Abstract
The SHP2 phosphatase plays a central role in a number of signaling pathways were it dephosphorylates various substrate proteins. Regulation of SHP2 activity is, in part, achieved by an intramolecular interaction between the PTP domain of the protein, which contains the catalytic site, and the N-SH2 domain leading to a “closed” protein conformation and autoinhibition. Accordingly, “opening” of the N-SH2 and PTP domains is required for the protein to become active. Binding of phosphopeptides to the N-SH2 domain is known to induce the opening event, while a number of gain-of-function (GOF) mutants, implicated in Noonan’s Syndrome and childhood leukemias, are thought to facilitate opening. In the present study a combination of computational and experimental methods are used to investigate the structural mechanism of opening of SHP2 and the impact of three GOF mutants, D61G, E76K, and N308D, on the opening mechanism. Calculated free energies of opening indicate that opening must be facilitated by effector molecules, possibly the protein substrates themselves, as the calculated free energies preclude spontaneous opening. Simulations of both wild type (WT) SHP2 and GOF mutants in the closed state indicate GOF activity to involve increased solvent exposure of selected residues, most notably Arg362, which in turn may enhance interactions of SHP2 with its substrate proteins and thereby aid opening. In addition, GOF mutations cause structural changes in the phosphopeptide-binding region of the N-SH2 domain leading to conformations that mimic the bound state. Such conformational changes are suggested to enhance binding of phosphopeptides and/or decrease interactions between the PTP and N-SH2 domains thereby facilitating opening. Experimental assays of the impact of effector molecules on SHP2 phosphatase activity against both small molecule and peptide substrates support the hypothesized mechanism of GOF mutant action. The present calculations also suggest a role for the C-SH2 domain of SHP2 in stabilizing the overall conformation of the protein in the open state, thereby aiding conformational switching between the open active and closed inactive states.
Keywords: Noonan’s syndrome, Childhood leukemia, CHARMM, Molecular dynamics, Potential of mean force, normal mode analysis, phosphatase assay
Introduction
SHP2 is a widely-expressed Src homology 2 (SH2) domain-containing protein tyrosine phosphatase (PTP) that is important for normal cell development. It plays a critical role in a variety of signal transduction pathways, including growth factor mediated cell proliferation1–7. The SHP2 protein contains 525 residues with two SH2 domains (N-SH2 and C-SH2) at the N terminus that include residues 1 to 103 and 111 to 213, respectively, with the PTP domain at the C-terminal region that contains the catalytic site. The PTP domain of the SHP2 protein has a low basal activity due to auto-inhibition by its own N-SH2 domain. This can be seen from the X-ray crystal structure8 that shows an extensive intramolecular interaction between the PTP and N-SH2 domains. Based on this structure it is evident that the protein must undergo a significant conformational change, with the N-SH2 domain moving away from the PTP domain (i.e. “opening”), in order for protein substrates of SHP2 to interact with the catalytic site.
Of particular interest is the presence of a number of mutants that have enhanced catalytic activity. These gain-of-function (GOF) mutations have been shown to contribute to the inherited developmental disorder Noonan syndrome (NS)9,10 and also to contribute to the pathogenesis of childhood leukemias11,12. The increased catalytic activity of these SHP2 mutants is presumably due to the ability of the protein to open more readily, leading to an autoactivated state. Increased activity is also induced by a number of phosphotyrosine (pY) containing proteins13,14, including SHP2 substrates, upon their interaction with either the N-SH2 domain or simultaneously with the N- and C-SH2 domains. Towards understanding this latter phenomena, crystal structures of the N-SH2 domain both with and without bound pY-peptides show the pY binding region to assume an open conformation in the region between the BG (residues 89 to 92) and EF (residues 66 to 68) loops15,16, while in the full SHP2 crystal structure this region of the N-SH2 domain is in a closed conformation and therefore spatially inaccessible to pY-peptides8. Thus, there is clearly conformational communication between the N-SH2 and PTP domains, with the conformations of the domains coupled to their interaction.17
Computational studies, based on molecular dynamics (MD) simulations indicate that opening of the BG loop spontaneously occurs in the absence of the PTP domain even though it is not in direct contact with the PTP domain.18 In contrast, opening of the EF loop does not occur spontaneously, and it was predicted that Tyr66 acts as a conformational switch18 controlling the conformation of the EF loop. These studies indicate a model where communication between the PTP domain and the pY binding region of the N-SH2 domain occur despite their not being in direct contact and that binding of pY effector peptides to the N-SH2 domain leads to conformational changes that facilitate sampling of the activated, open state of the full SHP2 protein. However, the nature of the structural changes that occur upon opening of SHP2 to allow access of protein substrates to the catalytic site on the PTP domain is not known. In addition, the molecular roles that GOF mutants of SHP2 play in facilitating SHP2 activation are unclear.
To gain insights into these phenomena we have undertaken an MD simulation-based study. This includes extended MD simulations of the wild-type (WT) form of the full-length SHP2 protein and of three GOF mutants D61G, E76K and N308D. In addition, calculation of the free energy associated with opening of the N-SH2 domain from the PTP domain was performed using potential of mean (PMF) force calculations in the four species. The three GOF mutants were selected for study based on their different locations in SHP2. The D61G and E76K mutants are located on the N-SH2 domain and are involved in direct interactions with the PTP domain, but are spatially removed from each other. In contrast, the N308D mutation is in the PTP domain, but is not in direct contact with the N-SH2 domain. In addition, the mutations each represent changes in the charge of the amino acid. By selecting these mutations it was anticipated that insights into the nature of their facilitation of sampling of the activated state of SHP2 could be achieved, thereby yielding molecular-detail models of their GOF activity.
Based on the molecular details obtained from the MD simulations, a model was developed in which the GOF mutations impact the binding of pY effector proteins to the N-SH2 domain, thereby facilitating opening. This observation was tested by performing experimental assays on wild-type (WT), E76K, and D61G SHP2 using both low molecular weight and protein substrates. Results from the experiments supported the presented model. It is anticipated that this model, as well as additional observations from the present study concerning the C-SH2 domain, will stimulate additional experimental work to understand the molecular details of this biologically essential protein.
Materials and Methods
Computational
Molecular modeling and simulations were performed with the program CHARMM19, with selected simulations performed with NAMD, version 2.620. Calculations were performed with the CHARMM22 all-atom additive protein force field21 with the CMAP extension22. To initiate the calculations the crystal structure of SHP2 [PDB:2SHP]8 was downloaded from Protein Data Bank. It contained two copies of self-inhibited SHP2 phosphatase in the asymmetric unit; of these the A monomer was used to initiate the present studies. The crystal structure of SHP2 has coordinates missing for residues 1, 156–160, 236–245, 295–301, and 313–323, three mutated residues Thr2Lys, Phe41Leu, and Phe513Ser and is missing coordinates for the sidechains of Lys235 and Asp294. In order to construct the missing loop residues, loop modeling23 as implemented in MODELLER 9v324 software was used. This technique predicts the best loop conformation that corresponds to the lowest energy conformation among 500 independent molecular dynamics and simulated annealing optimization runs for each loop. The loops were built in a way that residue 1 was modeled first, with the selected conformation included in the initial structure for modeling of loop 156–160 and so on.
When the loop modeling was completed the three mutations, Lys2, Leu41 and Ser513, present in the crystal structure were converted to their wild-type residues Thr2, Phe41, and Phe513 and the missing coordinates of Lys235 and Asp294 were constructed using version c35b2 of the CHARMM software19. The missing sidechain atoms of Thr2, Phe41, Lys235, and Asp294, were built using database statistics25, while the sidechain of Phe513 was built using χ angles from Phe509 of the crystal structure of SHP-1 catalytic domain. Hydrogen atoms were built based on the internal coordinates in CHARMM22 force field21. Harmonic restraints were imposed on all heavy atoms with a force constant of 10 kcal/mol/Å except for those involving the five sidechains, and the geometry of the system was optimized with 1000 steps of steepest descent (SD)26 followed by 1000 steps of conjugate gradient (CG)27. During the minimizations all energy terms were turned on except for the electrostatics. Finally, the Reduce software28 was used to optimize the conformations of the following hydrogens: OH, SH, NH3+, Met-CH3, and the orientation of Asn, Gln, and His sidechains.
Since a major goal of the study was investigating the effect of GOF mutations on the conformational properties of the SHP2 protein, we also constructed the following three mutated analogues: 2SHP-D61G, 2SHP-E76K, and 2SHP-N308D. Mutations were performed by changing Asp61 to Gly61 in the modeled WT structure and minimizing the structure for 1000 SD steps. A similar procedure was followed for the mutation Glu76 to Lys76 and Asn308 to Asp308. The four systems will be referred as WT, D61G, E76K, and N308D in the remainder of the text.
Following modeling to obtain the initial structures of the WT and mutant proteins, as described above, the crystallographic water molecules within 5 Å of the SHP2 were added to each coordinate set and each system was energy optimized with 5000 SD steps followed by 5000 CG steps. Next, a pre-equilibrated cubic box of TIP3P water molecules29,30 was superimposed on the protein + crystallographic water structures. The size of the box was chosen such that there was at least a 12 Å layer of water between the solute and the edge of the box. Water molecules within 4.1 Å of the protein were deleted, and sodium counterions were placed at random positions around the solute to neutralize the net charge. The following four systems were generated – 118,793 atoms including 3 counterions for WT SHP2 in a cubic box of size 108 Å, 135,180 atoms including 3 counterions for D61G SHP2 in a cubic box of size 113 Å, 135,469 atoms including 1 counterion for E76K SHP2 in a cubic box of size 113 Å and 135,181 atoms including 4 counterions for N308D SHP2 in a cubic box of size 113 Å. Periodic boundary conditions31 were used and Coulomb interactions were treated with the particle mesh Ewald method32 with a real space cutoff of 10 Å, a kappa value of 0.34, order 6 B-spline interpolation, and a grid spacing of approximately 1.0 Å. Lennard-Jones interactions were truncated from 8 Å to 10 Å with force switching33, and a LJ long-range correction31 was applied to account for the effect of Lennard-Jones interactions beyond the truncation distance. The SHAKE34 algorithm was applied to all covalent bonds involving hydrogen atoms. All protein structures were initially harmonically restrained with a force constant of 50 kcal/mol/Å2, to allow the water and counterions to minimize for 2000 SD steps. Subsequently the restraints were relaxed to 5 kcal/mol/Å2 and the whole solute+water+ions system was minimized for 1000 CG steps.
