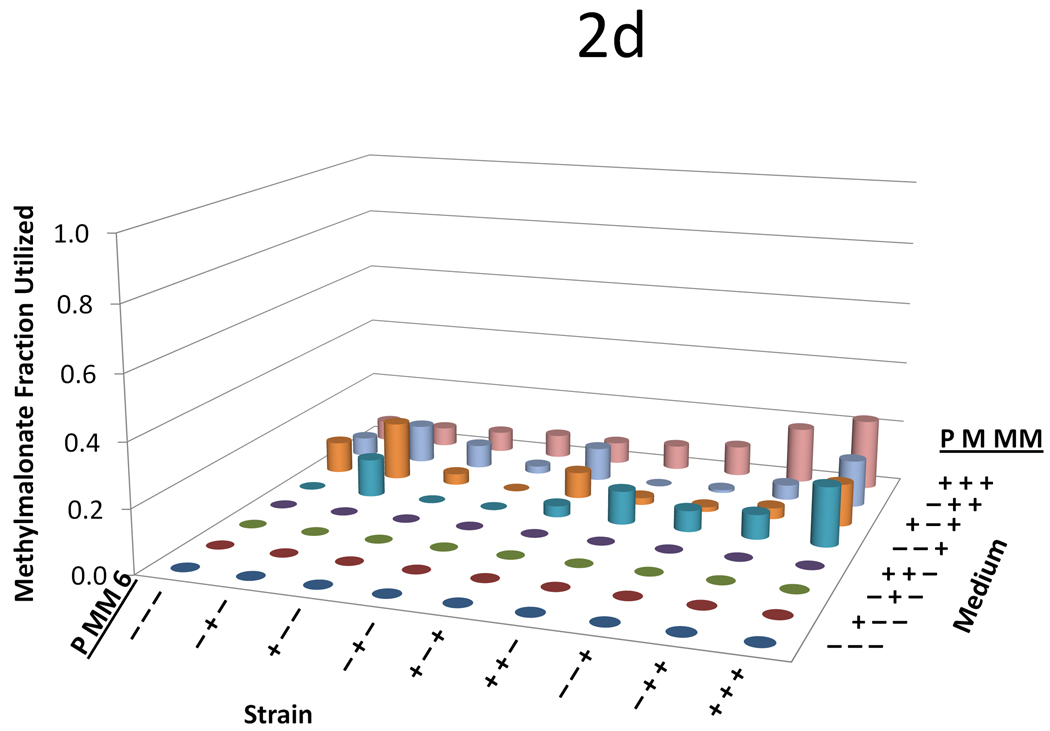
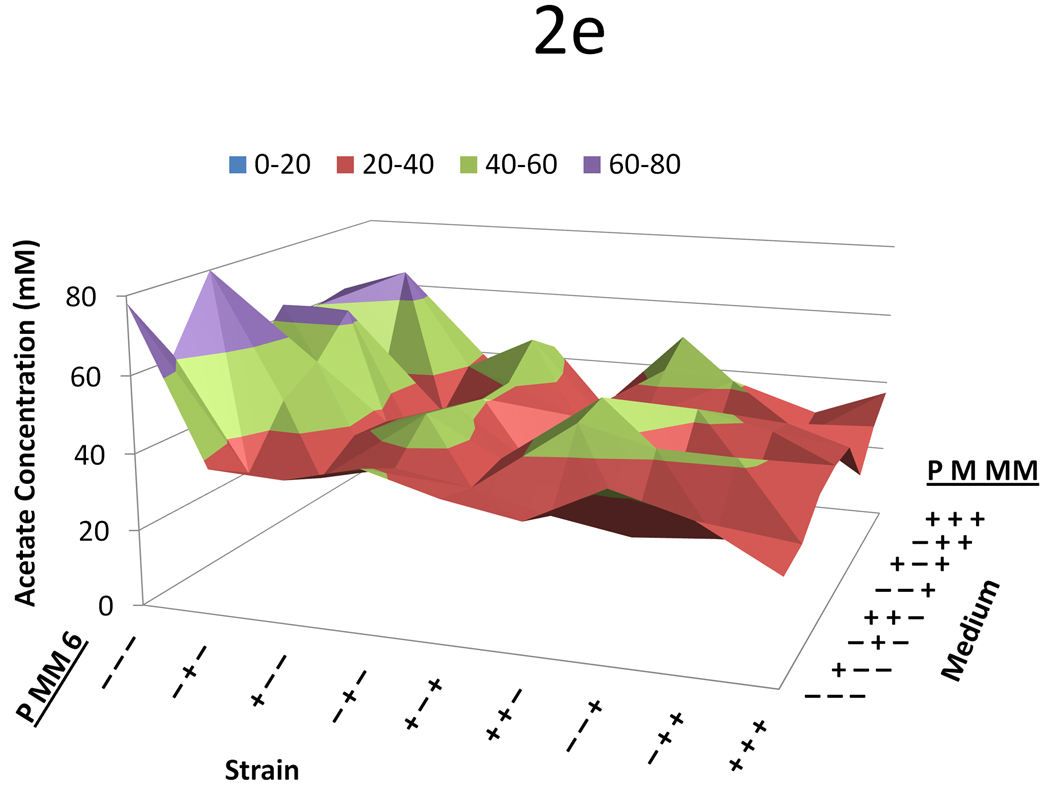
Data from the three variable (propionate, malonate, and methylmalonate) two-level (0 mM or 20 mM) full-factorial supplementation experiment across nine strains with a variety of plasmid combinations. For all figures, on the left-hand axis −’s and +’s indicate whether the propionate pathway, the methylmalonate pathway, or the DEBS complex is included, respectively. On the right hand axis, −’s and +’s indicate whether propionate, malonate, or methylmalonate is supplemented in the medium at a concentration of 0 or 20mM, respectively. The y-axis shows (a) 6-dEB titer, (b) fraction of propionate utilized, (c) fraction of malonate utilized, (d) fraction of methylmalonate utilized, and (e) amount of acetate produced as a function of these two parameters.
In terms of precursor consumption, propionate was favored in the strains that did not contain the MatBC pathway; whereas when MatBC was present, malonate consumption was preferred and increased dramatically even in the presence of multiple substrates (Figure 2b and Figure 2c). Propionate uptake did not change dramatically in any of the conditions tested, even in the cases where 6-dEB production was observed (Figure 2a). As can be seen in Figure 2d, in general, methylmalonate consumption was minimal (often <2 mM consumed) in all cases, although the MatBC pathway did stimulate consumption slightly (to approximately 4 mM in some cases). Acetate overflow was significant, reaching approximately 80 mM in some cases; however, this overflow metabolism was retarded with the addition of propionate, malonate, and methylmalonate (Figure 2e).
Propionate Pathway Engineering
The propionate pathway has previously been designed to provide the precursors for 6-dEB production in E. coli (Wang et al. 2007; Zhang et al. 2010). In the current production system, atoC, encoding an activator of E. coli short-chain fatty acid metabolism, is natively expressed from the chromosome. Whereas, prpE, encoding a propionyl-CoA synthetase, is inducibly expressed from the chromosome and the pcc genes, encoding a propionyl-CoA carboxylase, are expressed from a multi-copy plasmid (15–20 copies per cell). As a result of the variation in expression design, this pathway is likely ‘unbalanced’ for 6-dEB heterologous production as prpE and atoC are expected to be expressed to a smaller extent than the pcc genes. Balancing of expression levels may then improve production. Three different vectors were generated to over-express E. coli’s native prpE and atoC separately and in combination. These constructs were then co-transformed with the 6-dEB production plasmids (pBP130 and pBP144) in two strains (BAP1 and TB3).
