Abstract
Members of the cyanovirin-N homolog (CVNH) lectin family are found in bacteria, fungi and plants. As part of our ongoing work on CVNH structure-function studies, we determined the high-resolution NMR solution structure of the homolog from the wheat head blight disease causing ascomycetous fungus Gibberella zeae (or Fusarium graminearum), hereafter called GzCVNH. Like cyanovirin-N (CV-N), GzCVNH comprises two tandem sequence repeats and the protein sequence exhibits 30% identity with CV-N. The overall structure is similar to those of other members of the CVNH family, with the conserved pseudo-symmetric halves of the structure, domains A and B, closely resembling recently determined structures of Tuber borchii, Neurospora crassa and Ceratopteris richardii CVNH proteins. Although GzCVNH exhibits a similar glycan recognition profile to CV-N and specifically binds to Manα(1–2)Manα, its weak carbohydrate binding affinity to only one binding site is insufficient for conferring anti-HIV activity.
Keywords: NMR solution structure, CVNH, lectin, antiviral protein, HIV
Introduction
Lectins are mono- and oligosaccharide binding proteins found in a wide range of organisms, including animals, plants and microorganisms.1 They are structurally highly diverse and carry out a plethora of biological functions. Animal lectins play a role in innate immunity by recognizing pathogen-associated glycans, mediate cell-cell communication and function in intra-cellular trafficking. In most cases, one lectin molecule possesses two or more carbohydrate binding sites, enabling cross-linking of targets by binding the displayed sugars.2 The various roles attributed to plant lectins are nutrient storage, recognition of symbionts and defense reactions against pathogens and insects. Lectins from higher fungi are less well studied, although knowledge about their molecular properties has grown substantially in recent years. Many fungi are known to secrete lectins, however their functional roles are still unclear.3 Given their highly specific sugar binding properties, interest in lectins is steadily increasing, particularly for biotechnological and medical applications, such as their use as diagnostic tools.
Recently, a new structural class of lectins, the so-called CVN homolog (CVNH) family, has been identified, with cyanovirin-N (CV-N) as the class defining member.4 All CVNH proteins share a common three-dimensional fold similar to the one previously considered to be unique to CV-N.5,6 Cyanovirin-N was originally isolated from an extract of the cyanobacterium Nostoc ellipsosporum7 and was found to exhibit antiviral activity against a variety of viruses, most prominently against HIV-1. The protein’s activity is mediated by binding to the terminal Manα(1-2)Manα units on the D1 and D3 arms of high-mannose oligosaccharides (Man-8 and Man-9) on the viral surface envelope glycoprotein gp120, thereby blocking viral entry into the cell.8 Extensive biochemical and structural studies have established that multi-valent and multi-site interactions are necessary for antiviral activity. Variants of CV-N that contain only one binding site do not exhibit any antiviral activity, irrespective of whether this binding site is present on domain A or B.9–12
In fungi, CVNH sequences have been identified in the genomes of Neurospora crassa, a model organism, in pathogenic fungi such as Magnaporthe grisea, Gibberella zea, Botrytis cinerea and Aspergillus fumigatus, and in symbiotic fungi of great commercial interest such as truffles. Fungal CVNHs therefore may present important opportunities for developing environmental biocontrol strategies aimed at interfering with (pathogens) or promoting (symbionts) their interactions with other organisms.