MD simulations for all the systems were performed in the isobaric-isothermal ensemble. Prior to the production MD calculations, each solute+water+ions system was minimized and equilibrated using CHARMM. For equilibration the equations of motion were integrated with the 'leap-frog' algorithm35 with a 2 fs integration timestep to propagate the system. The temperature was maintained at 298 K by a Nosé-Hoover heat bath36,37, with the thermal piston parameter of 10,000 kcal/mol/ps2. A constant pressure of 1 atm was maintained using the Langevin piston method38 with a mass of 1,000 amu. The initial 100 ps of water-ion equilibration was performed by imposing 5 kcal/mol/Å2 harmonic restraints on the solute, followed by 500 ps of unrestrained dynamics of the entire system. Finally, 20 ns Langevin dynamics simulations were performed using NAMD2.6 software with the CHARMM22/CMAP force field. Langevin dynamics employed a coupling coefficient of 1/ps with a temperature bath of 298 K, and the Langevin piston method was used to maintain system pressure at 1 atm.
Calculation of the potential of mean force (PMF) for opening of the N-SH2 domain with respect to the PTP domain was calculated using the MMFP module in CHARMM. The reaction coordinate was defined as the distance between the center of mass (COM) of the two domains. A restraining harmonic potential, E(r) = k·(r−r0)2, was applied between the COMs of the two interacting domains, N-SH2 (residues 1–103) and PTP (residues 221–525) and was used to “pull” the domains apart to the fully open state. The value of r represents the equilibrium center of mass distance and the value of k was set to be 20.0 kcal/mol/Å2. To obtain initial structures for the PMF calculations r was first set to 30 Å corresponding to the distance in the WT SHP2 crystal structure. Each system was then equilibrated for 100 ps in the constant volume and temperature (NVT) ensemble. The final structure was then used as the starting structure for the adjacent window by changing r by ±1 Å, performing a 100 ps equilibration and using the final structure to initiate the next window. This process was performed decreasing r to 25 Å and increasing r to 60 Å, representative of the fully open state. Data for the PMF were then generated by running two adjacent windows from these equilibrated structures. For example, the final structure from the r = 30 Å equilibration was used to initiate windows at 30 and 30.5 Å. PMF sampling was performed in the constant temperature and pressure (NPT) ensemble, with the coordinates saved every 2ps and the COM distances every MD step for calculation of the free energy surface. The free energies were calculated from the weighted-histogram procedure (WHAM)39,40. The final PMFs were calculated from 27 to 50 Å due to the WHAM procedure not converging at distances shorter than 27 Å and not all PMFs being fully sampled out to 60 Å. This was not deemed an issue as the free-energy profiles showed asymptotic convergence with increasing r by the point where r = 50 Å.
Normal mode analysis (NMA) was performed using the VIBRAN module in CHARMM. Quasiharmonic normal modes were calculated from the atomic fluctuations obtained from the unrestrained MD simulations over the 5 to 20 ns range. The atom position fluctuation tensor was generated, mass weighted and diagonalized for all Cα atoms of the WT, D61G, E76K, and E308D systems. The significant difference of the quasi-harmonic method from the conventional normal modes calculation is the presence of anharmonic terms. Energy minimized average coordinates of each system from the 15ns simulations were set as the reference. The temperature was set 298 K. Also the overall translational and rotational motions were removed from the dynamics trajectory to properly study the internal motions. 1575 normal mode vectors for each system were generated and analyzed.
Experimental
In Vitro and In Vivo Phosphatase Assay
WT and GOF mutant SHP2 GST fusion proteins were generated and purified following standard procedures. A phospho-peptide corresponding to the surrounding sequence of pTyr1018 in EGF receptor (Asp-Ala-Asp-Glu-Tyr[PO3H2]-Leu-Ile-Pro-Gln-Gln-Gly) (pY-EGFR) or a low molecular weight compound p-nitrophenyl phosphate (pNPP) were used as substrates. When pY-EGFR was used as the substrate, GST-SHP2 fusion proteins (1 µg) were incubated with pY-EGFR peptide (0.5 mM) in 40 µL assay buffer (25 mM Tris-HCl, PH7.4, 50 mM NaCl, 5 mM DTT, and 2.5 mM EDTA) at 30°C for 30 min. 50 µl of Malachite Green solution was then added and OD620 was measured after 5min. When pNPP was used as the substrate, purified GST-fusion proteins (1 µg) were incubated with pNPP (3.3 mg/ml) at 37 °C for 1 hour in a reaction buffer containing 50 mM imidazole (pH 7.5) and 5 mM dithiothreitol. The reaction was stopped by the addition of 0.1 N NaOH, and the pNPP hydrolysis was measured by absorbance at 410 nm (OD410).
For the in vivo phosphatase assay, WT and SHP2D61G/D61G mutant mouse embryonic fibroblasts derived from WT and SHP2 D61G knock-in mice41 were starved in serum-free medium overnight and then stimulated with 15% fetal bovine serum (FBS) for 10 min. The cells were harvested and lysed in RIPA buffer (50 mM Tris-HCl pH 7.4, 1% NP-40, 0.25% Na-deoxycholate, 150 mM NaCl, 1 mM EDTA, 1 mM NaF, 10 µg/mL leupeptin, 10 µg/mL aprotin, and 1 mM PMSF). Whole cell lysates (500 µg) prepared were immunoprecipitated with 1 µg of anti-SHP2 antibody. Immunoprecipitates were washed three times with HNTG buffer (20 mM Hepes pH 7.5, 150 mM NaCl, 1% Glycerol, and 0.1% Triton X-100) and assayed for SHP2 catalytic activity using pY-EGFR or pNPP as substrates as described above.
Results and Discussion
Molecular dynamic simulations are emerging as an important tool in elucidating motions and conformational properties of biologically important molecules on the nanosecond timescales. They can provide detailed information on the changes occurring in the molecule that are difficult to observe using crystallographic, NMR or other experimental approaches. To understand structural changes associated with the opening of SHP2 leading to its activated state and to elucidate molecular details of the impact of selected GOF mutations of SHP2 that lead to enhanced biological activity of SHP2 we undertook calculations on the WT and three mutant proteins.
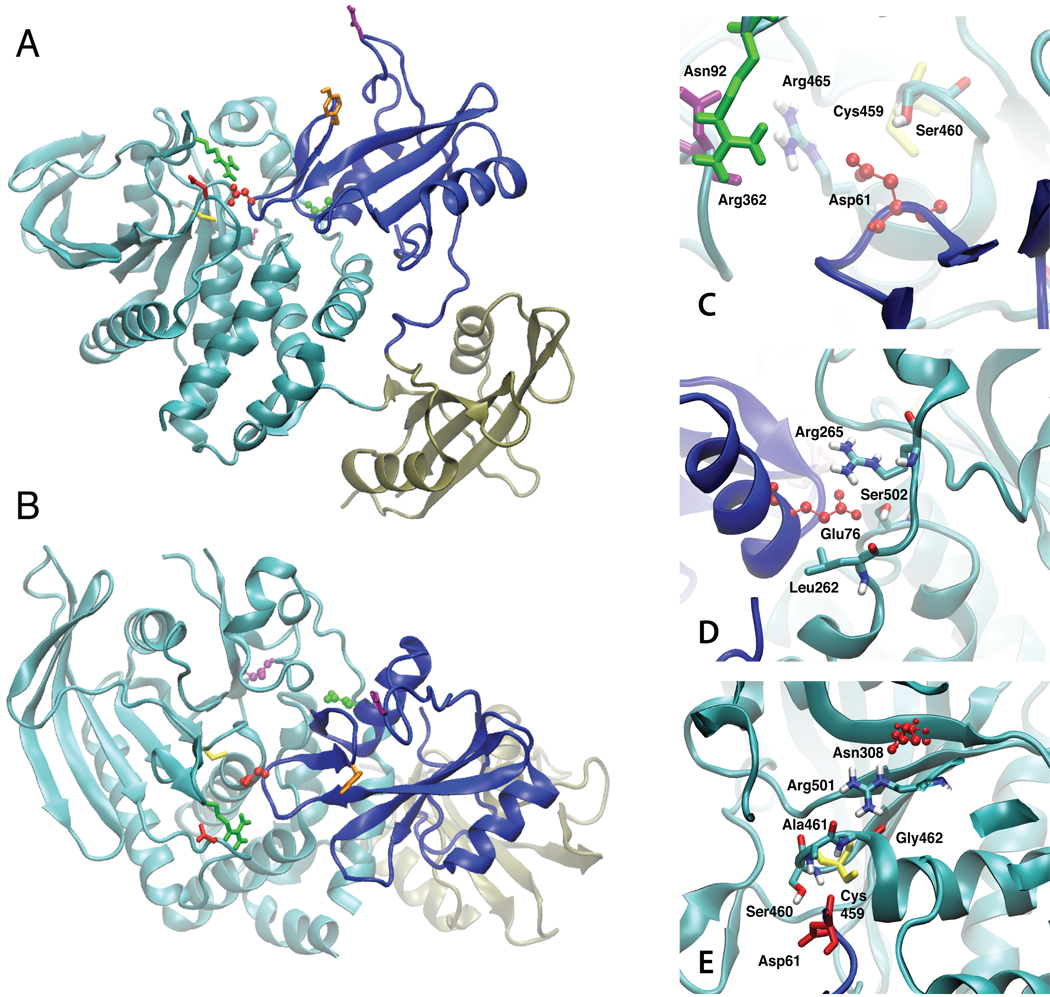
In the crystal structure of the SHP2 protein (Fig. 1 A and B) the PTP domain (residues 221–525, cyan) interacts with its N-SH2 domain (residues 1–103, blue), an interaction that involves a large protein-protein interface. In contrast, the C-SH2 domain (residues 112–216, tan) is tethered to both the N-SH2 and PTP domains but does not have a significant interface with either. The large interface between the PTP and N-SH2 domains makes the catalytic site inaccessible, thereby leading to the inactive form of the protein. Opening of the N-SH2 domain away from the PTP domain is suggested to lead to an open, catalytically competent state, based on a number of experimental studies42,43 showing that SHP2 truncation mutants lacking the N-SH2 domain have significantly increased activity.
FIGURE 1.
Images of the crystal structure of the SHP2 protein in cartoon representation. A and B represents two orientations of the protein. The PTP domain is cyan, the N-SH2 domain is blue and the C-SH2 domain is tan. Residues shown include Cys459 (yellow), Asn92 (purple, on BG loop), Tyr66 (orange, on EF loop), Arg362 (green, on FH loop) and Asp425 (red, on LF loop) in licorice and the three mutant residues in CPK are Asp61 (red, D61G), Glu76 (green, E76K) and Asn308 (purple, N308D). Images of the crystal structure in the region of the C) D61G, D) E76K, and E) N308D mutations. The respective residues undergoing mutation are in red CPK format and selected residues in the vicinity of the mutated residues are in licorice, atom type coloring with the exception of Cys459 in yellow licorice in C and E, Asp61 in red licorice in E, Asn92 in purple licorice in C and Arg362 in green in C.