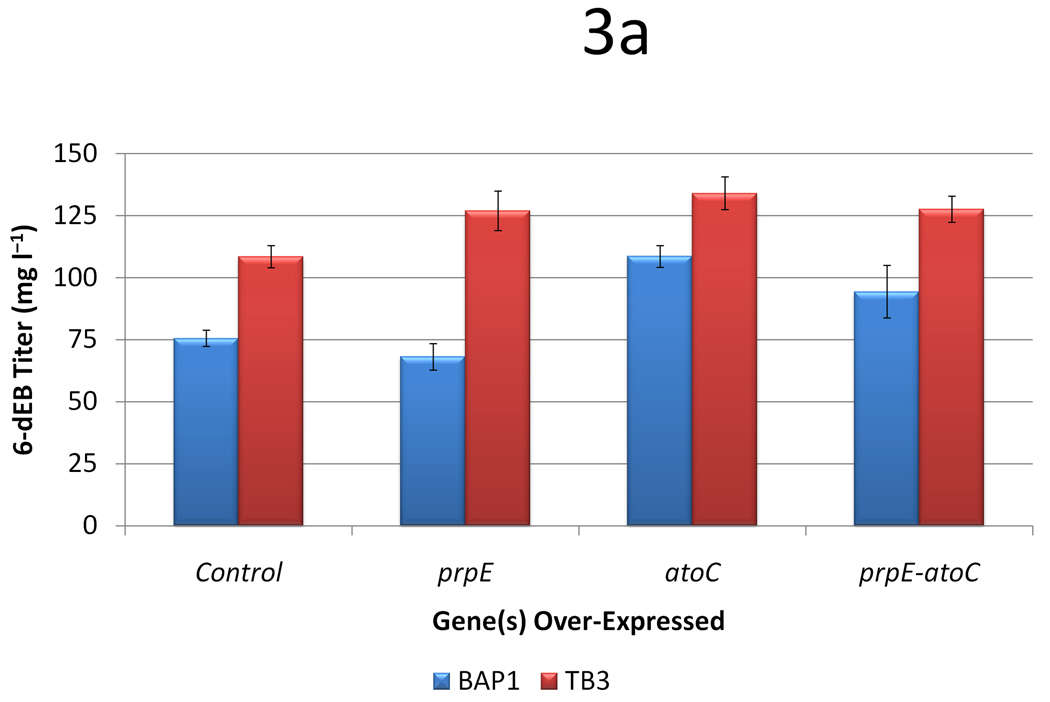
In BAP1, over-expression of prpE by itself had no effect on 6-dEB production (p = 0.214). Over-expression of atoC improved 6-dEB production 1.44-fold (Figure 3a). Similar titers were observed when both prpE and atoC were over-expressed. Compared to BAP1, TB3 improved titer approximately 1.5-fold (a slightly lower improvement than observed previously (Zhang et al. 2010), which is likely due to a different inoculation method). In TB3, all three plasmid constructs improved 6-dEB production similarly (ANOVA p = 0.416). In this case, the over-expression of prpE alone did not help improve the 6-dEB production in strain TB3 when compared to the co-expression of prpE and atoC. This is probably because the deletion of ygfH in TB3 resulted in sufficient propionyl-CoA and further accumulation through PrpE activity did not aid 6-dEB biosynthesis. The 6-dEB titers presented here increased from 75.6 ± 3.2 mg l−1 to 134.1 ± 6.6 mg l−1 after varying these plasmid and strain systems.
Figure 3.
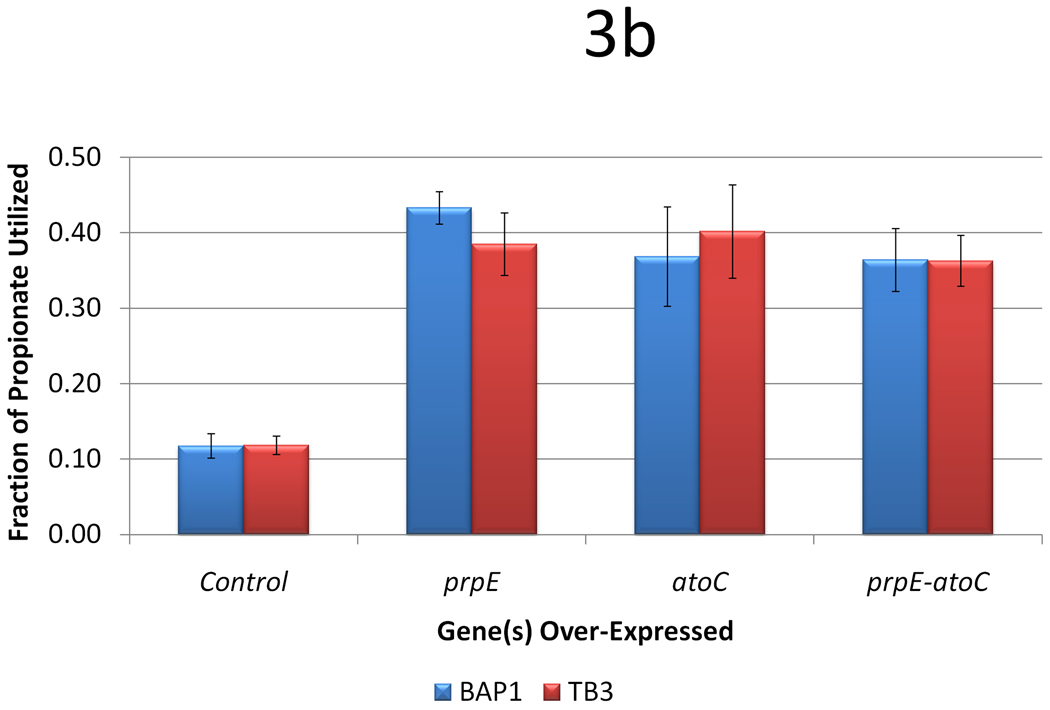
Data from the propionate pathway engineering study, as a function of two strains (BAP1 and TB3) and four plasmid systems (a control with only pBP130 and pBP144, an additional pACYCDuet-prpE, an additional pACYCDuet-atoC, and an additional pACYCDuet-prpE-atoC). Panel (a) shows the 6-dEB titer while panel (b) shows the fraction of propionate uptake as a function of these cellular parameters. Error bars represented ± one standard deviation from three replicates.
Figure 3b shows the fraction of propionate consumed by the cells after 120 hr of shake-flask culture across all of the conditions tested. The atoC gene encodes for the regulatory controller of the ATO operon (responsible for short-chain fatty acid degradation (Jenkins and Nunn 1987)), and prpE expression is needed for the production of propionyl-CoA, a direct precursor for 6-dEB biosynthesis. It was shown that over-expression of prpE and atoC both stimulate propionate uptake to the same effect. For both BAP1 and TB3, regardless of whether prpE and atoC were over-expressed separately or together, the fraction of propionate utilized increased between 3–4 fold (ANOVA p = 0.653). In all plasmid systems, propionate uptake between BAP1 and TB3 was the same (p > 0.05), meaning that the yield of 6-dEB on propionate was higher in the TB3 strain as TB3 produced higher 6-dEB titers.
