Abstract
Objective
The withdrawal of cerivastatin involved an uncommon but serious adverse reaction, rhabdomyolysis. The bimodal response--rhabdomyolysis in a small proportion of users-- points to genetic factors as a potential cause. We conducted a case-control study to evaluate genetic markers for cerivastatin-associated rhabdomyolysis.
Methods
The study had two components: a candidate gene study to evaluate variants in CYP2C8, UGT1A1, UGT1A3, and SLCO1B1; and a genome-wide association (GWA) study to identify risk factors in other regions of the genome. 185 rhabdomyolysis cases were frequency matched to statin-using controls from the Cardiovascular Health Study (n=374) and the Heart and Vascular Health Study (n=358). Validation relied on functional studies.
Results
Permutation test results suggested an association between cerivastatin-associated rhabdomyolysis and variants in SLCO1B1 (p = 0.002), but not variants in CYP2C8 (p = 0.073) or the UGTs (p = 0.523). An additional copy of the minor allele of SLCO1B1 rs4149056 (p.Val174Ala) was associated with the risk of rhabdomyolysis (OR: 1.89, 95% CI: 1.40 to 2.56). In transfected cells, this variant reduced cerivastatin transport by 40% compared with the reference transporter (p < 0.001). The GWA identified an intronic variant (rs2819742) in the ryanodine receptor 2 gene (RYR2) as significant (p=1.74E-07). An additional copy of the minor allele of the RYR2 variant was associated with a reduced risk of rhabdomyolysis (OR: 0.48; 95% CI: 0.36 to 0.63).
Conclusion
We identified modest genetic risk factors for an extreme response to cerivastatin. Disabling genetic variants in the candidate genes were not responsible for the bimodal response to cerivastatin.
Keywords: Genetics, drugs, epidemiology, rhabdomyolysis
Introduction
High-profile drug withdrawals have often involved serious adverse reactions such as liver failure in troglitazone users or heart-valve damage in dexfenfluramine users [1, 2]. The occurrence of an extreme adverse reaction to a new drug sets a small number of people apart from the rest of users who have few if any side effects. Although the cause may be environmental, such as a drug-drug interaction, the bimodal response points to genetic factors as a potential cause [3, 4].
Cerivastatin, an HMG-CoA reductase inhibitor (statin), was withdrawn from the market in 2001 because of a pronounced increase in the risk of rhabdomyolysis, which causes muscle pain and weakness and sometimes renal failure and death [5]. About half of the rhabdomyolysis cases reported to the FDA occurred in subjects who had taken both cerivastatin and the fibrate, gemfibrozil [5, 6]. In one study, combined cerivastatin-fibrate use was associated with a 1411 fold higher risk of rhabdomyolysis than the use of other statin monotherapies [7]. In pharmacokinetic studies, gemfibrozil inhibited the major disposal pathways for cerivastatin, not only the drug-metabolizing enzymes, CYP2C8, UGT1A1, and UGT1A3 [8,9], but also the transporter, OATP1B1, responsible for uptake of cerivastatin into hepatic cells [10].
The pronounced effect of gemfibrozil on these candidate gene products suggested that genetic variants, including rare ones, might play a major role in this adverse effect, especially in subjects who did not use gemfibrozil. We performed a case-control study to evaluate common, novel, and rare genetic risk factors for cerivastatin-associated rhabdomyolysis. This study of cerivastatin-associated rhabdomyolysis had two components: (1) a candidate gene study to evaluate the association of variants in CYP2C8, UGT1A1, UGT1A3, and SLCO1B1 (the gene that encodes OATP1B1); and (2) a genome-wide association (GWA) study to identify risk factors in other regions of the genome. The primary purpose of the study was to test the hypothesis that common or rare disabling variants in the genes encoding the metabolizing enzymes or the transporter would have a gemfibrozil-like effect on the incidence of this severe idiosyncratic adverse drug reaction.
Methods
Setting
Cases were recruited through attorneys who had represented cerivastatin users with rhabdomyolysis and had settled their cases with the manufacturer. Attorneys were offered reimbursement for their recruitment efforts. Because the identification of the optimal control group, a random sample of cerivastatin users who did not develop rhabdomyolysis, was infeasible, we selected as controls statin-using subjects from two ongoing population-based studies: the Heart and Vascular Health Study (HVH) [11-13] and the Cardiovascular Health Study (CHS) (see Methods, Supplemental Digital Content 1, Setting) [14]. The recruitment of the case subjects and the use of the CHS subjects were approved by the University of Washington Institutional Review Board. The Group Health Human Subjects Review Committee approved the use of the HVH study subjects. Subjects provided written informed consent.
Case Definition, Recruitment, and Data Collection
Eighteen attorneys screened their clients for interest in the study and provided contact information for potential participants to study coordinators (See Figure 1, Supplemental Digital Content 1, Rhabdomyolysis case subject recruitment). Case subjects who participated in the study were demographically similar to those who did not (see Table 1, Supplemental Digital Content 1, Characteristics of case subjects who did and did not participate in the study). For consenting cases, study staff conducted a telephone interview and obtained copies of medical records from attorneys, doctors, and hospitals. Trained abstracters used the medical records to validate the rhabdomyolysis event and collect information about participants' medical history. Definite rhabdomyolysis was defined as muscle pain or weakness associated with creatine kinase levels greater than 10 times the upper limit of laboratory normal. Six subjects who would likely have been definite cases had their medical history been complete were included in the primary analysis. Subjects collected buccal cell DNA using a swish and spit mouthwash kit. Information on race was obtained from the telephone interview, medical records, and attorneys.
Selection of Controls
CHS participants selected as controls reported using a statin at an annual study visit between 1989 and 2002. They were frequency matched to the rhabdomyolysis cases by sex at a ratio of 2 controls per case. CHS controls were free of inpatient diagnostic codes for rhabdomyolysis in the year before their qualifying statin-use visit (n=0 excluded) [15]. HVH controls used a statin at the time they were selected as a control for the HVH studies (between 1995 and 2005). They were frequency matched to the rhabdomyolysis cases by sex and age by decade at a ratio of 2 to 1 (See Methods, Supplemental Digital Content 1, Selection of controls). HVH controls with a creatine kinase value >10 times the upper limit of laboratory normal in the year before their qualifying statin-use date were excluded (n=2).