Our initial assumption in designing the present study was that the GOF mutants simply lead to either a lowering of the barrier for the conformational change from the closed to the open state or a change in the equilibrium between the closed and open states. To address these possibilities, potential of mean force (PMF) calculations, in which the change in the free energy as a function of a selected reaction coordinate is obtained, were undertaken. These efforts required first modeling of residues missing from the crystallographic structure of the full-length SHP2 protein8, modeling of the mutations into the structure and equilibration of the 4 systems before undertaking the PMF calculations. Results from the PMF calculations lead to the conclusion that GOF impact on activity was related to alterations of the closed state of SHP2. Accordingly, extensive analysis of unbiased simulations of the closed state was performed yielding a model for the mechanism of activation of SHP2, including the impact of the GOF mutations. Subsequent experimental studies were consistent with this model.
Modeling of structures
Initial calculations involved building loops on the WT protein that were not observed in the crystal structure followed by creation of the three mutants and the appropriate equilibration of the structures. This was followed by unrestrained 20 ns MD simulations on all four systems. Overall behavior of the simulations was monitored based on the root-mean-square deviations (RMSD) and fluctuations (RMSF) of the backbone Cα atoms. In the simulations the structures relax during the initial 2.5 ns (see Fig. S1, supporting information), followed by fluctuations around approximately stable values for the remainder of the simulations. RMSD values averaged over the final 15 ns of the simulations are presented in Table I. As is evident, the magnitude of the RMSD for the full protein is dominated by the C-SH2 domain (Table IA), while those of the PTP and N-SH2 domains are significantly less. The larger RMSD values of the C-SH2 domain are likely due the minimal amount of direct interactions it has with either the PTP or N-SH2 domains (Fig. 1). However, its intramolecular conformation relatively stable, as based on RMSD values of 2.7 Å or less following alignment based on its own Cα atoms (Table IB). With the PTP domain, the RMSD is dominated by residues in the vicinity of 298 and 319 (Fig. S2 of the supplemental material), both of which are regions that were not resolved in the experimental structure, requiring their modeling in the present study. These regions as well as a number of regions in the C-SH2 domain, also undergo significant fluctuations during the MD simulations (Fig. S3 of the supplemental material) suggesting their intrinsic flexibility, results that are consistent with their experimental B factors (Fig. S3). However, as these regions are not adjacent to the N-SH2 domain, their flexibility is anticipated to not significantly impact the remainder of the protein. Overall, these results indicate that the applied simulation methodology yields stable simulations suitable for analysis of the impact of the mutations on the protein structure and dynamics.
TABLE I.
Average RMSD values of Ca (Å) of the entire protein from the 15 ns MD simulations. RMSD is calculated for the entire protein A) based on the alignment to the full protein and B) based on the alignment of individual domains (N-SH2, C-SH2 and PTP) to themselves. Averages and standard deviations in parentheses were calculated using 3000 ps block averages.
| WT | D61G | E76K | N308D | |
|---|---|---|---|---|
| A) Full | ||||
| SHP2 | 2.5 (0.1) | 2.6 (0.2) | 2.2 (0.2) | 2.2 (0.2) |
| N-SH2 | 1.6 (0.1) | 2.0 (0.1) | 1.8 (0.4) | 1.8 (0.1) |
| C-SH2 | 3.6 (0.3) | 4.2 (0.4) | 3.4 (0.5) | 3.2 (0.2) |
| PTP | 2.4 (0.0) | 2.0 (0.1) | 1.7 (0.2) | 1.9 (0.1) |
| B) Each domain | ||||
| N-SH2 | 1.2 (0.1) | 1.3 (0.1) | 1.3 (0.2) | 1.3 (0.1) |
| C-SH2 | 2.7 (0.3) | 2.2 (0.2) | 2.2 (0.3) | 1.9 (0.2) |
| PTP | 2.1 (0.2) | 1.5 (0.1) | 1.5 (0.2) | 1.6 (0.1) |
SHP2 opening potential of mean force calculations
To investigate the pathway of opening of the SHP2 N-SH2 domain from the PTP domain and estimate the free energy associated with that opening event PMF calculations were performed on the four systems. The reaction coordinate for the PMF was the distance between the center of mass (COM) of all atoms in the N-SH2 domain and the COM of all atoms in the PTP domain. Based on this definition a COM distance of approximately 30 Å represents the closed conformation as observed in the crystal structure of full SHP2. The use of a biasing potential on this degree of freedom allows for full relaxation of both the individual domains and their relative orientation as a function of opening distance. The PMF calculation was performed over the range of 27–50 Å; the shorter distances were performed to artificially “push” the PTP and N-SH2 domains together to achieve an increase in energy at short distances thereby allowing for a minimum in the surfaces to be identified. The force constant, k=20 kcal/mol/Å2, on the COM restraint was chosen such that the COM distance histograms for the individual windows had adequate overlap (Fig. S4 of the supporting information) as required for generating a continuous and reliable free-energy profile. However, at several distances a force constant of 40 kcal/mol/Å2 was used to obtain the appropriate sampling; during calculation of the PMF using WHAM39,40 the data was weighted appropriately.
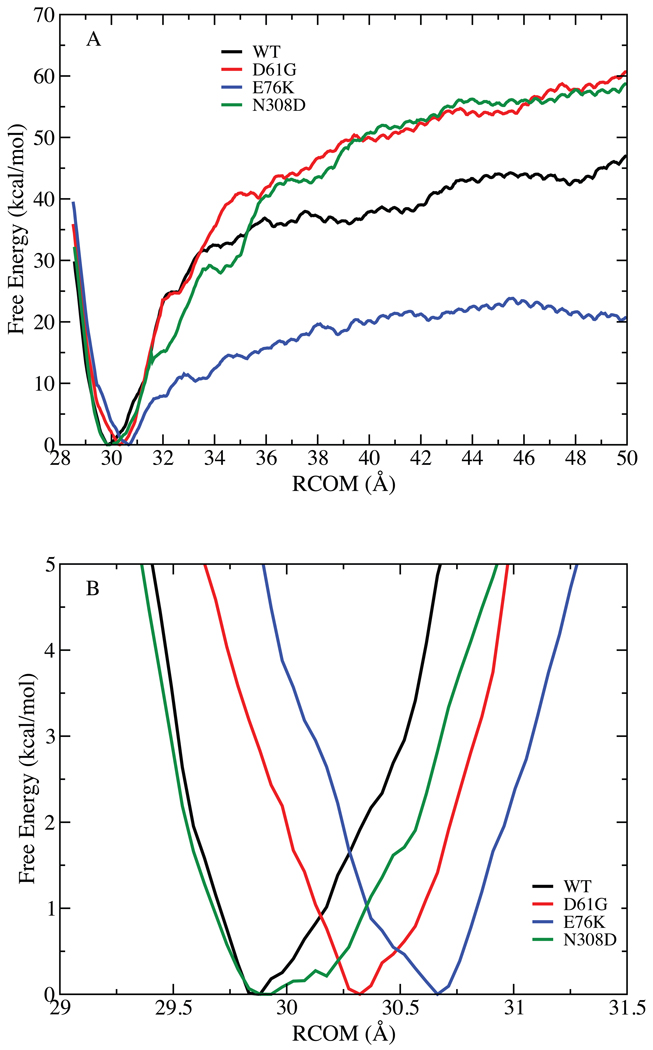
The resulting free energy profiles are presented in Fig. 2. The overall shape of the curves for all four systems is similar. Going to distances shorter than the COM distance in the WT crystal structure of 30 Å leads to an increase in energy as the two domains press against each other. As opening occurs there is a rapid rise in energy, reaching an approximate plateau in the vicinity of 30 to 50 kcal/mol at a COM distance of 45 Å. The impact of the extent of sampling on the computed PMF was initially performed by calculating the PMFs over 500 – 1000, 1000 – 1500 and 1500 – 2000, and 2000 – 2500 ps sampling windows (Fig. S5 and S6 of the supporting information). Increased sampling leads to a decrease in the maximum energy at 50 Å by over 50 kcal/mol upon going from sampling from 500 – 1000 ps to 2000 – 2500 ps, indicating the importance of extended sampling to obtain satisfactory convergence in the PMFs. In the case of the E76K mutant an additional 500 ps of sampling was performed (Fig. S5, supporting information), indicating that the 2000 – 2500 ps results are adequately converged. While the present results appear to be converged the possibility that additional sampling will lead to alterations in the energy surfaces cannot be excluded. Towards this end the results of Banavali and Roux are of note44. The authors undertook the PMF calculations on Src kinase, a protein of 270 residues, determining the free energy profile for opening of the 10 N-terminal residues. In their case 210 ps of equilibration followed by 280 ps of sampling per window was deemed to be sufficient. Other studies associated with flipping of a base out of a DNA duplex45,46 indicate that 200 ps of sampling per window is adequate. However, it may be expected that different types of structural changes will require different amounts of sampling, leading to extended amount of sampling for the opening of SHP2 used in the present study.
FIGURE 2.
Potential of mean force for opening of the N-SH2 domain from the PTP domain. A) Free energy (kcal/mol) as a function of distance (RCOM, Å) between the center of mass the PTP and N-SH2 domains as calculated from the last 500ps of sampling in each window. B) Expanded view of the minima in the PMF for all 4 systems.
To obtain an estimate of the error in the calculated PMFs additional analysis of the WT PMF was undertaken. This involved calculating PMFs for five 200ps blocks over the final 1 ns of sampling (1500 to 2500 ps) from which averages and standard deviations over the five block values were calculated. Results from these calculations are shown in Figure S5.1. This analysis indicates the standard deviation in the free energy profiles to be 1 kcal/mol in the vicinity of the minimum in the RCOM free energy surface and increase to approximately 10 kcal/mol in the region from 32 Å to 50 Å. It should be noted that such an analysis assumes that the five windows represent independent samples. A more rigorous error analysis would require repeating the full PMF calculations multiple times, a computational effort that is beyond the scope of the present study.