Methylmalonate Pathway Engineering in the Presence of Propionate
The initial screening study revealed that methylmalonate supplementation could improve 6-dEB production in the presence of propionate, prompting us to further investigate this interaction. It was first hypothesized that the lack of improvement upon incorporation of the MatBC pathway (even with the improved methylmalonate uptake) was because the malonyl-CoA synthase generated the (2R) stereoisomer of methylmalonyl-CoA; whereas, the DEBS complex only accepts the (2S) isomer of methymalonyl-CoA. Because a methylmalonyl-CoA epimerase has not been previously identified in E. coli, we introduced a heterologous methylmalonyl-CoA epimerase gene to allow for the interconversion between isomers and to determine its effect on 6-dEB production.
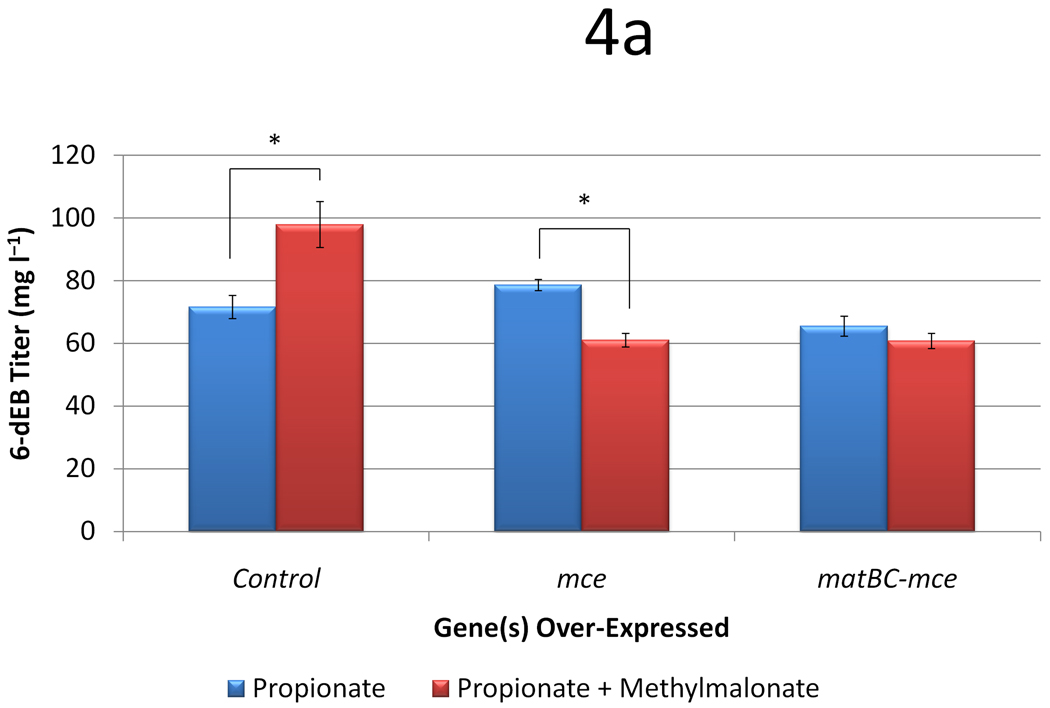
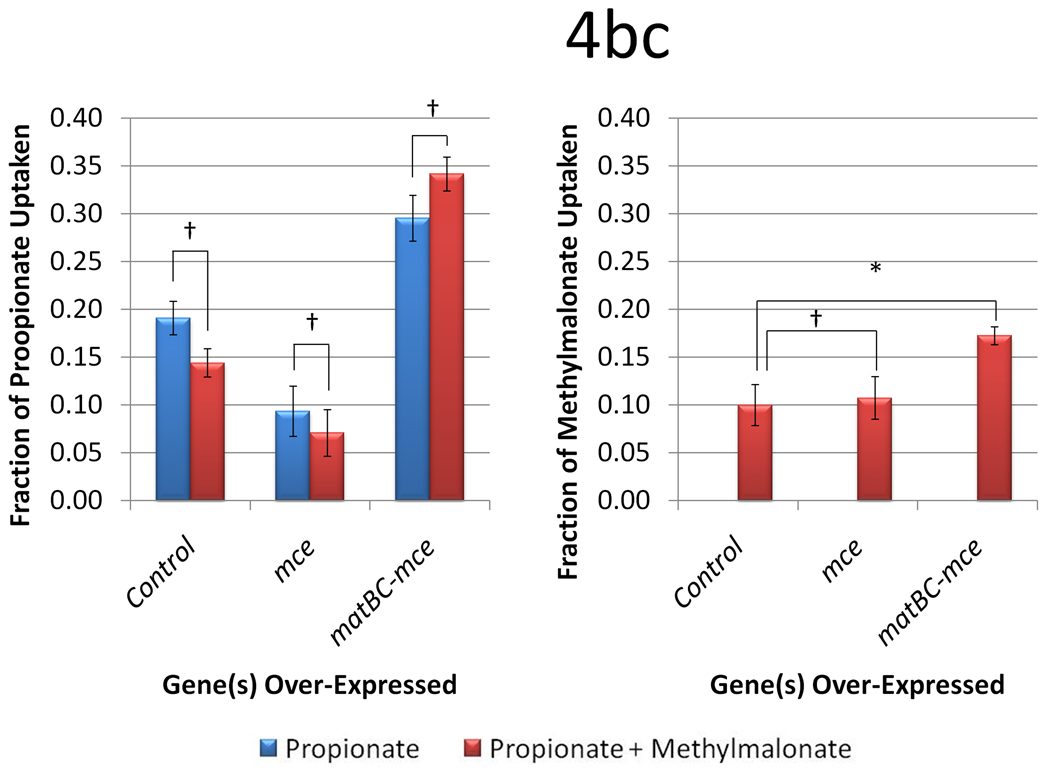
We first tested the effects of propionate and methylmalonate supplementation in BAP1 in the presence of: 1) no extra plasmids, 2) a plasmid over-expressing the S. coelicolor A3(2) methylmalonyl-CoA epimerase gene (mce), and 3) a plasmid over-expressing mce and the matBC genes. Consistent with the small-scale cultures in the initial screening study, methylmalonate supplementation improves 6-dEB production in the presence of propionate in the shake-flasks from 71.7 ± 3.74 mg l−1 to 97.9 ± 7.32 mg l−1 (Figure 4a). However, 6-dEB production decreases to 61.1 ± 2.18 mg l−1 and 60.9 ± 2.43 mg l−1 when both carbon sources are used in the presence of mce and matBC-mce, respectively. As shown in Figure 4b, propionate uptake is decreased when mce is used. When matBC is included, propionate uptake is stimulated, indicating that this MatC carrier is not specific and may facilitate propionate transport as well.
Figure 4.
Data from engineering the methylmalonate pathway in the presence of propionate for BAP1. Cultures were fed either 20 mM propionate or 20 mM propionate and 20 mM methylmalonate. Three different plasmid systems were analyzed (a control with only pBP130 and pBP144, an additional pCDFDuet-mce, and pACYCDuet-matBC/pCDFDuet-mce). Panel (a) shows the 6-dEB titer, (b) shows the fraction of propionate uptake, and (c) shows the fraction of methylmalonate uptake as a function of these parameters. Error bars represented ± one standard deviation from three replicates. * indicates statistically significant results (p < 0.05), while † indicates statistically insignificant results (p > 0.05) between the comparisons shown.