Selection of Single Nucleotide Polymorphisms in Candidate Gene Analysis
To identify novel genetic variants in the four candidate genes, we re-sequenced the DNA from the first 126 rhabdomyolysis cases recruited (See Methods and Tables 2-4, Supplemental Digital Content 1, PCR and sequencing methods, and primers and conditions). Re-sequenced regions included exons, intron-exon boundaries, the promoter and the untranslated regions of each gene. The resequencing effort identified 45 novel variants, which were submitted to dbSNP. To identify other variants of interest, we used HapMap CEU data and publicly available resequencing data to select two other sets of single nucleotide polymorphisms (SNPs): 1) tagSNPs that captured information on common variants (minor allele frequency >= 2.5%) [16]; and 2) SNPs predicted to be functional (F-SNPs) by the bioinformatics program F-SNP (http://compbio.cs.queensu.ca/F-SNP/).
In vitro functional data for key SLCO1B1 variant
The functional effect of the SLCO1B1 variant rs4149056 (c.521T>C; p.Val174Ala) on cerivastatin uptake was measured in stable HEK293 (human embryonic kidney epithelial) cells expressing the reference and variant transporter (See Methods, Supplemental Digital Content 1, in vitro functional assessment of SLCO1B1 rs4149056 variant).
Genome-Wide Association Screen
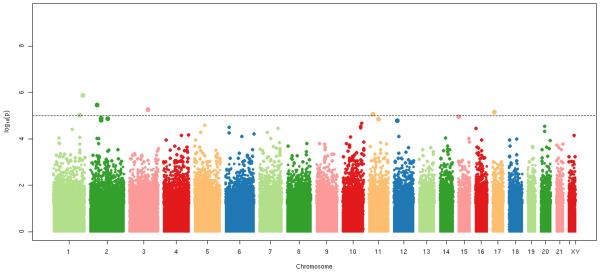
We used the Illumina Infinium 370 CNV chip to obtain GWA data on the cases (See Methods, Supplemental Digital Content 1, Genome-wide association screen: subjects and selection of SNPs). We used existing GWA data on statin-using CHS controls who had been genotyped on the same Illumina platform. (See Table 5, Supplemental Digital Content 1, Characteristics of GWA screen study subjects). Because the genotyping was done at two different times in two different laboratories (See Table 5, Supplemental Digital Content 1, Characteristics of GWA screen study subjects), we re-genotyped GWA high signal markers in the rhabdomyolysis cases and controls selected for the candidate gene work. The most significant SNP from each of the 6 high-signal loci (SNPs with p-values <1E-05) (Figure 1), was re-genotyped to exclude drift, batch, or laboratory effects.
Figure 1.
Stripe plot of genome-wide association screen among rhabdomyolysis cases (N=175) and CHS statin-using controls with GWA data (n=645). Directly genotyped SNPs with minor allele frequencies ≥ 5% = 292,461. The analysis was adjusted for age and sex. Six loci with minor allele frequencies ≥5% fell below the p<1E-05 threshold for re-genotyping.
Genotyping in cases and controls
Re-genotyped SNPs included (1) the novel SNPs discovered by resequencing, and (2) the high-signal markers identified by the GWA screen. The other sources of SNPs for genotyping were the candidate gene tagSNPs and F-SNPs (See Table 6, Supplemental Digital Content 1, SNPS selected for genotyping by gene, type, platform, and outcome). Genotyping methods included an Illumina Goldengate custom panel and, for failed or undesignable SNPs, the Taqman 5' nuclease discrimination assay. SNPs with poor Taqman results underwent pyrosequencing on the PyroMark Q96MD platform. For each SNP, all cases and controls were genotyped on the same platform (See Methods, Supplemental Digital Content 1, Re-genotyping in cases and controls). SNPs were in Hardy-Weinberg equilibrium in controls of European descent, except for two UGT F-SNPs, rs17862867 and rs6431625.
Statistical Analysis
We pooled the CHS and HVH controls for the analysis. Rare functional SNPs (<1% minor allele frequency) within each gene were collapsed to assess the association between case status and having at least one rare variant [17]. Rare (<1% minor allele frequency) novel SNPs within each gene were also collapsed. All other SNPs were modeled additively. Logistic regression models were fit using robust standard errors and were adjusted for age at statin use, sex, and race (See Methods, Supplemental Digital Content 1, Statistical analysis). We used a permutation test to provide a gene level p-value that accounted for the multiple SNPs tested within each candidate gene [18]. UGT1A1 and UGT1A3, which share exons 2-5, were analyzed together. A candidate gene was considered significant if the permutation p-value was <0.05. High signal GWA markers were considered significant if their p-value after re-genotyping was < 4E-07 (1/2.5million tests). Sensitivity analyses included an analysis restricted to controls and definite cases and an analysis restricted to subjects who did not use gemfibrozil.
Results
Subjects
Among the 215 eligible rhabdomyolysis cases recruited to the study, 27 were deceased, 2 did not have usable DNA, and 1 failed genotyping (See Figure 1, Supplemental Digital Content 1, Rhabdomyolysis case subject recruitment). Most cases experienced both muscle pain and weakness; creatine kinase values indicated significant muscle damage; 81.6% were hospitalized; and 21.6% had renal failure (top part of Table 1). The prescribed daily dose of cerivastatin was high; approved doses ranged between 0.2mg and 0.8mg.
Table 1.
Characteristics of Rhabdomyolysis Cases and Controls
| Rhabdomyolysis Case Characteristics (N=185) | |
|---|---|
| Muscle pain, % | 91.9 |
| Muscle weakness, % | 84.9 |
| Creatine kinase* U/L, mean (std. dev) | 55,166.2 (78,373.9) |
| Creatine kinase* U/L, median (range) | 30,200 (1341 – 720,000) |
| Hospitalized rhabdomyolysis, % | 81.6 |
| Days of hospitalization, mean (std. dev.) | 6.7 (6.2) |
| Creatinine† μmol/L, mean (std. dev.) | 176.8 (212.2) |
| Creatinine† μmol/L, median (range) | 88.4 (17.7 – 1211.1) |
| Renal Failure, % | 21.6 |
| Hemodialysis, % | 7.0 |
| Cerivastatin dose‡ mg, mean (std. dev.) | 0.6 (0.2) |
| Cerivastatin dose‡ mg, median (range) | 0.8 (0.2 – 1.6) |
| Case and Control Characteristics | |||
|---|---|---|---|
| Rhabdomyolysis Cases | CHS§ controls | HVH∥ controls | |
| Subjects with any genotype data (N=917) | N=185 | N=374 | N=358 |
| Female, % | 61.1 | 61.0 | 48.0 |
| Age, mean (std) | 63.5 (10.6) | 73.6 (4.1) | 64.5 (9.4) |
| Age, range | 34-89 | 65-89 | 36-89 |
| European descent, % | 90.8 | 84.5 | 89.4 |
| Location | US, Canada | CA, MD, PA, NC | Western WA |
| Statin user#, % | 100.0 | 100.0 | 100.0 |
| Gemfibrozil user, % | 63.8 | 0.0 | 1.4 |
| Subjects with Illumina genotype data (n=910) | N=181 | N=372 | N=357 |
| Subjects with Taqman genotype data (n=904) | N=183 | N=366 | N=355 |
Eight values recorded as ‘out of range’ were not used in this summary. Upper limits of normal ranged from 115 to 400 U/L.