Extent of opening of SHP2 required for substrate-protein binding
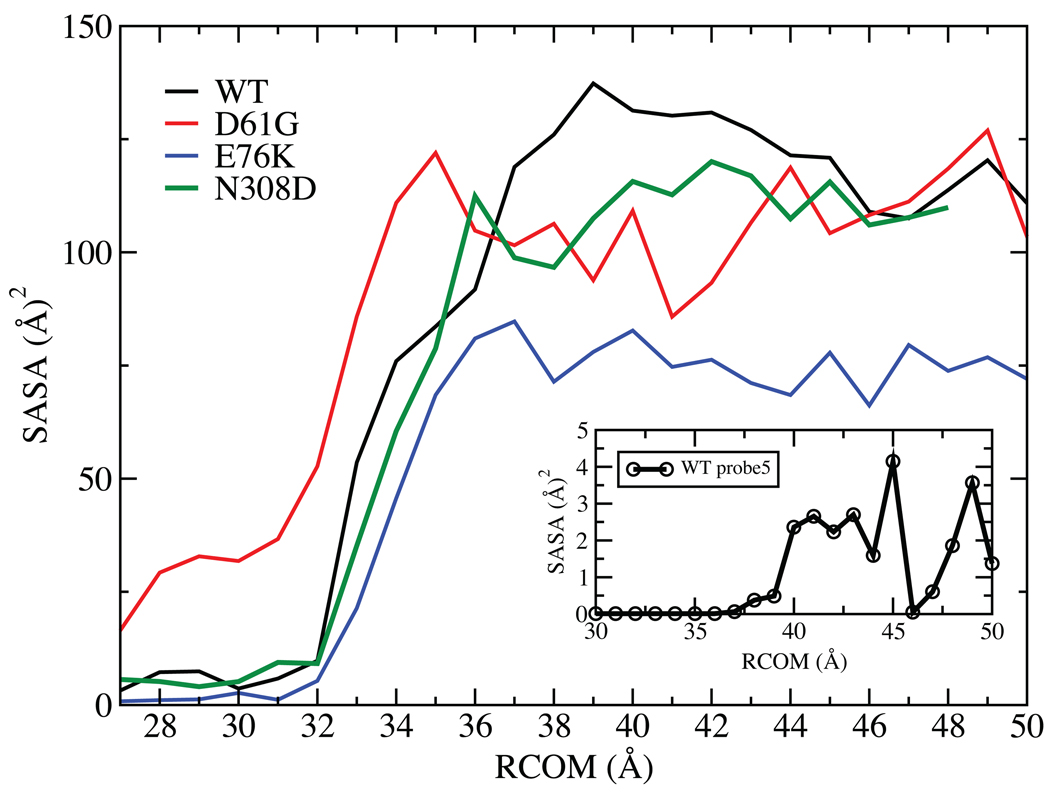
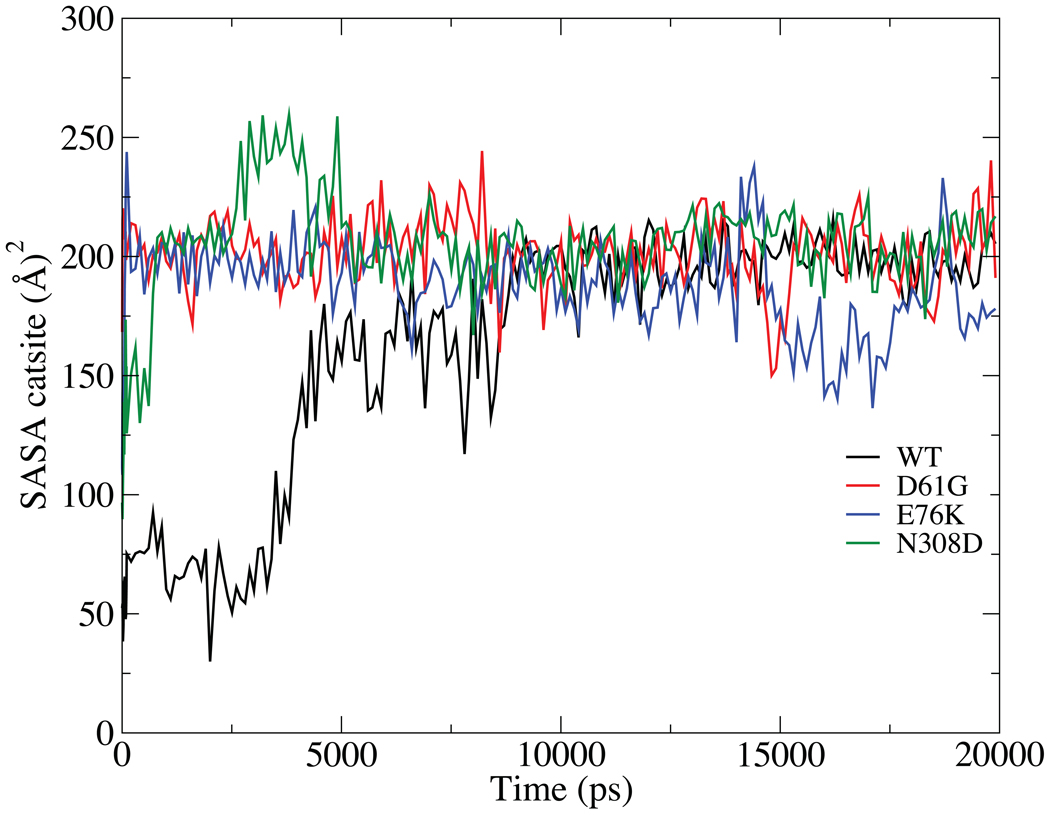
While absolute convergence of the energy surfaces in Fig. 2 cannot be assured the results do suggest that opening of SHP2 alone to achieve its activated state does not occur spontaneously. In the WT and E76K simulations, free energies of 10 kcal/mol are encountered within 2 Å of the energy minimum associated with the closed state as opening occurs. This is before significant opening has occurred as evidenced by the lack of change in the solvent accessible surface area (SASA) of the catalytic residues in the PTP domain until 32.5 Å (Fig. 3). The E76K mutant has a lower SASA after about 37 Å then the WT and the two other mutants. Analysis of individual residues revealed that Ile 463, Gly 464 and Agr 465 in the catalytic site were responsible for this decrease (data not shown). This is another indication that the changes associated with E76K mutations altering the catalytic site are more pronounced than in other two mutants, leading to the drastic changes of the solvent accessibility at the RCOM greater than 37 Å. In addition, to mimic the opening required to accommodate a “protein” interacting with the catalytic sites, the SASA calculation was repeated using a probe size of 5 Å on the WT protein. The resulting change in SASA, shown in the inset of Fig. 3, indicates that adequate opening required for interaction of a phosphoprotein with the catalytic site does not occur until > 37 Å. In addition, it is clear that there is no single stable open state as the free energy profiles continue to increase out to 50 Å with no significant energy minimum being observed. These results indicate that it is unlikely that opening of the N-SH2 domain from the PTP domain occurs spontaneously. Accordingly, the present results suggest that opening of SHP2 to attain an activated state is a facilitated process that may involve interactions with the substrate protein or other effector molecules.
FIGURE 3.
Solvent accessible surface area (SASA, Å2) with the 1.4 Å probe sphere radius of the catalytic site (residues 458 to 466) as a function of RCOM (Å) from the four PMF calculations. The inset is the SASA calculated for the WT with the 5 Å probe sphere radius as a function of RCOM (Å). The values are calculated from the last 500ps of sampling in each window.
Role of C-SH2 domain in SHP2 opening
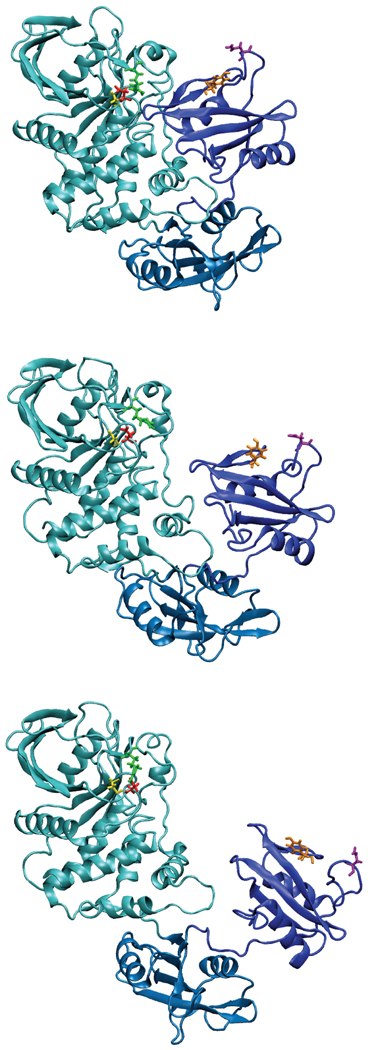
Images of the opening of WT SHP2 are presented in Fig. 4 and Fig. S7 of the supplemental material. As may be seen, the movement of the N-SH2 domain relative to the PTP domain involves a sliding motion, moving that domain over the C-SH2 domain. While some motion of the C-SH2 domain occurs, it is relatively small. From the images it is evident how the motion of the N-SH2 domain readily allows the protein substrates of SHP2 access to the catalytic site. Further, given that the motion involves sliding of the N-SH2 domain onto the C-SH2 domain, it may be hypothesized that the role of the C-SH2 domain is to act as a platform to stabilize the overall protein structure while in the open state, potentially facilitating the protein switching back to the closed, inactive state. In the absence of the C-SH2 domain, the N-SH2 domain may lose structural organization making it more difficult to revert to the closed conformation.
FIGURE 4.
Images of the opening of WT SHP2 taken from the PMF calculation at 30 top), 40 (middle) and 50 (bottom) Å using the time frame at the first 500 ps from the respective windows. The PTP (cyan), N-SH2 (blue) and C-SH2 (blue2) domains are shown in cartoon format. Residues shown include Cys459 (yellow), Asn92 (purple, on BG loop), Tyr66 (orange, on EF loop), Arg362 (green, on FH loop) and Asp425 (red, on LF loop).
Analysis of the conformation of SHP2 and its mutants in the closed state
Importantly, the energy changes in the PMFs calculated for the mutants all show a significant increase in energy upon opening. With the D61G and N308D mutants the energies are higher than those of the WT, while lower energies occur with opening in the E76K. However, in all cases it is evident that the energy required to open the protein enough to allow protein substrates to bind is 20 kcal/mol or more, indicating that these mutations do not lead to spontaneous opening. Thus, while significant increases in activity occur with the GOF mutants due to increased access to the open, activated state of the protein, the present calculations indicate that this is not due to the protein simply opening more easily. In combination with the conclusion that opening of SHP2 requires interactions with additional molecules, these results suggest that the change in activity associated with the GOF mutants is due to their impact on the closed state thereby facilitating interactions with the substrate proteins or other effector molecules. Motivated by this model, subsequent analyses focused on the closed state of the WT and GOF mutants.