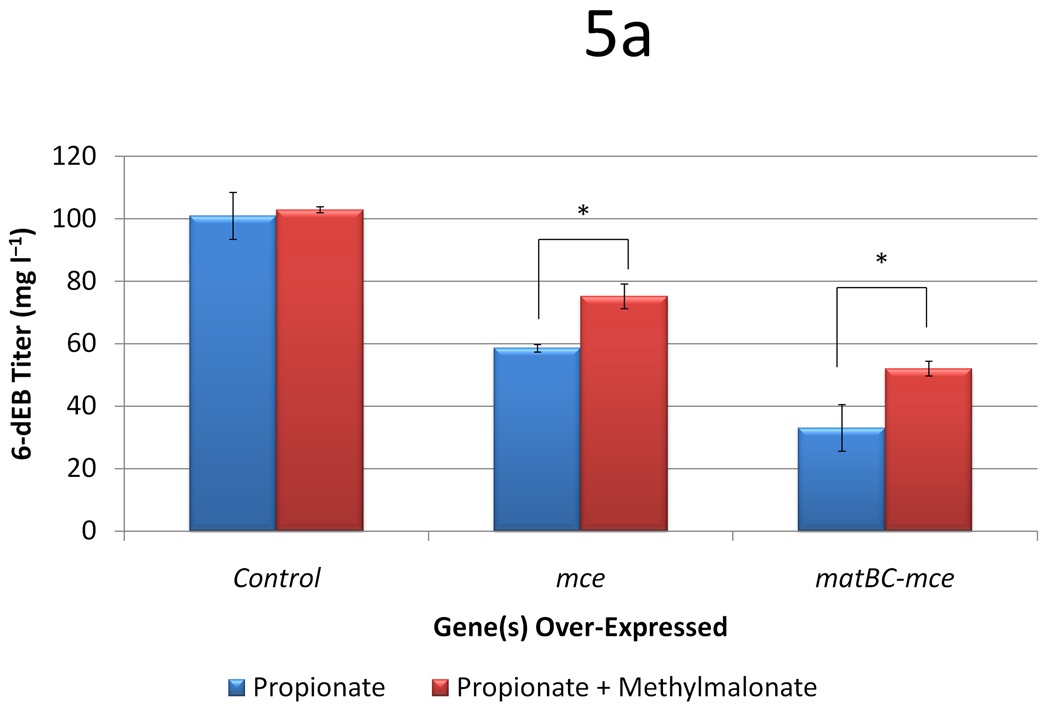
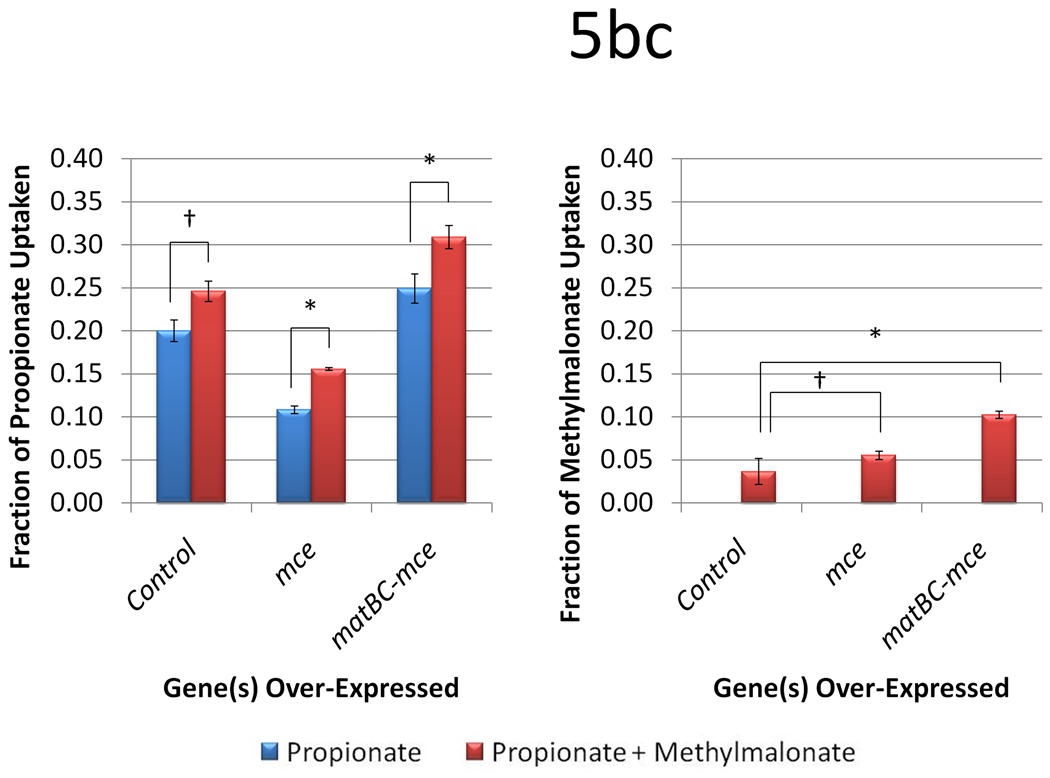
The same expression and production studies were then conducted in TB3, yielding different results. As established previously, TB3 showed higher titers of 6-dEB from propionate (101.0 ± 7.5 mg l−1), however, when methylmalonate was supplemented, the titer was unchanged (p = 0.730; Figure 5a). When over-expressing mce or both matBC-mce, the titers decrease significantly to 58.6 ± 1.19 mg l−1 and 33.1 ± 7.4 mg l−1 with only propionate, and to 75.2 ± 3.9 mg l−1 and 52.1 ± 2.4 mg l−1 with both substrates present. This departs from the trends observed in BAP1, where when either mce or matBC-mce was expressed, 6-dEB production decreased in the presence of methylmalonate. As can be seen in Figure 5b and Figure 5c, propionate and methylamlonate uptake was not drastically different than what was observed in BAP1. Again, expression of matBC-mce stimulated methylmalonate uptake, while expression of mce alone did not.
Figure 5.
Data from engineering the methylmalonate pathway in the presence of propionate for TB3. Cultures were fed either 20 mM propionate or 20 mM propionate and 20 mM methylmalonate. Three different plasmid systems were analyzed (a control with only pBP130 and pBP144, an additional pCDFDuet-mce, and pACYCDuet-matBC/pCDFDuet-mce). Panel (a) shows the 6-dEB titer, (b) shows the fraction of propionate uptake, and (c) shows the fraction of methylmalonate uptake as a function of these parameters. Error bars represented ± one standard deviation from three replicates. * indicates statistically significant results (p < 0.05), while † indicates statistically insignificant results (p > 0.05) between the comparisons shown.