Upper limits of normal for creatinine ranged from 88.4 to 150.3.
Dose information was missing for 33 case subjects
Cardiovascular Health Study
Heart and Vascular Health Study
Cases used cerivastatin. CHS Controls used lovastatin (43.9%), simvastatin (18.7%), atorvastatin (12.6%), fluvastatin (5.6%), pravastatin (18.5%), or cerivastatin (1.1%). HVH controls used lovastatin (25.7%), simvastatin (69.0%), atorvastatin (3.9%), or pravastatin (1.4%).
Genotype data were available on 185 cases, 374 controls from CHS, and 358 controls from HVH (bottom part of Table 1). The cases were mostly female (61%) and ranged widely in age (34-89). The CHS controls were mostly women (61%), were elderly (mean age 74 years), and resided in four US communities. The HVH controls, from western Washington State, included fewer female statin users, but were similar in age to the rhabdomyolysis cases. Cases used cerivastatin, and 64% were concomitant users of gemfibrozil. The CHS and HVH controls used lovastatin, simvastatin, atorvastatin, fluvastatin, pravastatin or cerivastatin. The distribution of statin use reflects the Group Health formulary preferences for HVH controls and the time period over which the CHS controls were sampled. Few controls used gemfibrozil.
Candidate-gene results
Permutation test results suggested strong evidence for an association between cerivastatin-associated rhabdomyolysis and variants in SLCO1B1 (p = 0.002), but not CYP2C8 (p = 0.073) or the UGTs (p = 0.523). Table 2 summarizes the key results for SLCO1B1 in the top section and for CYP2C8 in the bottom section. The entire and largely null set of results for SLCO1B1, CYP2C8, and for the UGT candidate gene complex are described in Results and Tables 7, 8, and 9, Supplemental Digital Content 1.
Table 2.
Selected findings for the candidate genes SLCO1B1 and CYP2C8. Models adjusted for age at statin use, sex, and race.
| Modeled Allele | Minor Allele Frequency (Controls) N=732 |
Minor Allele Frequency (Cases) N=185 |
Odds ratio (95% confidence interval) |
p-value | |
|---|---|---|---|---|---|
| SLCO1B1 | |||||
| TagSNPs* | |||||
| rs4149033 (G/A) | # copies A allele | 0.210 | 0.270 | 1.41 ( 1.06 to 1.87) | 0.02 |
| FSNPs † | |||||
| rs4149056 (G/A) | # copies A allele | 0.140 | 0.240 | 1.89 ( 1.40 to 2.56) | 3.62E-05 |
| Gene-wide permutation test result p=0.002 | |||||
| CYP2C8 | |||||
| F-SNPs † | |||||
| rs1058930 (*4 allele) (C/G) | # copies G allele | 0.054 | 0.041 | 0.67 ( 0.36 to 1.26) | 0.22 |
| rs11572080 (*3 allele) (C/T) | # copies T allele | 0.097 | 0.087 | 0.73 ( 0.47 to 1.13) | 0.16 |
| rs11572103 (*2 allele) (A/T) | # copies T allele | 0.013 | 0.006 | 0.68 ( 0.15 to 3.12) | 0.62 |
| rs10509681 (*3 allele) (A/G) | # copies G allele | 0.100 | 0.081 | 0.64 ( 0.42 to 0.98) | 0.04 |
| Gene-wide permutation test result p=0.07 | |||||
Single nucleotide polymorphism
Functional single nucleotide polymorphism
For SLCO1B1 (Table 2), the primary finding involved the functional, nonsynonymous SNP rs4149056. An additional copy of its minor allele was associated with an increased risk of rhabdomyolysis (OR: 1.89, 95% CI: 1.40 to 2.56). Carriers of two copies of the minor allele had a four-fold higher risk of rhabdomyolysis compared with carriers of two copies of the major allele (OR: 4.34; 95% CI: 1.86 to 10.10).
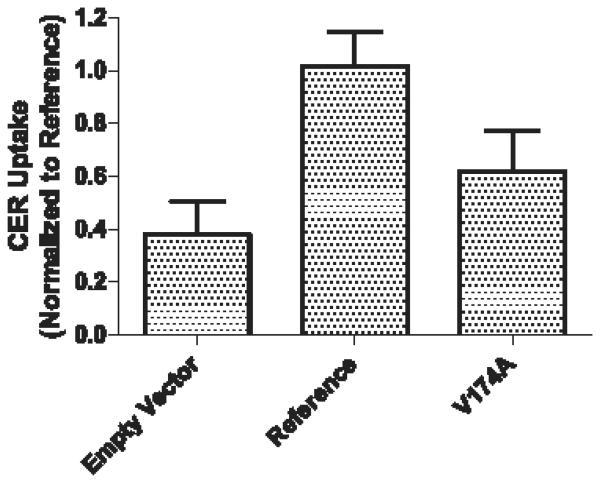
In functional studies, transport of cerivastatin into the transfected reference cells was 2.7-fold higher than the empty vector cells (1.01 ± 0.12 vs. 0.38 ± 0.12; p < 0.001), indicating the presence of OATP1B1-specific transport (Figure 2). Compared with the reference transporter, the transporter with the SLCO1B1 rs4149056 variant showed a 40% reduction in cerivastatin uptake (0.61 ± 0.15 vs. 1.01 ± 0.12; p < 0.001).
Figure 2.