As the present results predict that opening of SHP2 does not occur spontaneously and that opening may be facilitated by the interaction of the substrate protein with SHP2, simulations in the vicinity of the closed state were analyzed in more detail. Evident from the PMF calculations are differences in the vicinity of the minima in the WT and mutant free energy surfaces, where there is a small shift in the location of the minima (Fig. 2B and Fig. S6 of the supporting information for the impact of sampling time). In the case of the WT, the minimum is located at 29.8 Å, in the both the D61G and N308D mutants it shifts outward 30.2 Å while a larger shift to 30.7 Å occurs in the E76K mutants. While these shifts are quite small and may be an artifact of sampling, they support the hypothesis that changes in the protein conformation may be occurring in the closed state due to the mutations. Such subtle conformational changes may allow for the mutants to more readily achieve open, activated states as compared to WT SHP2 via interactions with substrate or effector molecules.
The D61G and E76K mutants are located on the surface of the N-SH2 domain directly adjacent to the catalytic region of the PTP domain (Fig. 1). Asp61 is located such that it mimics the location of the phosphate moiety of the normal substrate, as well as being involved in additional hydrogen bond interactions with Ser460 and ionic interactions with Arg362. Glu76 is involved in an ionic interaction with Arg265 and a hydrogen bond with Ser502. Accordingly, the D61G and E76K mutants will directly impact the strength of the PTP-N-SH2 domain interactions, an effect that appears to lead to the outward shift of the location of their minima (Fig. 2B). However, the N308D mutation is not involved in direct interactions with the N-SH2 domain and further analysis, discussed below, indicates that the D61G and E76K mutants may have additional impact on the structure of SHP2 contributing to their GOF. Further analysis focused on the simulations around the minimum COM distance at 29.5, 30.0 and 30.5 Å used for the PMF calculation and on the final 15 ns of the 20 ns simulations performed without restraints.
Comparison of SASA in WT and mutant SHP2
Presented in Tables SI and SII are the solvent accessible surface area (SASA) values and differences relative to WT for selected residues in the N-SH2 and PTP domains, respectively, in the closed state. Residues include those on the N-SH2 domain at the interface with the PTP domain (Table SI) and PTP residues in the catalytic site (458–466) and in three loops containing Asp425 (LF loop), Arg362 (FH loop), and Arg278 (D’B loop) involved in catalysis and phosphate binding (Table SII). In the case of the N-SH2 domain changes in the SASA for the individual residues are similar in the WT and mutants. The only significant differences occur with Thr2, Ser3, Pro9 and Asn10, which are located on the side of the N-SH2 domain adjacent to the C-SH2 domain. The location of these residues suggest that the differences in SASA may not be associated with the change in activity of the mutants, though a possible role for these residues in impacting interactions between the N- and C-SH2 domains that may effect opening cannot be excluded. The only other significant change is with Glu76 in the E76K mutant. This is due to a shift in the mutated sidechain towards the protein surface consistent with the location of Arg265 adjacent to Glu76 in WT SHP2 (Fig. 1D).
Larger differences in SASA between the WT and mutants are evident in the PTP domain (Table SII). Leu261 and Leu262 show a significant increase in exposure in the E76K and N308D mutants. However, the largest difference is seen with Arg362, which has significantly larger SASA in the three mutants. Notably, the SASA of Arg362 is low (38 Å2) in the WT crystal structure. Additional analysis of the SASA of Arg362 as a function of time was performed using the 20 ns MD simulations (Fig. 5). This shows the SASA to increase instantaneously in the D61G and E76K mutants, followed by the N308D mutant and with the change occurring over almost 9 ns in the WT simulation. Given that a positive charge at position 362 is highly conserved in the SHP phosphatases8, further analysis was performed to understand the nature of this differential behavior and its possible link to GOF in the mutants.
FIGURE 5.
Solvent accessible surface area (SASA, Å2) for Arg362 as calculated over the course of the 20ns MD simulations.
Shown in Fig. S8 of the supplemental material is the WT crystal structure, including surrounding image proteins in the crystal environment. As may be seen, Arg362 has an interaction with Asp61 at a distance of approximately 8 Å. While not in direct contact, the ionic nature of the interaction would be anticipated to be favorable such that mutation of the aspartate to a glycine in D61G would lead to the loss of this stabilizing interaction and subsequent movement of Arg362 leading to the increase in its SASA. Both Glu76 and Asn308 are more than 20 Å from Arg362 and their impact on the orientation of Arg362 appears to be indirect in nature (see below, residues not shown in Fig. S8). Interestingly, the sidechain of Arg362 is close to the carboxylate of Asp451’, a residue on a symmetry related molecule in the crystal that is in direct contact with the Arg362 guanidinium group. The presence of this interaction is suggested to stabilize the Arg362 sidechain in the conformation seen in the crystal structure; the loss of the interaction when SHP2 is in solution leads to the shift in the position of this residue in the WT simulation. However, the motion does not occur until after 5 ns in the WT simulation, suggesting that the lower SASA conformation of Arg362 is more stable in the WT versus the mutants. Indeed analysis of the 500 ps timeframe of the PMF 30 Å window of the WT systems shows the guanidinium moiety of Arg362 to be in direct contact with the sidechain of Asp61, participating in an ionic interaction (not shown). That the mutants impact the orientation of Arg362 as well as the conformation of the region around that residue stimulated additional analysis of this residue.
Role of EF, BG, FH, and LF loops on SHP2 conformation
Arg362 is located in an interesting region on the protein (Fig. 1). It is on the FH loop, which is on the surface of SHP2 in the closed state, lying over a portion of the N-SH2 loop on which Asp61 is situated. As stated above, Asp61 mimics the interaction of the substrate phosphate moiety with the catalytic site in the PTP domain. Its location is such that as the N-SH2 domain moves away from the PTP domain, opening initially occurs in the vicinity of the FH loop (See Fig. 4). In the crystal structure (WT, Fig. S8) the Arg362 sidechain is in an orientation in which it is largely excluded from solvent and, as stated above, can participate in ionic interactions with Asp61. Indeed, during initial stages of opening of the WT protein, there is an ionic interaction between Arg362 and Asp61. In addition, Arg362 is spatially adjacent to Asp425, located in the LF loop, another highly conserved residue that is also on the surface of the protein in the region where opening initially occurs (Fig. 4). Though the conformational change of Arg362 is ultimately observed in WT SHP2, the presence of that change along with the conserved nature of the charge associated with that residue and of Asp425, suggest that this region of the protein may play a role in initiating the opening of SHP2 required for interactions with substrate proteins. Further, given the negative charge of the pY residue on the SHP2 substrate proteins, it is tempting to hypothesize that the conserved, positively charged Arg362 sidechain may participate in initial interactions with substrates, interactions that may then facilitate opening of SHP2 to its active form. In the D61G mutant, the loss of negative charge would lead to a loss of ionic interactions, allowing the residue to interact more with the surrounding environment, thereby facilitating opening of SHP2 by more readily interacting with the substrate proteins. With E76K, its location on the B helix is such that the mutation could impact the small β sheet that includes strands D’, E and F, thereby impacting Asp61 on the D’E loop and facilitating the conformational change in Arg362. A long range communication network leading to Asp61, as discussed below, may be responsible for the change in Arg362 that occurs with the N308D mutation, though the slower change in the conformation with this mutation indicates that the change may be solely due to the loss of the crystal contact discussed above for the WT.
The SASA analysis showed a significant increase in the accessibility of Leu262 in the E76K mutant (Table SII). Analysis of the region around Glu76 shows interactions with several residues in WT SHP2, including Leu262, Arg265 and Ser502 (Fig. 1D). The Oγ atom of Ser502 is hydrogen bonded to the carboxylate of Glu76, being 2.80 Å from the Oε1 atom in the crystal structure, the guanidinium moiety of Arg265 is approximately stacked with the Glu76 carboxylate (Arg265 Cζ and Nη2 atoms are 3.94 and 3.96 Å, respectively, from Glu76 Oε1 and the Glu76 Cγ atom is adjacent to the Leu262 Cδ2, at a distance of 3.63 Å). The E76K mutation significantly disrupts these interactions, leading to the increased solvent exposure of Leu262 (Table SII), which allows the sidechain of Lys76 to move freely. The average calculated RMS fluctuations of Lys 76 sidechain over the 15 ns simulation are 1.88 Å for WT and 1.96 Å for D61G, 3.99 Å for E76K and 1.24 Å for N308D mutants respectively. The increased motion of Lys76 is translated to the motion of the αB helix (residues 74 to 85). Alterations in the αB helix, which ends at residue 85, may potentially impact the BG loop on the N-SH2 domain (residues 89 to 92), thereby impacting the phosphopeptide binding region (see below). Moreover, the loss of the interactions of Glu76 with the residues on the PTP domain listed above will have a direct, adverse impact on the interaction between the N-SH2 and PTP domains, contributing to the shift in the location of the minima in the PMF and possibly to the overall lowering of the energy of opening in this mutant (Fig. 2).