Methylmalonate Pathway Engineering in the Absence of Propionate
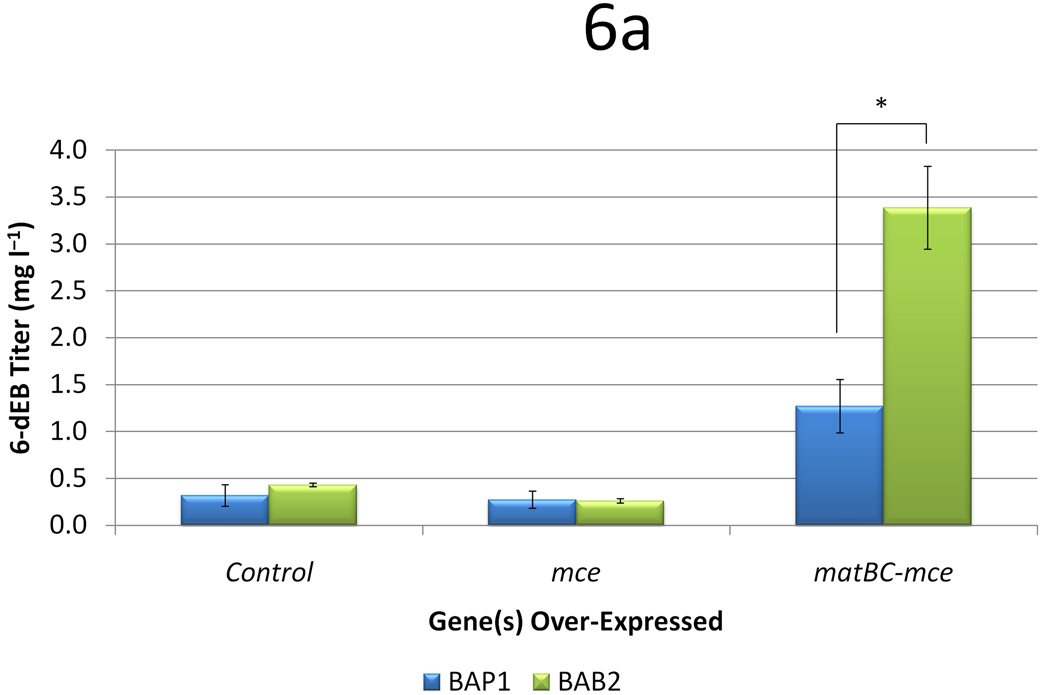
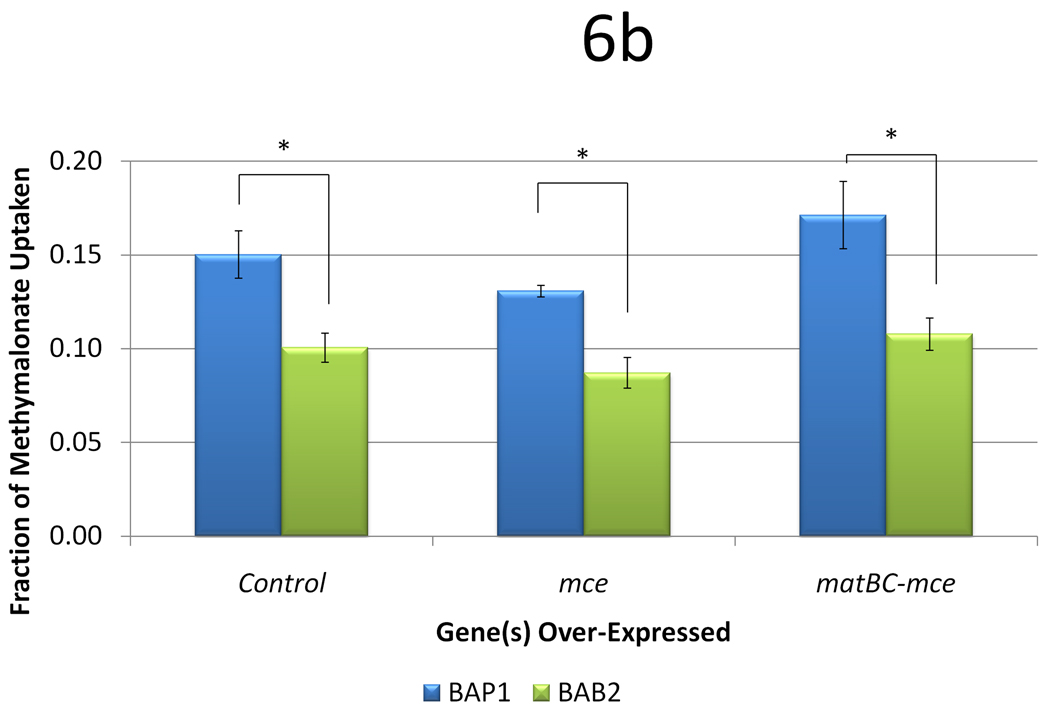
It has been found that BAP1 could produce 6-dEB in the absence of propionate and methylmalonate (Zhang et al. 2010). While it still remains unclear as to why methylmalonate supplementation in the presence of propionate improves 6-dEB production, we sought to analyze the effects of these strains and plasmids in the absence of propionate. With the utilization of mass spectrometry, the limit of detection of 6-dEB in the culture is approximately 0.1 mg l−1, allowing 6-dEB production to be quantified in the absence of propionate supplementation. BAP1 produced 6-dEB at a titer of 0.32 ± 0.11 mg l−1 from 20 mM methylmalonate (Figure 6a). However, the uptake of methylmalonate was still minimal at 3.0 ± 0.2 mM (Figure 6b). The inclusion of the mce pathway had no effect on 6-dEB titer (p = 0.636) or methylmalonate uptake (p = 0.142). However, when the MatBC pathway was also expressed, 6-dEB titer improved to 1.27 ± 0.29 mg l−1 (p = 0.037) but methylmalonate uptake was not significantly different (p = 0.261).
Figure 6.
Data from engineering the methylmalonate pathway in the absence of propionate, as a function of three strains (BAP1, TB3, and BAB2) and three plasmid systems (a control with only pBP130 and pBP144, an additional pCDFDuet-mce, and pACYCDuet-matBC/pCDFDuet-mce). Panel (a) shows the 6-dEB titer, while panel (b) shows the fraction of methylmalonate utilized as a function of these cellular parameters. Error bars represented ± one standard deviation from three replicates. * indicates statistically significant results (p < 0.05) between the comparisons shown.
In an effort to understand the effect of mce and matBC-mce on 6-dEB production from methylmalonate, we constructed a strain that lacked the ability to convert polyketide precursors to metabolites to be used for growth (encoded by the ygf operon). In the ygf operon mutant of BAP1 (BAB2), 6-dEB production from methylmalonate was not significantly different when compared to BAP1 (p = 0.350) or BAP1 with mce (p = 0.770). Methylmalonate uptake was the lowest in BAB2 in all cases. Interestingly, with the inclusion of the MatB-MatC pathway, 6-dEB titer improved almost 8-fold to 3.39 ± 0.74 mg l−1, even with decreased methylmalonate uptake.