Intracellular accumulation of CER in HEK293 cells expressing Empty Vector, OATP1B1 Reference and the p.Val174Ala variant of OATP1B1. All values are expressed relative to OATP1B1 Reference and are shown as mean ± S.D of 11 replicates measured in three separate experiments. Significant differences were detected by one-way ANOVA followed by Bonferroni correction for multiple testing and post hoc multiple comparison testing: Reference and p.Val174Ala variant (P<0.001), Reference and Empty Vector (P<0.001), Empty Vector and p.Val174Ala variant (P<0.01).
A second potential SLCO1B1 signal was the intronic tagSNP, rs4149033 (OR = 1.41; 95% CI = 1.06 to 1.87). In an analysis that excluded the gemfibrozil users, the association was especially strong (OR: 2.17; 95% CI: 1.47 to 3.21; p-value=0.00011). Rs4149033 was weakly correlated with rs4149056 (r2=0.19). The associations remained when both rs4149033 and rs4149056 were included in a model limited to subjects who did not use gemfibrozil (p=0.029 and p=0.012, respectively).
The nonsynonymous variant, rs2306283 (c.388A>G, p.Asn130Asp), which often co-exists with rs4149056 was not associated with an increased risk of rhabdomyolysis (OR: 0.96; 95% CI: 0.75 to 1.24; p-value 0.78). The results for rs2306283 are shown in Table 7 of the supplementary material.
Although the permutation test was not significant (p=0.073) for CYP2C8 (bottom of Table 2) and (Table 8, Supplemental Digital Content 1, CYP2C8 tagSNP, FSNP and novel SNP analysis), three tag SNPs and one F-SNP had uncorrected p-values <0.05. The well-characterized CYP2C8*2, *3, and *4 alleles were less common in cases than controls, though not significantly so. Results were similar when analyses were limited to definite cases, and non-users of gemfibrozil.
GWA results
The 6 high-signal markers identified by GWA analyses were re-genotyped in the cases and both HVH and CHS controls (Table 3). After re-genotyping, one intronic variant (rs2819742) in the ryanodine receptor 2 gene (RYR2) was significant at the a priori p-value threshold of 4E-07 (p=1.74E-07). An additional copy of the minor allele of the RYR2 variant was associated with a reduced risk of cerivastatin-associated rhabdomyolysis (OR: 0.48; 95% CI: 0.36 to 0.63). Carriers of two copies of the minor allele had a smaller risk of rhabdomyolysis than carriers of two copies of the major allele (OR: 0.24; 95% CI: 0.13 to 0.44). The SLCO1B1 variant rs4149056 was not genotyped in the GWA, but a SNP in LD with it (rs4363657, r2 = 0.79) had similar results (OR: 2.30; p-value=1.62E-05) to those observed for rs4149056 in the candidate gene work.
Table 3.
Results from re-genotyping high signal markers from the GWA* screen on the TaqMan platform in candidate gene subjects. Models adjusted for age at statin use, sex, and race.
| Gene | SNP† | Modeled Allele | p-value from GWA screen |
Minor allele frequency (Controls) N=721‡ |
Minor allele frequency (Cases) N=183‡ |
Odds ratio (95% confidence interval) |
p-value |
|---|---|---|---|---|---|---|---|
| Intergenic | rs10049478 (G/A) | # copies A allele | 5.32E-06 | 0.230 | 0.160 | 0.58 ( 0.43 to 0.80) | 7.48E-04 |
| Intergenic | rs1519414 (A/C) | # copies C allele | 8.48E-06 | 0.300 | 0.230 | 0.64 ( 0.47 to 0.85) | 0.002 |
| Intergenic | rs6703753 (C/T) | # copies T allele | 9.23E-06 | 0.260 | 0.190 | 0.70 ( 0.51 to 0.95) | 0.02 |
| Intergenic | rs7556683 (T/C) | # copies C allele | 3.50E-06 | 0.380 | 0.280 | 0.60 ( 0.45 to 0.78) | 1.70E-04 |
| NTN1 | rs1880646 (A/G) | # copies G allele | 6.83E-06 | 0.300 | 0.210 | 0.62 ( 0.46 to 0.82) | 8.06E-4 |
| RYR2 | rs2819742 (G/A) | # copies A allele | 1.30E-06 | 0.380 | 0.270 | 0.48 ( 0.36 to 0.63) | 1.74E-07 |
Genome-wide association
Single nucleotide polymorphism
Number of subjects successfully re-genotyped on the TaqMan/pyrosequencing platform.
Exploratory Analyses
Table 4 displays the results of the significant candidate gene and GWA variants in strata defined by gemfibrozil use. The association of the RYR2 variant with cerivastatin-associated rhabdomyolysis was similar in strata defined by gemfibrozil use and there was no evidence of interaction with gemfibrozil use (p=0.439). Among the gemfibrozil users, there was little evidence of association of the SLCO1B1 rs4149056 variant with rhabdomyolysis (OR: 0.68, 95%CI: 0.24 to 1.98). Among non-users, the association was strong (OR: 2.45, 95% CI: 1.61 to 3.75) and the interaction p-value was significant (p=0.018). A similar pattern was evident for rs4149033. For all other SNPs, results in analyses limited to non-gemfibrozil users were similar to the results of the primary analysis.
Table 4.
Analysis of top findings stratified by gemfibrozil use. Models adjusted for age at statin use, sex, and race.
| Concomitant gemfibrozil use | No gemfibrozil use | |||||||
|---|---|---|---|---|---|---|---|---|
| Minor allele frequency Controls (n=5) |
Minor allele frequency Cases (n=118) |
Odds ratio (95% confidence interval) |
p- value |
Minor allele frequency Controls (N=716) |
Minor allele frequency Cases (N=65) |
Odds ratio (95% confidence interval) |
p-value | |
| RYR2 | ||||||||
| rs2819742 | 0.50 | 0.27 | 0.47 (0.12, 1.88) | 0.29 | 0.378 | 0.269 | 0.52 (0.34, 0.80) | 0.003 |
| Interaction p-value = 0.44 | ||||||||
| SLCO1B1 | ||||||||
| rs4149056 | 0.30 | 0.22 | 0.68 (0.24 to 1.98) | 0.49 | 0.136 | 0.277 | 2.45 (1.61 to 3.75) | 3.11E-05 |
| Interaction p-value = 0.02 | ||||||||
| SLCO1B1 | ||||||||
| rs4149033 | 0.40 | 0.22 | 0.41 (0.07 to 2.25) | 0.30 | 0.213 | 0.354 | 2.17 (1.47 to 3.21) | 1.08E-04 |
| Interaction p-value = 0.03 | ||||||||
To evaluate if findings change when we control for gemfibrozil use, we re-analyzed our top two findings adjusted for gemfibrozil use. The point estimates remained similar, though precision was reduced due to the small number of controls using gemfibrozil (n=5). Adjusting for gemfibrozil, the point estimate for the significant RYR2 variant was 0.50 (95% CI: 0.33 to 0.75) and the point estimate for SLCO1B1 rs4149056 was 2.23 (95% CI 1.46 to 3.43).