Normal mode analyses of loop flexibility
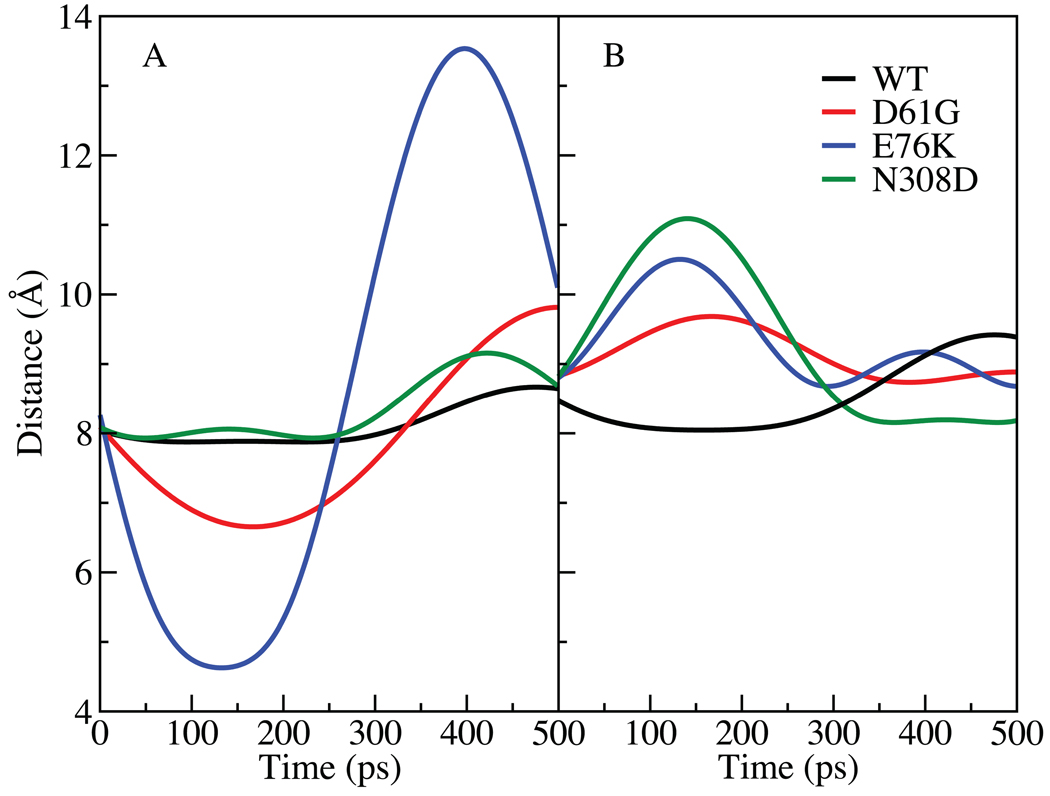
Given the important roles of the EF, BG, FH, and LF loops on the activity of SHP2, normal mode analysis was undertaken to further investigate changes in the flexibilities of the loops in the GOF mutations. Quasiharmonic normal mode analysis is a useful technique for studying vibrational and thermal properties of molecules; in the present study the goal was to identify motions that may be associated with the changes in SHP2 associated with the D61G and E76K mutants discussed above and identify alterations associated with the N308D mutant that may lead to its GOF activity. In quasiharmonic normal mode analysis the fluctuations of the atoms, presently the Cα atoms, from the MD simulations is used to create an atom-atom fluctuation matrix that is diagonalized from which a series of eigenvalues, or frequencies, and their associated eigenvectors, which describe the collective motions of groups of atoms, are obtained. Low frequency modes, which represent the largest scale motions, are then analyzed to identify interesting motions, in the present case being motions of the EF, BG, FH and LF loops. Accordingly, quasiharmonic vibrational analysis was performed for the WT and the three mutants at room temperature (298K). Systematic analysis of the low frequency modes was then undertaken, from which the lowest frequency mode was observed to contain significant contributions from the four loops in the mutant structures. The COM distances as a function of time between the BG and EF loops and FH and LF loops associated with the lowest frequency mode were than calculated and are presented in Fig. 6. From Fig. 6 it is clear that the three mutants demonstrate increased fluctuations between both pairs of loops relative to the WT. In the case of the BG and EF loops located on the N-SH2 domain the largest changes occur with the mutations in the N-SH2 domain, though the motion is slightly larger in the N308D mutant as compared to the WT. The change is especially pronounced for E76K, with the distance between the loops fluctuating between 4.7 Å and 13.5 Å (Fig. 6A). Significant motions also occur with the D61G mutant. In the case of the relative motion of the FH and LF loops (Fig. 6B) the largest amplitude motion occurs with the N308D mutant while somewhat larger amplitude motions occur with the D61G and E76K mutants. Verification of the changes in the amplitudes of the motions was performed by directly analyzing the variation of the distance between the BG and EF loops and FH and LF loops as a function of time from the 20 ns simulations (Fig. S9 of the supplemental material). This analysis showed differences in the fluctuations for the different systems consistent with those calculated from the quasiharmonic analysis.
FIGURE 6.
COM distances as calculated form the trajectory generated from the lowest mode1575 between A) the BG loop, residues 91–93 and EF loop, residues 66–68 and B) the FH loop, residues 361–363 and LF loop, residues 425–427. The WT SHP2 is represented in black and the mutants are: D61G in red, E76K in blue and N308D in green.
Role of BG and EF loops of the N-SH2 domain in SHP2 opening
The BG and EF loops represent the region on the N-SH2 domain directly involved in interactions with the pY moiety of phosphopeptides targeting that domain. In the crystal structure of the full SHP2, these residues are in a closed state, being 8.2 Å apart based on the Cα atoms of Gly67 and Asn92 versus 14.3 Å in the open state observed in the crystal structure of the N-SH2 domain alone. Previous studies in our laboratory have analyzed this interaction in detail, indicating an important role for Tyr66 in controlling the opening of this region18. From the quasiharmonic analysis presented above, it is clear that additional motion is occurring in the BG and EF loops in the D61G and E76K mutants. This motion appears to lead to these mutants sampling conformations of the BG and EF loops associated with the open state observed in the crystal structure of the N-SH2 domain alone. Average distances between the BG and EF loops (Table II) are consistent with these results and suggest that the N308D mutant may also favor the open state. Notably, the open state of the N-SH2 domain represents a conformation that disrupts the PTP recognition surface as well as favoring binding of phosphopeptides16. Accordingly such sampling may favor the equilibrium towards active form of SHP2 both by facilitating the binding of pY peptides to the N-SH2 domain and causing additional perturbations to the interaction between the PTP and N-SH2 domains.
TABLE II.
Average distance between the BG and EF loops based on the Cα atoms of residues Gly67 and Asn92 from the average structure calculated from the 15 ns MD simulations and the values from the crystals structures of full SHP2 and the N-SH2 domain alone. The averages and standard deviations in parentheses were calculated from 3000 ps block averages.
| Structure | Distance (Å) |
|---|---|
| WT simulation | 7.3 (0.4) |
| D61G simulation | 9.6 (0.7) |
| E76K simulation | 11.8 (0.9) |
| N308D simulation | 7.9 (1.2) |
| Full SHP2 crystal structure | 8.2 |
| N-SH2 domain crystal structure | 14.3 |
With the D61G mutant, its impact on the BG/EF loops can be via direct covalent connectivity. Asp61 is located on the D’E loop, located before β-strand E leading into the EF loop. Thus, mutation of Asp61 may be directly communicated to the EF loop, thereby impact the region of the N-SH2 domain involved in binding of phosphopeptide. In addition, Asp61 is involved in additional interactions with the PTP domain (Fig. 1C). As mentioned above its carboxylate is approximately 7 Å from the guanidinium of Arg362 and the Oδ1 atom of Asp61 is 3.83 Å from the Cδ atom of the invariant Arg465, which is part of the phosphate-binding cradle47, and 3.81 Å from the Oγ atom of Ser460. The mutation of residue 61 from an Asp to a Gly will lead to a loss of these favorable interactions, contributing to the shifting of the minimum in the PMF (Fig. 2) to distances longer than those in the WT SHP2, analogous to the situation with the E76K mutation.
While with the D61G and E76K mutants there are clear, direct lines of communication between those residues and the BG and EF loops in the N-SH2 domain leading to the changes discussed above, the mechanism by which N308D causes its effects is not evident. The distance between the Cα atom of Asn308 and Arg362 in the crystal structure is 27.1 Å and the distance to the Asp425 Cα atom is 25.4 Å. However, analysis of the crystal structure of SHP2 suggests a possible line of communication (Fig. 1E). The Oδ1 atom of Asn308 is 2.97 Å from the Nε atom of Arg501, whose NΗ1 atom is 2.55 and 3.28 Å from the carbonyl oxygens of Ala461 and Gly462. Notably, these residues are adjacent to the catalytic nucleophile, Cys459 in the primary sequence. In addition, the intervening residue, Ser460, has a direct interaction with the carboxylate group of Asp61 (Ser460 Oγ to Asp61 Oδ1 distance = 3.81 Å). These interactions are maintained in the WT simulation (not shown). As discussed above, perturbation of Asp61 can directly affect the FH loop via its interactions with Arg362 and the EF loop via direct covalent connectivity via β-strand E, such that the N308D mutation, in the closed state, may affect the region of the N-SH2 domain that interacts with phosphoprotein.
Impact of N308D mutation on the FH and LF loops
As discussed above, the N308D mutant has the largest impact on the relative position of the FH and LF loops. The FH loop, composed of residues 361–371 and the LF loop, comprised of residues 421–431, are located at the PTP domain above the catalytic site. Accordingly, changes in the structure and dynamics of these loops would directly impact the catalytic nucleophile, Cys459, potentially facilitating its catalytic efficiency. In addition, the FH loop is in direct contact with the D’E region of the N-SH2 domain. This includes the interaction between Arg362 and Asp61 discussed above. The N308D mutation may therefore decrease the favorable interactions between the PTP and N-SH2 domains as well as favor sampling of the open state of the N-SH2 domain that favors phosphopeptide binding, thereby further facilitating SHP2 accessing its active state. In addition, the N308D mutation may alter the conformation of the FH and LF loops that may impact interactions of SHP2 while in its closed state with substrate proteins, changes that may potentially facilitate opening.
Experimental Results
The present results, whereby enhanced opening in GOF mutants by increasing the ability of substrate and activator peptides to facilitate opening via either interactions with the PTP domain and/or the pY peptide binding site of the N-SH2 domain, are consistent with experimental studies by Kontaridis et al48 on the E76K and D61G mutants. In that work it was shown that both mutants had increased activity with both a protein substrate, lysozyme, and the low molecular weight substrate p-nitrophenolphosphate (pNPP), and the E76K mutant had nearly full activity against both substrates in the absence of additional phosphopeptides (IRS-1-derived peptide Tyr(P)-1172 or Tyr(P)-1222 peptide). To corroborate these results and further investigate the proposed model additional experiments were undertaken. The experiments were designed to identify if the presence of activator peptides leads to a greater enhancement of activity for the mutants relative to WT SHP2, with emphasis on the D61G mutant. This involved measuring the phosphatase activity of SHP2 using both pNPP and pY-EGFR, a peptide derived from tyrosine phosphorylated epidermal growth factor receptor (EGFR), as substrates. Since phosphopeptide substrates such as pY-EGFR can also act as activators via binding to the N-SH2 domain, inclusion of pNPP as a substrate is designed to avoid this effect. Similarly, if interactions of the substrates with the PTP domain via Arg362 on the FH loop can facilitate opening such an effect may be enhanced in a peptide substrate, though the presence of the phosphate on both pNPP and pY-EGFR makes differential effects in this case less likely.
Role of pY-EGFR substrate on the activity of SHP2
The initial experiment measured the activity of the two substrates with full-length SHP2, only the PTP domain of SHP2, the D61G full-length SHP2 mutant and the E76K full-length SHP2 mutant. With pY-EGFR as the substrate (Fig. 7A) significant activity is present in full length SHP2 due to the substrate potentially acting as an activator by direct interactions with the N-SH2 domain. However, some gains in activity are observed in the PTP domain alone as opening is no longer required, an observation consistent with a number of previous studies42,43. As expected the level of activity in the D61Y and E76K mutants is higher then WT, with the activity in the E76K mutant close to that of the PTP domain alone. The enhanced activity of the two mutants may be due to both the proposed increased facilitation of opening due to interactions with residues on the PTP domain as well as facilitation of binding of the pY substrate to the N-SH2 domain, as it still remains unclear whether these phosphorylated proteins can target both the N-SH2 and the PTP domains. Interestingly, the enhanced activity of the E76K mutant over the D61G mutant is consistent with the MD simulations indicating the E76K mutant to assume a conformation of the pY binding site in the N-SH2 domain more similar to that of the open state (Table II) and also to experience less of a barrier to full opening (Fig. 2).