Methylmalonyl-CoA Mutase-Epimerase Pathway Engineering
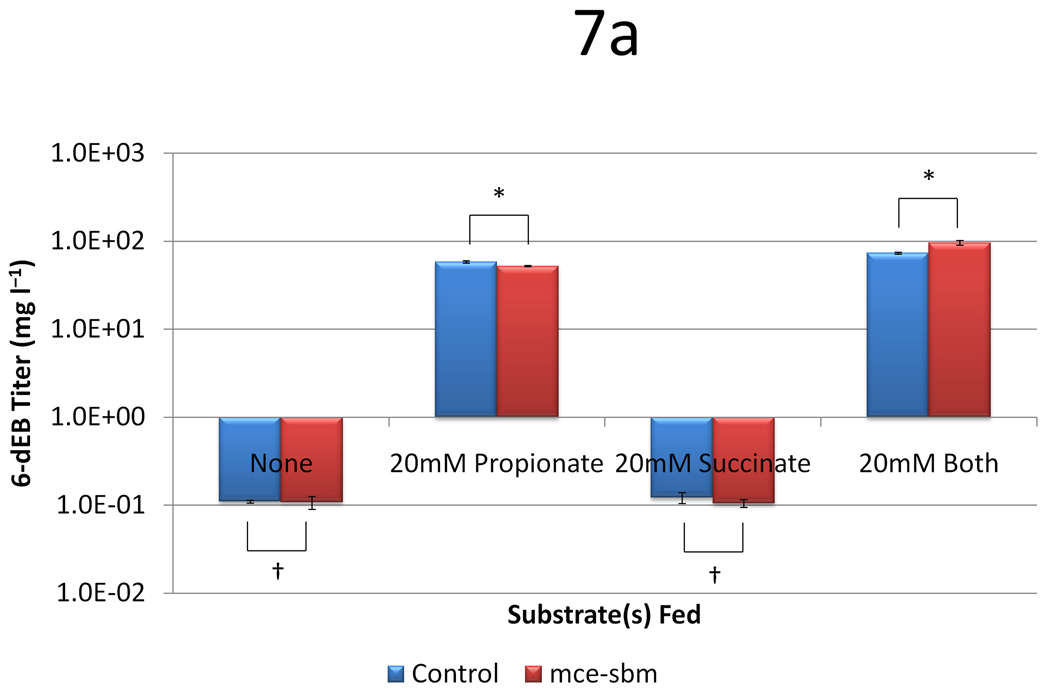
Last, we aimed to further understand the role of Sbm and E. coli’s native succinate-to-propionate conversion cycle in 6-dEB production. We previously over-expressed and deleted sbm in BAP1 in separate experiments, to find that neither genetic modification had an influence on 6-dEB production (Zhang et al. 2010). We hypothesized the lack of effect of these sbm expression studies on 6-dEB production was due to a lack of methylmalonyl-CoA epimerase activity, since Sbm has been shown to generate the (2R) isomer of methylmalonyl-CoA; whereas, DEBS accepts only the (2S) isomer (see Figure 1). In this set of experiments, we over-expressed sbm with the mce gene previously described, thereby fully connecting the succinate pathway with the 6-dEB production pathway. We then tested this combination in four different medium compositions which included 20 mM propionate, 20 mM succinate, 20 mM both substrates, and no substrates.
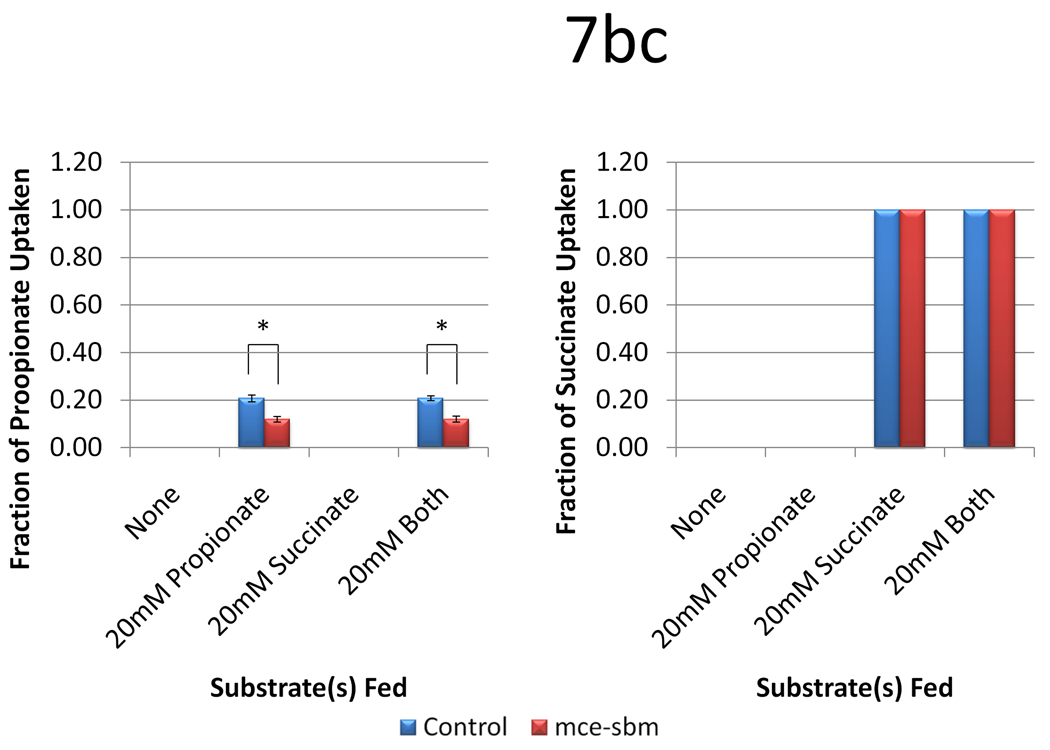
In the absence of either substrate, the titers with and without the Sbm-MCE were roughly 0.1 mg l−1, consistent with what was observed previously (Zhang et al. 2010) (Figure 7a). When propionate was utilized as the sole substrate, the inclusion of the Sbm-MCE pathway slightly decreased 6-dEB titer (p = 0.046). When succinate was utilized as the sole substrate, 6-dEB production decreased to levels observed in absence of a substrate; the Sbm-MCE pathway had no effect (p = 0.280). When both propionate and succinate were utilized, the titer without the Sbm-MCE pathway increased to 73.4 ± 2.0 mg l−1, while the inclusion of the Sbm-MCE pathway further increased titer to 96.4 ± 5.6 mg l−1. As can be seen in Figure 7b, the provision of succinate in the medium decreased propionate uptake in both the control and with expression of sbm-mce. In all cases, succinate was absent in the medium at the end of the culture period (Figure 7c).
Figure 7.
Data from engineering the methylmalonyl-CoA mutase-epimerase pathway as a function of two plasmid systems (a control with only pBP130 and pBP144, and pCDFDuet-mce-sbm) and four medium formulations (no substrates, 20 mM propionate, 20 mM succinate, or 20 mM both substrates). Panel (a) shows the 6-dEB titer, panel (b) shows the fraction of propionate utilized, and panel (c) shows the fraction of succinate utilized as a function of these cellular parameters. Error bars represent ± one standard deviation from three replicates. * indicates statistically significant results (p < 0.05), while † indicates statistically insignificant results (p > 0.05) between the comparisons shown.
Discussion
Heterologous polyketide biosynthesis presents a significant challenge in recombinant protein production and metabolic pathway engineering. Polyketides, being significant sources of therapeutic compounds, and PKS’s, being complex enzymes, are attractive systems for chemists, biologists, and engineers. This study focused on the metabolic engineering of multiple pathways for substrate provision for heterologous polyketide biosynthesis in E. coli. The ultimate goals of which are to 1) understand the interactions of these pathways, 2) identify the rate-limiting steps in polyketide biosynthesis, and 3) rationally engineer a system with improved titer. We applied a multi-scale engineering strategy through systematic gene over-expression and deletion experiments and feeding experiments to better understand the interplay of native propionyl-CoA and methylmalonyl-CoA metabolism as well as heterologous methylmalonate and native succinate metabolism. While 6-dEB was used as a means of examining the effect of these pathways and substrates, similar strategies could be applied to improving the titer of other polyketides which use the same starter or extended acyl-CoA units. For example, Streoptomyces hygroscopicus uses seven (2S)-methylmalonyl-CoA extender units for making the immunosuppressant rapamycin, while Mycobacterium tuberculosis uses (2S)-methylmalonyl-CoA for the biosynthesis of mycolic acids (Chan et al. 2009).