To evaluate the net effect of the RYR2 variant rs2819742 and the SLCO1B1 variant rs4149056, we included both in the same regression model. The odds ratio for SLCO1B1 rs4149056 adjusting for RYR2 rs2819742 was 1.86 (95% CI 1.37 to 2.53) and the odds ratio for RYR2 rs2819742 adjusting for SLCO1B1 rs4149056 was 0.48 (95% CI 0.37 to 0.64). These results suggest that the RYR2 and SLCO1B1 associations are independent.
Selecting users of statins other than cerivastatin as controls will introduce bias if the decision to prescribe one statin versus another is related to the subjects' genotypes. We evaluated this assumption and found no significant association between the type of statin used and genotype among controls for our two key SNP findings (See Table 10, Supplemental Digital Content 1, Distribution of genotype by type of statin among controls).
Discussion
We used candidate-gene re-sequencing and other methods to search comprehensively for known, novel, and rare genetic variants that might underlie cerivastatin-associated rhabdomyolysis. We identified two variants with significant associations: rs2819742 in the RYR2 gene and rs4149056 in the SLCO1B1 gene. Functional studies corroborated the SLCO1B1 finding. For the other candidate genes, CYP2C8 and the UGTs, there was little evidence for an association with rhabdomyolysis. The modest associations observed in this study suggest that the manner in which genetic factors affect rhabdomyolysis may be complex, involving multiple genes, interactions, or rare variants outside the candidate pharmacokinetic genes.
Though our findings support the hypothesis that underlying genetic factors contribute to extreme adverse drug reactions, we had anticipated finding a number of rare or disabling variants that affected drug-metabolizing enzymes and drug transporters. The CYP2C8 gene is predominantly responsible for the first pass oxidative metabolism of cerivastatin [8] and contains variant alleles (CYP2C8*2, *3, and *4) which alter the function of the enzyme for some substrates [19, 20, 21-24]. The UGT1A1 and 1A3 glucuronidation is an alternate to the CYP-oxidation pathway for cerivastatin metabolism. Gemfibrozil, which inhibits the cerivastatin oxidation and glucuronidation pathways, was strongly associated with cerivastatin-associated rhabdomyolysis; yet in this study, neither known nor novel genetic variation in UGT1A1 or 1A3 was associated with the risk of rhabdomyolysis and the CYP2C8*2, *3, and *4 alleles were less common in cases than controls, though not significantly so. For cerivastatin, these excellent candidate genes did not explain the high incidence of rhabdomyolysis.
OATP1B1, the product of SLCO1B1, is a transporter that mediates the hepatic uptake of cerivastatin as well as several other statins [10, 25]. The nonsynonymous coding variant rs4149056 variant slows the uptake of other statins in vitro [26, 27]. In the STRENGTH study, rs4149056 was associated with mild statin induced side effects (including muscle symptoms) among users of simvastatin, atorvastatin and pravastatin [28]. A GWA study of myopathy among high-dose simvastatin users [29] found an additional copy of the minor allele of rs4149056 was associated with a 4.5 fold increase in the risk of myopathy (95% CI 2.6 to 7.7). Our study results extend the SLCO1B1 rs4149046 findings to cerivastatin-associated rhabdomyolysis, and our functional studies provide additional support for a causal association.
Our study results suggest there may be an interaction between SLCO1B1 rs4149056 and gemfibrozil use (p=0.02). Analyses limited to subjects using gemfibrozil were null with wide confidence intervals while analyses excluding subjects who used gemfibrozil showed a strong association between rs4149056 and the risk of rhabdomyolysis (OR: 2.45; 95% CI 1.61 to 3.75). While it's difficult to draw strong conclusions about the association between rs4149056 and rhabdomyolysis among gemfibrozil users because there were so few controls using gemfibrozil (n=5), we suspect the exceedingly large effect of gemfibrozil obscured the much smaller genetic association. In our study, gemfibrozil use was associated with a 278 fold increased risk of rhabdomyolysis (95% CI: 106-733). Another study suggested that combined cerivastatin-gemfibrozil use relative to statin monotherapy was associated with a 1411 fold increased risk of rhabdomyolysis [7]. The role of SLCO1B1 variants was minor relative to the risk associated with the drug-drug interaction.
The RYR2 gene encodes the ryanodine receptor type 2. The ryanodine receptors are intracellular calcium release channels that are expressed in diverse tissues [30]. Three genes encode the different isoforms. RYR1 is expressed predominantly in skeletal muscles where it contributes to Ca2+ signaling and muscle contraction. Variants in RYR1 are associated with malignant hyperthermia and central core disease [31]. RYR3 but not RYR1 expression was found to be upregulated in the skeletal muscle of patients with statin-associated structural muscle injury. The expression of RYR2 was not investigated [32]. RYR2 is expressed in cardiac muscle and mutations in RYR2 are associated with arrhythmogenic right ventricular cardiomyopathy type 2 and stress induced polymorphic ventricular tachycardia [33-35]. RYR2 is also expressed in the brain [36, 37] and neonatal skeletal muscle [33]. A study in rabbits showed that RYR2 expression is upregulated and RYR1 expression is downregulated in conditioned muscle [33]. In mouse cardiomyocytes, RYR2 splice variants can modulate apoptosis, with certain variants reducing Ca2+ release and preventing apoptosis [38]. An apoptotic effect of cerivastatin on skeletal muscle through Ca2+ release [39, 40] in the presence of genetic variants that either alter RYR2 expression or disrupt splice variants suggests potential mechanisms by which RYR2 variation might contribute to cerivastatin-associated rhabdomyolysis.