FIGURE 7.
In vitro phosphatase activity of full-length SHP2, the PTP domain of SHP2, the E76K full-length SHP2 mutant and the D61G full-length SHP2 mutant with both A) pY-EGFR and B) pNPP as substrates. Phosphatase activity of SHP2 in WT and SHP2D61G/D61G mutant mouse embryonic fibroblasts stimulated with 15% fetal calf serum. Both C) pY-EGFR and D) pNPP were used as substrates.
Role of pNPP substrate on the activity of SHP2
Results for the in vitro phosphatase assays using pNPP as a substrate are shown in Fig. 7B. The low activity of full length SHP2 with the small molecule substrate in combination with a significant increase in activity for the PTP domain alone is consistent with the lack of spontaneous WT opening, though residual activity is evident, as seen by Kontaridis et al48. As expected, significant increase of activity occurs in the PTP domain alone, with significant gains also evident in the two mutants. The increase in activity of the E76K mutant over the D61G mutant observed with the pY-EGFR is still present. This suggests that facilitated binding of activator peptide to the N-SH2 domain is not the dominant mechanism of activation in the mutants. However, the extent of activation relative to PTP alone for pNPP as a substrate is less than that for pY-EGFR as substrate for the two mutants, suggesting that facilitated binding of activator peptide to the N-SH2 domain is contributing to increased activity of the mutants, consistent with the proposed model.
Another consideration is the possible impact of the interaction of substrate with Arg362 and other residues thereby facilitating opening. The proposed mechanism of opening could play a role with both the peptide and small molecule substrates. If such interactions are dominated by the phosphate moiety, it would occur with both substrates. Indeed this may contribute to the residual activity with pNPP in WT SHP2 in Fig. 7B. However, the activity of WT against pNPP is about 30 fold lower as compared to pY-EGFR. These results are consistent with previously published experiments in which Sugimoto et al49 showed that deletion of the N-SH2 domain increases kcat for pNPP from 0.046 to 49 1/s while for lysozyme as a substrate kcat is 0.11 1/s for full SHP2 and increases to 0.35 1/s in the absence of the N-SH2 domain. Thus, since kcat for pNPP is much less than kcat for lysozyme in the case of full SHP2, it appears that some interaction of the substrate protein with SHP2 is facilitating opening of the protein as required for catalysis. Such opening may be induced by pNPP, but the level is clearly significantly less that that with the protein substrates.
Impact of other activators on the activity of SHP2
To test the ability of additional activators to impact SHP2 activity in vivo experiments were performed in the absence and presence of 15% fetal bovine serum (FBS), which contains growth factors that activate SHP2 by binding to cell surface receptors and initiating intracellular signaling (Fig. 7 C and D). In the WT cells stimulation with serum led to a small increase in activity in WT SHP2 with pY-EGFR as the substrate, while in the D61G mutant cells a larger increase in SHP2 activity is induced by serum (Fig. 7C). These results are consistent with the mutation allowing for greater facilitation of opening by activators, though the dual roles of the pY-EGFR substrate significantly complicates interpretation of the results. Similar experiments were therefore performed with pNPP as substrate. The presence of serum led to a small increase in activity in WT SHP2 in WT cells. In the case of the D61G mutant cells, the addition of serum leads to a significant increase in activity of mutant SHP2 (2 to 3 fold). Notably, this increase is significantly greater than that occurring in WT cells consistent with the model that the mutation does increase the ability of activator peptides to facilitate opening of SHP2. These results, while not definitive, are consistent with the proposed model where SHP2 GOF mutations lead to increased activity by increasing the ability of activator proteins to facilitate opening.
The presented model of the opening of SHP2 may also explain experimental observations on the impact of pY peptides binding to the C-SH2 domain14. Binding of pY peptide to the C-SH2 domain alone does not increase the activity of SHP250. However, peptides that include two pY moieties, allowing them to bind simultaneously to both the N- and C-SH2 domains increase activity approximately 10-fold over pY peptides that bind to only the N-SH2 domain, yielding on overall increase in activity of 100 fold51. Based on the scenario for opening presented above, it may be hypothesized that a pY peptide binding simultaneously to both the N- and C-SH2 domains would optimize the interactions between the two domains, thereby facilitating the opening of the N-SH2 domain away from the PTP domain.
Summary
Presented are results from MD simulations, including potential of mean force calculations, associated with the opening of SHP2 in the WT protein and the D61G, E76K and N308D GOF mutants. Analysis of the PMF calculations on the WT protein shows the N-SH2 domain to undergo a sliding motion away from the PTP domain, with the N-SH2 domain sliding over the C-SH2 domain. This opening pathway suggests a role for the C-SH2 domain in stabilizing the open state of SHP2, thereby facilitating closing of the enzyme following catalysis. In addition, such a role of the C-SH2 domain may explain why peptides containing two pY moieties that simultaneously bind to the N- and C-SH2 domains activate SHP2 activity approximately 100-fold51 while phosphopeptides that bind only to the C-SH2 domain have no significant impact on activity50. The linking of the N- and C-SH2 domains may facilitate their interaction, thereby encouraging opening of the protein.
The D61G and E76K mutations alter direct interactions between the N-SH2 and PTP domains, thereby leading to less favorable interactions between those domains that can favor the open state. This is observed in the outward shift in the location of the minima in the PMF calculations of these mutants. However, the PMF calculations also suggest that spontaneous opening of SHP2 does not occur in the WT or mutant proteins, rather interactions with external factors, most likely the phosphoprotein substrates or other activator molecules, are required for this to occur. Analysis of the present results indicates that Arg362 may play a role in initial recognition of substrate proteins, thereby facilitating opening. Interestingly, while the calculated increase in free energy upon going to the open state is still large in the E76K mutant, it is significantly less than that of D61G as well as WT. Given the experiments showing increased activity of the E76K mutant in this work as well as the work by Kontaridis et al.48, the present work indicates that opening does occur more readily in this mutant, though interactions with the substrate, be it either a protein or small molecule, are needed to facilitate opening. However, given potential issues with sampling in the PMF calculations, as discussed above, the possibility of spontaneous opening of E76K cannot be totally excluded.
Phosphopeptide binding to the N-SH2 domain is known to activate SHP213,14,52. Notably, all three mutations studied lead to alterations of the N-SH2 domain in the region of the BG and EF loops. These changes lead to increased sampling of the open state of the N-SH2 domain. Such sampling would further destabilize the interactions between the N-SH2 and PTP domains. In addition, increased sampling of the open state would facilitate binding of phosphopeptides to the N-SH2 domain, further facilitating opening of SHP2.
In the case of the N308D mutation, significant alteration of the motion of the FH and LF loops in the PTP domain is observed. This perturbation of the structure would potentially alter the catalytic activity of SHP2 via alterations of the active site properties. In addition, the changes may facilitate opening through alterations of this region that impact direct interactions with the substrates that may lead to opening as well as direct interactions between the PTP and N-SH2 domain. A potential network from residue 308 to the relevant regions of the protein by which these changes may be communicated is presented.
Experimental studies were subsequently performed motivated by the above observations. While not definitive, they, as well as previous studies48, are consistent with a model by which substrates play an active role in leading to the opening of SHP2 as required for activity. They also are consistent with the model by which the mutants favor binding of activator peptides to the N-SH2 domain leading to additional activation of the protein. In particular, the experiments show larger activation in the E76K mutant, which correlates with the predicted ability of that mutant to induce a more bound-like state of the phosphopeptide binding region of the N-SH2 domain.
The present study offers a number of hypotheses concerning the activity of SHP2 and the impact of selected GOF mutants on the activity. While the present results are primarily based on computational methods, emphasis has been placed on interpretation of the results in the context of available experimental data as well as additional data generated as part of the present study. It is hoped that the models presented in this study will stimulate additional experimental work allowing more details of the molecular determinants of SHP2 regulation to be elucidated.
Supplementary Material
Acknowledgements
Financial support from the NIH (HL082670) and the University of Maryland, Baltimore, Computer-Aided Drug Design Center and computational support from the Department of Defense and NPACI Alliance are acknowledged.