Our initial screen was comprised of a three variable (propionate, malonate, and methylmalonate), two-level (0 mM or 20 mM) full-factorial supplementation experiment across nine different plasmid systems, all in the base strain of BAP1. Because propionate and methylmalonate are the deactivated forms of the direct precursors for 6-dEB biosynthesis, it is expected that increased intracellular levels would improve 6-dEB biosynthesis. Malonate was used to serve as a frame of reference for analyzing the effect of the MatBC pathway. As expected, the MatBC pathway had a stronger preference for malonate than for methylmalonate, as indicated by dramatic improvements in uptake when matBC was over-expressed. Malonate uptake was preferred over methylmalonate in general, presumably due to malonate being used a precursor for malonyl-CoA, the first committed step to essential fatty acid biosynthesis. However, no 6-dEB production was observed when malonate was used alone. The combination of malonate and propionate/methylmalonate did not improve production, leading us to believe that malonyl-CoA levels have no effect on propionyl-CoA or methylmalonyl-CoA levels.
Increasing the expression of prpE and/or atoC was identified as a metabolic engineering target considering that we previously identified ΔygfH as a target for improving 6-dEB production (Zhang et al. 2010) and both of these enzymes are connected to this metabolite node. It has been previously demonstrated that a constitutive mutation in E. coli’s atoC allowed for transcription of the ATO genes (Jenkins and Nunn 1987) and improved the propionate uptake roughly 10-fold in M9 minimal medium supplemented with 1% (wt vol−1) glucose and 10 mM propionate (Rhie and Dennis 1995). In our study, when prpE and/or atoC were over-expressed, propionate uptake increased roughly 4-fold, yet 6-dEB production only increased roughly 30%. This indicates that there is perhaps another significant sink of propionyl-CoA that is responsible for drawing from this metabolite pool, even in the absence of YgfH and PrpBCD. Possible sources could be enzymes that have preference for acetyl-CoA but are also promiscuous for propionyl-CoA. Moreover, PCC activity could be limiting in that there is not enough (2S)-methylmalonyl-CoA to accommodate the increasing pools of propionyl-CoA, in order to synthesize more 6-dEB.
This study presents the first production of 6-dEB solely from methylmalonate. Inclusion of the MatBC pathway in BAP1 increased the 6-dEB titer from 0.32 ± 0.11 mg l−1 to 1.27 ± 0.29 mg l−1, while deletion of the entire ygf operon further improved production to 3.39 ± 0.74 mg l−1. Previously, 6-dEB had been produced in only trace quantities (0.85 ± 0.2 mg l−1) from methylmalonate using matB and the Streptomyces coelicolor methylmalonyl-CoA mutase (mutAB), however 10 mM propionate was also added to the medium (Murli et al. 2003). The extremely low uptake rates of methylmalonate appear to be a barrier in improved 6-dEB production from this substrate. Even in the absence of propionate and in the presence of MatBC, methylamlonate uptake was never above 20% of the fed substrate (a total of 4 mM).Whereas other substrates, more commonly used carbon sources (propionate, succinate, and malonate), were uptaken at significantly higher rates. Interestingly, the rate of methylmalonate uptake was 21.8% that for malonate (as determined in the initial screening study), which is strikingly similar to a previous in vitro analysis of MatB, which demonstrated 20.4% activity utilizing methylmalonate when compared to malonate (An and Kim 1998). There is no known transporter dedicated to methylmalonate uptake, nor is there a known methylmalonyl-CoA synthetase or CoA-ligase specific for utilizing methylmalonate as a substrate. This specificity issue may be overcome by protein engineering of the R. trifolii matBC system to make it specific or strongly preferential for methylmalonate/methylmalonyl-CoA. In a slightly different manner, methylmalonate uptake could be improved by a laboratory evolution experiment utilizing methylmalonate as a sole carbon substrate.
Finally, another commonly utilized cellular metabolite, succinate, could be engineered into the 6-dEB backbone through the functional expression of a methylmalonyl-CoA mutase-epimerase pathway. In all the studied cases, propionate addition was needed for high-level production, indicating that propionate was the most favorable substrate for 6dEB biosynthesis in E. coli. When 20 mM succinate was fed (in addition to 20 mM propionate) without the introduced Sbm-MCE pathway, 6-dEB titer decreased. However, when the methylmalonyl-CoA mutase-epimerase pathway was expressed, 6-dEB titer increased, indicating that basal levels of Sbm (and lack of an epimerase) could not incorporate succinate into (2S)-methylmalonyl-CoA. This appears to contrast previous work where 6-dEB was only produced at roughly 1 mg l−1 from 5 mM propionate, 50 mM succinate, and 50 mM glutamate using a Propionibacterium shermanii methylmalonyl-CoA mutase and the same Streptomyces coelicolor methylmalonyl-CoA epimerase (Dayem et al. 2002). Combining this new information with previous information from computational modeling (Boghigian et al. 2010), leads us to believe that metabolic engineering of the succinate and succinyl-CoA metabolite nodes could be critical to further improve 6-dEB production (and production of other propionate-dependent polyketide products) by providing access to primary metabolism.
Acknowledgments
BAB was supported through NIH R01-GM085323. HZ was supported through NSF 0924699.
References
- An JH, Kim YS. A gene cluster encoding malonyl-CoA decarboxylase (MatA), malonyl-CoA synthetase (MatB) and a putative dicarboxylate carrier protein (MatC) in Rhizobium trifolii--cloning, sequencing, and expression of the enzymes in Escherichia coli. Eur J Biochem. 1998;257(2):395–402. doi: 10.1046/j.1432-1327.1998.2570395.x. [DOI] [PubMed] [Google Scholar]
- An JH, Lee GY, Jung JW, Lee W, Kim YS. Identification of residues essential for a two-step reaction by malonyl-CoA synthetase from Rhizobium trifolii. Biochem J. 1999;344(Pt 1):159–166. [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100050. 2006 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boghigian BA, Lee K, Pfeifer BA. Computational analysis of phenotypic space in heterologous polyketide biosynthesis--applications to Escherichia coli, Bacillus subtilis, and Saccharomyces cerevisiae. J Theor Biol. 2010;262(2):197–207. doi: 10.1016/j.jtbi.2009.10.006. [DOI] [PubMed] [Google Scholar]
- Cane DE, Walsh CT, Khosla C. Harnessing the biosynthetic code: combinations, permutations, and mutations. Science. 1998;282(5386):63–68. doi: 10.1126/science.282.5386.63. [DOI] [PubMed] [Google Scholar]
- Chan YA, Podevels AM, Kevany BM, Thomas MG. Biosynthesis of polyketide synthase extender units. J Nat Prod. 2009;26:90–114. doi: 10.1039/b801658p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158(1):9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- Clardy J, Fischbach MA, Walsh CT. New antibiotics from bacterial natural products. Nat Biotechnol. 2006;24(12):1541–1550. doi: 10.1038/nbt1266. [DOI] [PubMed] [Google Scholar]