Our study had several strengths. We successfully recruited subjects with a rare adverse event. The two control groups had complementary strengths: The CHS controls, like the cases, were geographically diverse; the HVH controls had an age distribution similar to that of the cases. We used multiple approaches--resequencing, tag-SNPs, and functional SNPs--to search for genetic variants of importance in the candidate genes. The resequencing effort had greater than 90% power to detect variants with a minor allele frequency of 1% in the rhabdomyolysis cases and 80% power to detect variants with a minor allele frequency of 0.65% [41]. Additionally, we used genome wide scans to identify genetic risk factors outside of the candidate genes.
The number of rhabdomyolysis cases, though large for a GWA study of an adverse event [42], is nonetheless small and provided limited power to detect associations. To our knowledge, no comparable group of rhabdomyolysis cases was available for replication. Our pre-specified replication plan for the candidate genes was in vitro functional studies (Figure 2) [43]. In one case,a novel CYP2C8 variant discovered in the re-sequencing effort introduced a frame shift (which changed the sequence of the last 22 amino acids of the CYP2C8 C-terminus and introduced an additional three amino acids), and this variant likely produced a non-functional protein [43]. Recombinant proteins containing two other non-synonymous variants discovered in the CYP2C8 re-sequencing had in- vitro kinetic values that were similar to wild type protein. Indeed, recombinant CYP2C8*3 and *4 proteins actually increased the clearance of cerivastatin relative to wild type as has been observed for some other substrates [21-24]. The findings of these functional studies may account for the observation that these variants occurred less frequently in cases than controls (ORs, 0.64 to 0.73, Table 2).
There were other limitations. Most of our controls did not use cerivastatin. The small number of cerivastatin users precluded a detailed examination of an interaction between cerivastatin dose and genotype on rhabdomyolysis risk. The cases selected for this study were not a random sample of rhabdomyolysis patients, but a select population of severe cases who went to litigation. Within this select population, the proportion participating was low. Insofar as the cases included in the study differ genetically or phenotypically from cases that were not, results may generalize poorly. We observed one finding from the GWA study at a p-value threshold at which one false positive is expected. Additional studies will be necessary to corroborate the RYR2 finding. The GWA study provided information about common, but not about rare variants, outside the candidate genes. If statin-induced rhabdomyolysis resembles a heterogeneous Mendelian disorder that has several forms caused by genetic variants at many loci in multiple genes - similar to malignant hyperthermia associated with inhalation anesthetics [44] or clinical phenylketonuria and dietary phenylalanine [45] - we would not have detected it. The focus of this paper was on genetic risk factors for rhabdomyolysis but other environmental exposures, including concomitant medication use, represent additional risk factors of interest.
For the SLCO1B1 rs4149056 variant, our results extend the findings for high-dose simvastatin-induced myopathy to cerivastatin-associated rhabdomyolysis. Despite a small sample size for the GWA study, our findings also suggest that disruptions in calcium signaling may be associated with the risk of cerivastatin-associated rhabdomyolysis. RYR2 is an interesting candidate for future investigations of statin-related muscle symptoms.
Supplementary Material
Acknowledgements
For their excellent work, we thank the attorneys and their staff. We also thank the dedicated staff members at the Cardiovascular Health Research Unit. We thank the rhabdomyolysis case participants, the participants at Group Health, and the participants from the Cardiovascular Health Study.
Funding Sources: The rhabdomyolysis case recruitment and the genotyping was supported in part by the grants HL078888, and HL085251 from the National Heart, Lung, and Blood Institute. The HVH research reported in this article was supported in part by the grants HL074745, HL43201, HL068639, HL068986, and HL73410 from the National Heart, Lung and Blood Institute. The CHS research reported in this work was supported in part by contract numbers N01-HC-85079 through N01-HC-85086, N01-HC-35129, N01-HC-15103, N01-HC-55222, N01-HC-75150, N01-HC45133 and grant numbers U01 HL080295 and R01HL087652 from the National Heart, Lung, and Blood Institute, with additional contribution from the National Institute of Neurological Disorders and Stroke. A full list of principal CHS investigators and institutions can be found at http://www.chs-nhlbi.org/pi.htm. The CHS DNA handling and genotyping was supported in part by National Center for Research Resources grant M01-RR00425 to the Cedars-Sinai General Clinical Research Center Genotyping core and National Institute of Diabetes and Digestive and Kidney Diseases grant DK063491 to the Southern California Diabetes Endocrinology Research Center. The SLCO1B1 functional work was supported in part by NIH grant GM61390 and Amgen Research Fellowship.
Footnotes
Financial Disclosures: One author, BMP, worked for plaintiffs' attorneys between 2002 and 2003. A complete statement of disclosure is available in reference 6. No other author reported having financial disclosures.
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cannistra LB, Davis SM, Bauman AG. Valvular heart disease associated with dexfenfluramine. N Engl J Med. 1997;337:636. doi: 10.1056/nejm199708283370912. [DOI] [PubMed] [Google Scholar]