References
- 1.Bennett AM, Hausdorff SF, O'Reilly AM, Freeman RM, Neel BG. Multiple requirements for SHPTP2 in epidermal growth factor-mediated cell cycle progression. Mol Cell Biol. 1996;16(3):1189–1202. doi: 10.1128/mcb.16.3.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xiao S, Rose DW, Sasaoka T, Maegawa H, Burke TR, Jr, Roller PP, Shoelson SE, Olefsky JM. Syp (SH-PTP2) is a positive mediator of growth factor-stimulated mitogenic signal transduction. J Biol Chem. 1994;269(33):21244–21248. [PubMed] [Google Scholar]
- 3.Rivard N, McKenzie FR, Brondello JM, Pouyssegur J. The phosphotyrosine phosphatase PTP1D, but not PTP1C, is an essential mediator of fibroblast proliferation induced by tyrosine kinase and G protein-coupled receptors. J Biol Chem. 1995;270(18):11017–11024. doi: 10.1074/jbc.270.18.11017. [DOI] [PubMed] [Google Scholar]
- 4.Yu M, Luo J, Yang W, Wang Y, Mizuki M, Kanakura Y, Besmer P, Neel BG, Gu H. The scaffolding adapter Gab2, via Shp-2, regulates kit-evoked mast cell proliferation by activating the Rac/JNK pathway. J Biol Chem. 2006;281(39):28615–28626. doi: 10.1074/jbc.M603742200. [DOI] [PubMed] [Google Scholar]
- 5.Neel BG, Gu H, Pao L. The 'Shp'ing news: SH2 domain-containing tyrosine phosphatases in cell signaling. Trends Biochem Sci. 2003;28(6):284–293. doi: 10.1016/S0968-0004(03)00091-4. [DOI] [PubMed] [Google Scholar]
- 6.Xu D, Qu CK. Protein tyrosine phosphatases in the JAK/STAT pathway. Front Biosci. 2008;13:4925–4932. doi: 10.2741/3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol. 2006;7(11):833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 8.Hof P, Pluskey S, Dhe-Paganon S, Eck MJ, Shoelson SE. Crystal structure of the tyrosine phosphatase SHP-2. Cell. 1998;92(4):441–450. doi: 10.1016/s0092-8674(00)80938-1. [DOI] [PubMed] [Google Scholar]
- 9.Eminaga S, Bennett AM. Noonan syndrome-associated SHP-2/Ptpn11 mutants enhance SIRPalpha and PZR tyrosyl phosphorylation and promote adhesion-mediated ERK activation. J Biol Chem. 2008;283(22):15328–15338. doi: 10.1074/jbc.M801382200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tartaglia M, Mehler EL, Goldberg R, Zampino G, Brunner HG, Kremer H, van der Burgt I, Crosby AH, Ion A, Jeffery S, Kalidas K, Patton MA, Kucherlapati RS, Gelb BD. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat Genet. 2001;29(4):465–468. doi: 10.1038/ng772. [DOI] [PubMed] [Google Scholar]
- 11.Tartaglia M, Niemeyer CM, Fragale A, Song X, Buechner J, Jung A, Hahlen K, Hasle H, Licht JD, Gelb BD. Somatic mutations in PTPN11 in juvenile myelomonocytic leukemia, myelodysplastic syndromes and acute myeloid leukemia. Nat Genet. 2003;34(2):148–150. doi: 10.1038/ng1156. [DOI] [PubMed] [Google Scholar]
- 12.Tartaglia M, Martinelli S, Cazzaniga G, Cordeddu V, Iavarone I, Spinelli M, Palmi C, Carta C, Pession A, Arico M, Masera G, Basso G, Sorcini M, Gelb BD, Biondi A. Genetic evidence for lineage-related and differentiation stage-related contribution of somatic PTPN11 mutations to leukemogenesis in childhood acute leukemia. Blood. 2004;104(2):307–313. doi: 10.1182/blood-2003-11-3876. [DOI] [PubMed] [Google Scholar]
- 13.Lechleider RJ, Sugimoto S, Bennett AM, Kashishian AS, Cooper JA, Shoelson SE, Walsh CT, Neel BG. Activation of the SH2-containing phosphotyrosine phosphatase SH-PTP2 by its binding site, phosphotyrosine 1009, on the human platelet-derived growth factor receptor. J Biol Chem. 1993;268(29):21478–21481. [PubMed] [Google Scholar]
- 14.Sugimoto S, Wandless TJ, Shoelson SE, Neel BG, Walsh CT. Activation of the SH2-containing protein tyrosine phosphatase, SH-PTP2, by phosphotyrosine-containing peptides derived from insulin receptor substrate-1. J Biol Chem. 1994;269(18):13614–13622. [PubMed] [Google Scholar]
- 15.Lee CH, Kominos D, Jacques S, Margolis B, Schlessinger J, Shoelson SE, Kuriyan J. Crystal-Structures of Peptide Complexes of the Amino-Terminal Sh2 Domain of the Syp Tyrosine Phosphatase. Structure. 1994;2(5):423–438. doi: 10.1016/s0969-2126(00)00044-7. [DOI] [PubMed] [Google Scholar]
- 16.Eck MJ, Pluskey S, Trub T, Harrison SC, Shoelson SE. Spatial constraints on the recognition of phosphoproteins by the tandem SH2 domains of the phosphatase SH-PTP2. Nature. 1996;379(6562):277–280. doi: 10.1038/379277a0. [DOI] [PubMed] [Google Scholar]
- 17.Barford D, Neel BG. Revealing mechanisms for SH2 domain mediated regulation of the protein tyrosine phosphatase SHP-2. Structure. 1998;6(3):249–254. doi: 10.1016/s0969-2126(98)00027-6. [DOI] [PubMed] [Google Scholar]
- 18.Guvench O, Qu C-K, MacKerell AD., Jr Tyr66 acts as a conformational switch in the closed-to-open transition of the SHP-2 N-SH2-domain phosphotyrosine-peptide binding cleft. BMC Structural Biology. 2007;7(1):14. doi: 10.1186/1472-6807-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brooks BR, Brooks CL, 3rd, Mackerell AD, Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30(10):1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacKerell AD, Jr, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiórkiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. Journal of Physical Chemistry B. 1998;102(18):3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 22.MacKerell AD, Jr, Feig M, Brooks CL., III Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. Journal of Computational Chemistry. 2004;25(11):1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 23.Fiser A, Do RKG, Sali A. Modeling of loops in protein structures. Protein Science. 2000;9(9):1753–1773. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sali A, Blundell TL. Comparative protein modeling by satisfaction of spatial restraints. Journal of Molecular Biology. 1993;234(3):779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 25.Lovell SC, Word JM, Richardson JS, Richardson DC. The penultimate rotamer library. Proteins: Structure Function and Genetics. 2000;40(3):389–408. [PubMed] [Google Scholar]
- 26.Levitt M, Lifson S. Refinement of protein conformations using a macromolecular energy minimization procedure. Journal of Molecular Biology. 1969;46(2):269–279. doi: 10.1016/0022-2836(69)90421-5. [DOI] [PubMed] [Google Scholar]
- 27.Fletcher R, Reeves C. Function minimization by conjugate gradients. Computer Journal. 1964;7:149–154. [Google Scholar]
- 28.Word JM, Lovell SC, Richardson JS, Richardson DC. Asparagine and glutamine: Using hydrogen atom contacts in the choice of side-chain amide orientation. Journal of Molecular Biology. 1999;285(4):1735–1747. doi: 10.1006/jmbi.1998.2401. [DOI] [PubMed] [Google Scholar]
- 29.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. Journal of Chemical Physics. 1983;79(2):926–935. [Google Scholar]
- 30.Durell SR, Brooks BR, Ben-Naim A. Solvent-induced forces between two hydrophilic groups. Journal of Physical Chemistry. 1994;98(8):2198–2202. [Google Scholar]
- 31.Allen MP, Tildesley DJ. Computer Simulation of Liquids. Oxford: Oxford University Press; 1987. [Google Scholar]
- 32.Darden T, York D, Pedersen L. Particle mesh Ewald: an N•log(N) method for Ewald sums in large systems. Journal of Chemical Physics. 1993;98(12):10089–10092. [Google Scholar]
- 33.Steinbach PJ, Brooks BR. New spherical-cutoff methods for long-range forces in macromolecular simulation. Journal of Computational Chemistry. 1994;15(7):667–683. [Google Scholar]
- 34.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. Journal of Computational Physics. 1977;23(3):327–341. [Google Scholar]
- 35.Hockney RW. The potential calculation and some applications. In: Alder B, Fernbach S, Rotenberg M, editors. Methods in Computational Physics. Volume 9. New York: Academic Press; 1970. pp. 136–211. [Google Scholar]
- 36.Hoover WG. Canonical dynamics: equilibrium phase-space distributions. Physical Review A. 1985;31(3):1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 37.Nosé S. A molecular dynamics method for simulations in the canonical ensemble. Molecular Physics. 1984;52(2):255–268. [Google Scholar]
- 38.Feller SE, Zhang YH, Pastor RW, Brooks BR. Constant pressure molecular dynamics simulation: the Langevin piston method. Journal of Chemical Physics. 1995;103(11):4613–4621. [Google Scholar]
- 39.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. The weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. Journal of Computational Chemistry. 1992;13(8):1011–1021. [Google Scholar]
- 40.Grossfield A. WHAM: an implementation of the Weighted Histogram Analysis Method. 2003 [Google Scholar]
- 41.Araki T, Mohi MG, Ismat FA, Bronson RT, Williams IR, Kutok JL, Yang W, Pao LI, Gilliland DG, Epstein JA, Neel BG. Mouse model of Noonan syndrome reveals cell type- and gene dosage-dependent effects of Ptpn11 mutation. Nat Med. 2004;10(8):849–857. doi: 10.1038/nm1084. [DOI] [PubMed] [Google Scholar]
- 42.Zhao Z, Larocque R, Ho WT, Fischer EH, Shen SH. Purification and characterization of PTP2C, a widely distributed protein tyrosine phosphatase containing two SH2 domains. J Biol Chem. 1994;269(12):8780–8785. [PubMed] [Google Scholar]
- 43.Qu CK, Shi ZQ, Shen R, Tsai FY, Orkin SH, Feng GS. A deletion mutation in the SH2-N domain of Shp-2 severely suppresses hematopoietic cell development. Mol Cell Biol. 1997;17(9):5499–5507. doi: 10.1128/mcb.17.9.5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Banavali NK, Roux B. The N-terminal end of the catalytic domain of SRC kinase Hck is a conformational switch implicated in long-range allosteric regulation. Structure. 2005;13(11):1715–1723. doi: 10.1016/j.str.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 45.Huang N, Banavali NK, MacKerell AD., Jr Protein-facilitated base flipping in DNA by cytosine-5-methyltransferase. Proc Natl Acad Sci U S A. 2003;100(1):68–73. doi: 10.1073/pnas.0135427100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Priyakumar UD, Mackerell AD., Jr NMR imino proton exchange experiments on duplex DNA primarily monitor the opening of purine bases. J Am Chem Soc. 2006;128(3):678–679. doi: 10.1021/ja056445a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jia Z, Barford D, Flint AJ, Tonks NK. Structural basis for phosphotyrosine peptide recognition by protein tyrosine phosphatase 1B. Science. 1995;268(5218):1754–1758. doi: 10.1126/science.7540771. [DOI] [PubMed] [Google Scholar]
- 48.Kontaridis MI, Swanson KD, David FS, Barford D, Neel BG. PTPN11 (Shp2) mutations in LEOPARD syndrome have dominant negative, not activating, effects. J Biol Chem. 2006;281(10):6785–6792. doi: 10.1074/jbc.M513068200. [DOI] [PubMed] [Google Scholar]
- 49.Sugimoto S, Lechleider RJ, Shoelson SE, Neel BG, Walsh CT. Expression, purification, and characterization of SH2-containing protein tyrosine phosphatase, SH-PTP2. J Biol Chem. 1993;268(30):22771–22776. [PubMed] [Google Scholar]
- 50.Barford D, Neel BG. Revealing mechanisms for SH2 domain mediated regulation of the protein tyrosine phosphatase SHP-2. Structure. 1998;6(3):249–254. doi: 10.1016/s0969-2126(98)00027-6. [DOI] [PubMed] [Google Scholar]
- 51.Pei D, Wang J, Walsh CT. Differential functions of the two Src homology 2 domains in protein tyrosine phosphatase SH-PTP1. Proc Natl Acad Sci U S A. 1996;93(3):1141–1145. doi: 10.1073/pnas.93.3.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cunnick JM, Mei L, Doupnik CA, Wu J. Phosphotyrosines 627 and 659 of Gab1 constitute a bisphosphoryl tyrosine-based activation motif (BTAM) conferring binding and activation of SHP2. J Biol Chem. 2001;276(26):24380–24387. doi: 10.1074/jbc.M010275200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.