- Cortes J, Haydock SF, Roberts GA, Bevitt DJ, Leadlay PF. An unusually large multifunctional polypeptide in the erythromycin-producing polyketide synthase of Saccharopolyspora erythraea. Nature. 1990;348(6297):176–178. doi: 10.1038/348176a0. [DOI] [PubMed] [Google Scholar]
- Dayem LC, Carney JR, Santi DV, Pfeifer BA, Khosla C, Kealey JT. Metabolic engineering of a methylmalonyl-CoA mutase-epimerase pathway for complex polyketide biosynthesis in Escherichia coli. Biochemistry. 2002;41(16):5193–5201. doi: 10.1021/bi015593k. [DOI] [PubMed] [Google Scholar]
- Dharmadi Y, Gonzalez R. A better global resolution function and a novel iterative stochastic search method for optimization of high-performance liquid chromatographic separation. J Chromatogr A. 2005;1070(1–2):89–101. doi: 10.1016/j.chroma.2005.02.075. [DOI] [PubMed] [Google Scholar]
- Donadio S, Staver MJ, McAlpine JB, Swanson SJ, Katz L. Modular organization of genes required for complex polyketide biosynthesis. Science. 1991;252(5006):675–679. doi: 10.1126/science.2024119. [DOI] [PubMed] [Google Scholar]
- Gokhale RS, Tsuji SY, Cane DE, Khosla C. Dissecting and exploiting intermodular communication in polyketide synthases. Science. 1999;284(5413):482–485. doi: 10.1126/science.284.5413.482. [DOI] [PubMed] [Google Scholar]
- Haller T, Buckel T, Retey J, Gerlt JA. Discovering new enzymes and metabolic pathways: conversion of succinate to propionate by Escherichia coli. Biochemistry. 2000;39(16):4622–4629. doi: 10.1021/bi992888d. [DOI] [PubMed] [Google Scholar]
- Harvey AL. Natural products in drug discovery. Drug Discov Today. 2008;13(19–20):894–901. doi: 10.1016/j.drudis.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Jenkins LS, Nunn WD. Regulation of the ato operon by the atoC gene in Escherichia coli. J Bacteriol. 1987;169(5):2096–2102. doi: 10.1128/jb.169.5.2096-2102.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao CM, Katz L, Khosla C. Engineered biosynthesis of a complete macrolactone in a heterologous host. Science. 1994;265(5171):509–512. doi: 10.1126/science.8036492. [DOI] [PubMed] [Google Scholar]
- Lau J, Tran C, Licari P, Galazzo J. Development of a high cell-density fed-batch bioprocess for the heterologous production of 6-deoxyerythronolide B in Escherichia coli. J Biotechnol. 2004;110(1):95–103. doi: 10.1016/j.jbiotec.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Leonard E, Yan Y, Fowler ZL, Li Z, Lim CG, Lim KH, Koffas MA. Strain improvement of recombinant Escherichia coli for efficient production of plant flavonoids. Mol Pharm. 2008;5(2):257–265. doi: 10.1021/mp7001472. [DOI] [PubMed] [Google Scholar]
- Menzella HG, Reid R, Carney JR, Chandran SS, Reisinger SJ, Patel KG, Hopwood DA, Santi DV. Combinatorial polyketide biosynthesis by de novo design and rearrangement of modular polyketide synthase genes. Nat Biotechnol. 2005;23(9):1171–1176. doi: 10.1038/nbt1128. [DOI] [PubMed] [Google Scholar]
- Murli S, Kennedy J, Dayem LC, Carney JR, Kealey JT. Metabolic engineering of Escherichia coli for improved 6-deoxyerythronolide B production. J Ind Microbiol Biotechnol. 2003;30(8):500–509. doi: 10.1007/s10295-003-0073-x. [DOI] [PubMed] [Google Scholar]
- Mutka SC, Bondi SM, Carney JR, Da Silva NA, Kealey JT. Metabolic pathway engineering for complex polyketide biosynthesis in Saccharomyces cerevisiae. FEMS Yeast Res. 2006;6(1):40–47. doi: 10.1111/j.1567-1356.2005.00001.x. [DOI] [PubMed] [Google Scholar]
- Newman DJ. Natural products as leads to potential drugs: an old process or the new hope for drug discovery? J Med Chem. 2008;51(9):2589–2599. doi: 10.1021/jm0704090. [DOI] [PubMed] [Google Scholar]
- Newman DJ, Cragg GM. Natural products as sources of new drugs over the last 25 years. J Nat Prod. 2007;70(3):461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- Paterson I, Anderson EA. Chemistry. The renaissance of natural products as drug candidates. Science. 2005;310(5747):451–453. doi: 10.1126/science.1116364. [DOI] [PubMed] [Google Scholar]
- Pfeifer BA, Admiraal SJ, Gramajo H, Cane DE, Khosla C. Biosynthesis of complex polyketides in a metabolically engineered strain of E. coli. Science. 2001;291(5509):1790–1792. doi: 10.1126/science.1058092. [DOI] [PubMed] [Google Scholar]
- Pfeifer BA, Khosla C. Biosynthesis of polyketides in heterologous hosts. Microbiol Mol Biol Rev. 2001;65(1):106–118. doi: 10.1128/MMBR.65.1.106-118.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieper R, Luo G, Cane DE, Khosla C. Cell-free synthesis of polyketides by recombinant erythromycin polyketide synthases. Nature. 1995;378(6554):263–266. doi: 10.1038/378263a0. [DOI] [PubMed] [Google Scholar]
- Quadri LE, Weinreb PH, Lei M, Nakano MM, Zuber P, Walsh CT. Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry. 1998;37(6):1585–1595. doi: 10.1021/bi9719861. [DOI] [PubMed] [Google Scholar]
- Rhie HG, Dennis D. Role of fadR and atoC(Con) mutations in poly(3-hydroxybutyrate-co-3-hydroxyvalerate) synthesis in recombinant pha+ Escherichia coli. Appl Environ Microbiol. 1995;61(7):2487–2492. doi: 10.1128/aem.61.7.2487-2492.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez E, Gramajo H. Genetic and biochemical characterization of the α and β components of a propionyl-CoA carboxylase complex of Streptomyces coelicolor A3(2) Microbiology. 1999;145(Pt 11):3109–3119. doi: 10.1099/00221287-145-11-3109. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Tang Y, Kim CY, Mathews II, Cane DE, Khosla C. The 2.7-Å crystal structure of a 194-kDa homodimeric fragment of the 6-deoxyerythronolide B synthase. Proc Natl Acad Sci U S A. 2006;103(30):11124–11129. doi: 10.1073/pnas.0601924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Textor S, Wendisch VF, De Graaf AA, Muller U, Linder MI, Linder D, Buckel W. Propionate oxidation in Escherichia coli: evidence for operation of a methylcitrate cycle in bacteria. Arch Microbiol. 1997;168(5):428–436. doi: 10.1007/s002030050518. [DOI] [PubMed] [Google Scholar]
- Wang Y, Boghigian BA, Pfeifer BA. Improving heterologous polyketide production in Escherichia coli by overexpression of an S-adenosylmethionine synthetase gene. Appl Microbiol Biotechnol. 2007;77(2):367–373. doi: 10.1007/s00253-007-1172-9. [DOI] [PubMed] [Google Scholar]
- Xue Q, Ashley G, Hutchinson CR, Santi DV. A multiplasmid approach to preparing large libraries of polyketides. Proc Natl Acad Sci U S A. 1999;96(21):11740–11745. doi: 10.1073/pnas.96.21.11740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Boghigian BA, Pfeifer BA. Investigating the role of native propionyl-CoA and methylmalonyl-CoA metabolism on heterologous polyketide production in Escherichia coli. Biotechnol Bioeng. 2010;105(3):567–573. doi: 10.1002/bit.22560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Wang Y, Boghigian B, Pfeifer BA. Probing the heterologous metabolism supporting 6-deoxyerythronolide B biosynthesis in Escherichia coli. Microbial Biotechnology. 2009;2(3):390–394. doi: 10.1111/j.1751-7915.2009.00099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Wang Y, Pfeifer BA. Bacterial hosts for natural product production. Mol Pharm. 2008;5(2):212–225. doi: 10.1021/mp7001329. [DOI] [PubMed] [Google Scholar]