- 2.Graham DJ, Drinkard CR, Shatin D. Incidence of idiopathic acute liver failure and hospitalized liver injury in patients treated with troglitazone. Am J Gastroenterol. 2003;98:175–179. doi: 10.1111/j.1572-0241.2003.07175.x. [DOI] [PubMed] [Google Scholar]
- 3.Wilke RA, Lin DW, Roden DM, Watkins PB, Flockhart D, Zineh I, Giacomini KM, Krauss RM. Identifying genetic risk factors for serious adverse drug reactions: current progress and challenges. Nat Rev Drug Discov. 2007;6:904–916. doi: 10.1038/nrd2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meyer UA. Pharmacogenetics and adverse drug reactions. Lancet. 2000;356:1667–1671. doi: 10.1016/S0140-6736(00)03167-6. [DOI] [PubMed] [Google Scholar]
- 5.Staffa JA, Chang J, Green L. Cerivastatin and reports of fatal rhabdomyolysis. N Engl J Med. 2002;346:539–540. doi: 10.1056/NEJM200202143460721. [DOI] [PubMed] [Google Scholar]
- 6.Psaty BM, Furberg CD, Ray WA, Weiss NS. Potential for conflict of interest in the evaluation of suspected adverse drug reactions: use of cerivastatin and risk of rhabdomyolysis. JAMA. 2004;292:2622–2631. doi: 10.1001/jama.292.21.2622. [DOI] [PubMed] [Google Scholar]
- 7.Graham DJ, Staffa JA, Shatin D, Andrade SE, Schech SD, La Grenade L, Gurwitz JH, Chan KA, Goodman MJ, Platt R. Incidence of hospitalized rhabdomyolysis in patients treated with lipid-lowering drugs. JAMA. 2004;292:2585–2590. doi: 10.1001/jama.292.21.2585. [DOI] [PubMed] [Google Scholar]
- 8.Muck W. Clinical pharmacokinetics of cerivastatin. Clin Pharmacokinet. 2000;39:99–116. doi: 10.2165/00003088-200039020-00002. [DOI] [PubMed] [Google Scholar]
- 9.Backman JT, Kyrklund C, Neuvonen M, Neuvonen PJ. Gemfibrozil greatly increases plasma concentrations of cerivastatin. Clin Pharmacol Ther. 2002;72:685–691. doi: 10.1067/mcp.2002.128469. [DOI] [PubMed] [Google Scholar]
- 10.Shitara Y, Hirano M, Sato H, Sugiyama Y. Gemfibrozil and its glucuronide inhibit the organic anion transporting polypeptide 2 (OATP2/OATP1B1:SLC21A6)-mediated hepatic uptake and CYP2C8-mediated metabolism of cerivastatin: analysis of the mechanism of the clinically relevant drug-drug interaction between cerivastatin and gemfibrozil. J Pharmacol Exp Ther. 2004;311:228–236. doi: 10.1124/jpet.104.068536. [DOI] [PubMed] [Google Scholar]
- 11.Psaty BM, Heckbert SR, Koepsell TD, Siscovick DS, Raghunathan TE, Weiss NS, Rosendaal FR, Lemaitre RN, Smith NL, Wahl PW, et al. The risk of myocardial infarction associated with antihypertensive drug therapies. JAMA. 1995;274:620–625. [PubMed] [Google Scholar]
- 12.Smith NL, Hindorff LA, Heckbert SR, Lemaitre RN, Marciante KD, Rice K, Lumley T, Bis JC, Wiggins KL, Rosendaal FR, Psaty BM. Association of genetic variations with nonfatal venous thrombosis in postmenopausal women. JAMA. 2007;297:489–498. doi: 10.1001/jama.297.5.489. [DOI] [PubMed] [Google Scholar]
- 13.Heckbert SR, Wiggins KL, Glazer NL, Dublin S, Psaty BM, Smith NL, Longstreth WT, Jr., Lumley T. Antihypertensive treatment with ACE inhibitors or beta-blockers and risk of incident atrial fibrillation in a general hypertensive population. Am J Hypertens. 2009;22:538–544. doi: 10.1038/ajh.2009.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fried LP, Borhani NO, Enright P, Furberg CD, Gardin JM, Kronmal RA, Kuller LH, Manolio TA, Mittelmark MB, Newman A, et al. The Cardiovascular Health Study: design and rationale. Ann Epidemiol. 1991;1:263–276. doi: 10.1016/1047-2797(91)90005-w. [DOI] [PubMed] [Google Scholar]
- 15.Andrade SE, Graham DJ, Staffa JA, Schech SD, Shatin D, La Grenade L, Goodman MJ, Platt R, Gurwitz JH, Chan KA. Health plan administrative databases can efficiently identify serious myopathy and rhabdomyolysis. J Clin Epidemiol. 2005;58:171–174. doi: 10.1016/j.jclinepi.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 16.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet. 2008;83:311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peter H. Resampling-Based Multiple Testing. Wiley; New York: 1993. Westfall SSY. [Google Scholar]
- 19.Bahadur N, Leathart JB, Mutch E, Steimel-Crespi D, Dunn SA, Gilissen R, Houdt JV, Hendrickx J, Mannens G, Bohets H, Williams FM, Armstrong M, Crespi CL, Daly AK. CYP2C8 polymorphisms in Caucasians and their relationship with paclitaxel 6alpha-hydroxylase activity in human liver microsomes. Biochem Pharmacol. 2002;64:1579–1589. doi: 10.1016/s0006-2952(02)01354-0. [DOI] [PubMed] [Google Scholar]
- 20.Dai D, Zeldin DC, Blaisdell JA, Chanas B, Coulter SJ, Ghanayem BI, Goldstein JA. Polymorphisms in human CYP2C8 decrease metabolism of the anticancer drug paclitaxel and arachidonic acid. Pharmacogenetics. 2001;11:597–607. doi: 10.1097/00008571-200110000-00006. [DOI] [PubMed] [Google Scholar]
- 21.Niemi M, Leathart JB, Neuvonen M, Backman JT, Daly AK, Neuvonen PJ. Polymorphism in CYP2C8 is associated with reduced plasma concentrations of repaglinide. Clin Pharmacol Ther. 2003;74:380–387. doi: 10.1016/S0009-9236(03)00228-5. [DOI] [PubMed] [Google Scholar]
- 22.Niemi M, Backman JT, Kajosaari LI, Leathart JB, Neuvonen M, Daly AK, Eichelbaum M, Kivisto KT, Neuvonen PJ. Polymorphic organic anion transporting polypeptide 1B1 is a major determinant of repaglinide pharmacokinetics. Clin Pharmacol Ther. 2005;77:468–478. doi: 10.1016/j.clpt.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 23.Kirchheiner J, Thomas S, Bauer S, Tomalik-Scharte D, Hering U, Doroshyenko O, Jetter A, Stehle S, Tsahuridu M, Meineke I, Brockmoller J, Fuhr U. Pharmacokinetics and pharmacodynamics of rosiglitazone in relation to CYP2C8 genotype. Clin Pharmacol Ther. 2006;80:657–667. doi: 10.1016/j.clpt.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 24.Tornio A, Niemi M, Neuvonen PJ, Backman JT. Trimethoprim and the CYP2C8*3 allele have opposite effects on the pharmacokinetics of pioglitazone. Drug Metab Dispos. 2008;36:73–80. doi: 10.1124/dmd.107.018010. [DOI] [PubMed] [Google Scholar]
- 25.Kalliokoski A, Niemi M. Impact of OATP transporters on pharmacokinetics. Br J Pharmacol. 2009;158:693–705. doi: 10.1111/j.1476-5381.2009.00430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pasanen MK, Neuvonen M, Neuvonen PJ, Niemi M. SLCO1B1 polymorphism markedly affects the pharmacokinetics of simvastatin acid. Pharmacogenet Genomics. 2006;16:873–879. doi: 10.1097/01.fpc.0000230416.82349.90. [DOI] [PubMed] [Google Scholar]
- 27.Kameyama Y, Yamashita K, Kobayashi K, Hosokawa M, Chiba K. Functional characterization of SLCO1B1 (OATP-C) variants, SLCO1B1*5, SLCO1B1*15 and SLCO1B1*15+C1007G, by using transient expression systems of HeLa and HEK293 cells. Pharmacogenet Genomics. 2005;15:513–522. doi: 10.1097/01.fpc.0000170913.73780.5f. [DOI] [PubMed] [Google Scholar]
- 28.Voora D, Shah SH, Spasojevic I, Ali S, Reed CR, Salisbury BA, Ginsburg GS. The SLCO1B1*5 genetic variant is associated with statin-induced side effects. J Am Coll Cardiol. 2009;54:1609–1616. doi: 10.1016/j.jacc.2009.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Link E, Parish S, Armitage J, Bowman L, Heath S, Matsuda F, Gut I, Lathrop M, Collins R. SLCO1B1 variants and statin-induced myopathy--a genomewide study. N Engl J Med. 2008;359:789–799. doi: 10.1056/NEJMoa0801936. [DOI] [PubMed] [Google Scholar]
- 30.Brini M. Ryanodine receptor defects in muscle genetic diseases. Biochem Biophys Res Commun. 2004;322:1245–1255. doi: 10.1016/j.bbrc.2004.08.029. [DOI] [PubMed] [Google Scholar]
- 31.Loke J, MacLennan DH. Malignant hyperthermia and central core disease: disorders of Ca2+ release channels. Am J Med. 1998;104:470–486. doi: 10.1016/s0002-9343(98)00108-9. [DOI] [PubMed] [Google Scholar]
- 32.Mohaupt MG, Karas RH, Babiychuk EB, Sanchez-Freire V, Monastyrskaya K, Iyer L, Hoppeler H, Breil F, Draeger A. Association between statin-associated myopathy and skeletal muscle damage. CMAJ. 2009;181:E11–18. doi: 10.1503/cmaj.081785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Froemming GR, Murray BE, Harmon S, Pette D, Ohlendieck K. Comparative analysis of the isoform expression pattern of Ca(2+)-regulatory membrane proteins in fast-twitch, slow-twitch, cardiac, neonatal and chronic low-frequency stimulated muscle fibers. Biochim Biophys Acta. 2000;1466:151–168. doi: 10.1016/s0005-2736(00)00195-4. [DOI] [PubMed] [Google Scholar]
- 34.Tiso N, Stephan DA, Nava A, Bagattin A, Devaney JM, Stanchi F, Larderet G, Brahmbhatt B, Brown K, Bauce B, Muriago M, Basso C, Thiene G, Danieli GA, Rampazzo A. Identification of mutations in the cardiac ryanodine receptor gene in families affected with arrhythmogenic right ventricular cardiomyopathy type 2 (ARVD2) Hum Mol Genet. 2001;10:189–194. doi: 10.1093/hmg/10.3.189. [DOI] [PubMed] [Google Scholar]
- 35.Laitinen PJ, Brown KM, Piippo K, Swan H, Devaney JM, Brahmbhatt B, Donarum EA, Marino M, Tiso N, Viitasalo M, Toivonen L, Stephan DA, Kontula K. Mutations of the cardiac ryanodine receptor (RyR2) gene in familial polymorphic ventricular tachycardia. Circulation. 2001;103:485–490. doi: 10.1161/01.cir.103.4.485. [DOI] [PubMed] [Google Scholar]
- 36.Otsu K, Willard HF, Khanna VK, Zorzato F, Green NM, MacLennan DH. Molecular cloning of cDNA encoding the Ca2+ release channel (ryanodine receptor) of rabbit cardiac muscle sarcoplasmic reticulum. J Biol Chem. 1990;265:13472–13483. [PubMed] [Google Scholar]
- 37.Nakai J, Imagawa T, Hakamat Y, Shigekawa M, Takeshima H, Numa S. Primary structure and functional expression from cDNA of the cardiac ryanodine receptor/calcium release channel. FEBS Lett. 1990;271:169–177. doi: 10.1016/0014-5793(90)80399-4. [DOI] [PubMed] [Google Scholar]
- 38.George CH, Rogers SA, Bertrand BM, Tunwell RE, Thomas NL, Steele DS, Cox EV, Pepper C, Hazeel CJ, Claycomb WC, Lai FA. Alternative splicing of ryanodine receptors modulates cardiomyocyte Ca2+ signaling and susceptibility to apoptosis. Circ Res. 2007;100:874–883. doi: 10.1161/01.RES.0000260804.77807.cf. [DOI] [PubMed] [Google Scholar]
- 39.Inoue R, Tanabe M, Kono K, Maruyama K, Ikemoto T, Endo M. Ca2+-releasing effect of cerivastatin on the sarcoplasmic reticulum of mouse and rat skeletal muscle fibers. J Pharmacol Sci. 2003;93:279–288. doi: 10.1254/jphs.93.279. [DOI] [PubMed] [Google Scholar]
- 40.Kobayashi M, Kagawa T, Narumi K, Itagaki S, Hirano T, Iseki K. Bicarbonate supplementation as a preventive way in statins-induced muscle damage. J Pharm Pharm Sci. 2008;11:1–8. doi: 10.18433/j33018. [DOI] [PubMed] [Google Scholar]
- 41.Eberle MA, Kruglyak L. An analysis of strategies for discovery of single-nucleotide polymorphisms. Genet Epidemiol. 2000;19(Suppl 1):S29–35. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI5>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 42.Daly AK. Genome-wide association studies in pharmacogenomics. Nat Rev Genet. 2010;11:241–246. doi: 10.1038/nrg2751. [DOI] [PubMed] [Google Scholar]
- 43.Kaspera R, Naraharisetti SB, Tamraz B, Sahele T, Cheesman MJ, Kwok PY, Marciante K, Heckbert SR, Psaty BM, Totah RA. Cerivastatin in vitro metabolism by CYP2C8 variants found in patients experiencing rhabdomyolysis. Pharmacogenet Genomics. 2010;20:619–629. doi: 10.1097/FPC.0b013e32833ecace. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Litman RS, Rosenberg H. Malignant hyperthermia: update on susceptibility testing. JAMA. 2005;293:2918–2924. doi: 10.1001/jama.293.23.2918. [DOI] [PubMed] [Google Scholar]
- 45.Scriver CR. The PAH gene, phenylketonuria, and a paradigm shift. Hum Mutat. 2007;28:831–845. doi: 10.1002/humu.20526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.