Here we report the solution structure of a CVNH protein from Gibberella zea, GzCVNH, that shares 30% sequence identity with CV-N (Figure 1). Gibberella zea or Fusarium graminearum is a ubiquitous pathogen of cereal crops causing scab disease on wheat and barley that reduces crop yield and grain quality.13 In addition to reducing seed mass and quality, the fungus contaminates grain with toxic metabolites, such as vomitoxin (deoxynivalenol), which in livestock and other animals causes a refusal to feed and lack of weight gain.14 Gibberella zea is among the most economically important plant pathogens world wide and representative of the most destructive and widely studied genus of plant pathogenic fungi. Gibberella's impact on agriculture and food safety and its growing role in human health, was identified as a priority for whole-genome sequencing by the Broad Institute’s Fungal Genome Initiative and in 2003 it became the second plant-pathogenic fungus for which the entire genome sequence was made publicly available (http://www.broad.mit.edu). Nearly all (99.8%) of the assembly of the genome was anchored into four chromosomes by genetically mapping markers derived from the genome sequence.15 Among these, two cvn homologous genes were identified in the Gibberella zeae PH-1 strain, one on chromosome 2 (accession number FG03554.1), and one on chromosome 3 (accession number FG04975.1). These two genes represent examples of different classes of proteins within the CVNH family. The gene found on chromosome 3 (accession number FG04975.1) encodes a type I-CVNH and is made up of two uninterrupted CVNH sequence repeats, while the protein encoded on chromosome 2 (accession number FG03554.1) codes for a type III-CVNH, in which a LysM module bisects the two CVNH repeats, with both modules connected by two short, glycine-rich linkers.4
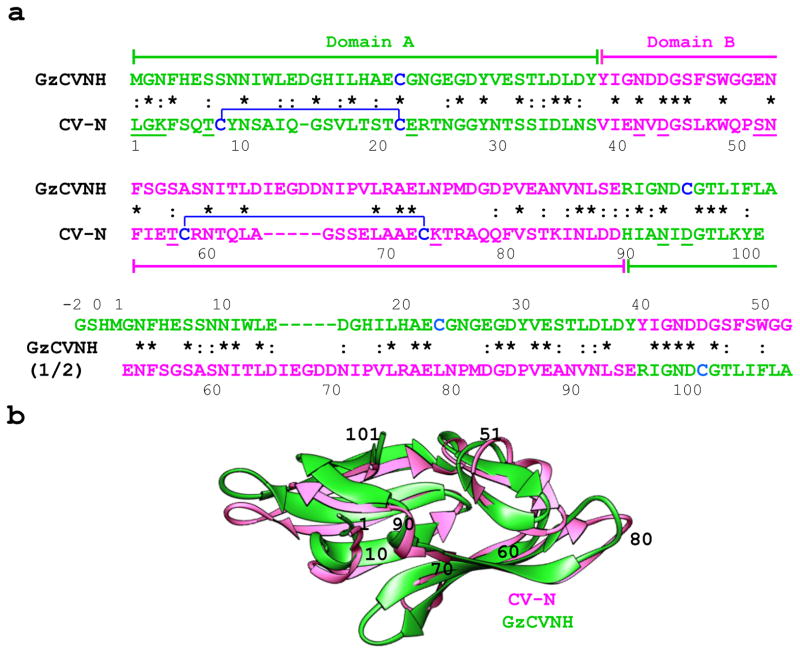
Figure 1. Sequence alignment and superposition of solution structures of CV-N from Nostoc ellipsosporum and GzCVNH from Gibberella zeae.
(a) Amino acid sequences of CV-N and GzCVNH. Domains A and B and the corresponding sequences are colored green and pink, respectively. The top sequence alignment is for CV-N and GzCVNH and the bottom compares the first and second sequence repeats of GzCVNH. Disulfide bonds are indicated by brackets and residues involved in protein-carbohydrate interactions are underlined in the CV-N sequence. Identical amino acids in the aligned sequence repeats are starred and conservative substitutions are marked by dashes. (b) Ribbon superposition of the NMR solution structures of GzCVNH (green) and wild type CV-N (pink). Amino acid positions for CV-N are indicated by numbers.
In order to provide a molecular framework for understanding sequence and conformational similarities at the atomic level as well as the relationship of specific structural features with function, we determined the high-resolution structure of the protein encoded on chromosome 3 (FG04975.1). Carbohydrate glycoarray screening and NMR sugar titration studies were employed to examine the carbohydrate binding specificity and to structurally map the sugar binding site.
Our results show that: (i) like CV-N, the GzCVNH structure exhibits a pseudo-symmetric, bilobal architecture; (ii) GzCVNH exhibits similar carbohydrate specificities as CV-N, however the binding affinity for Manα(1–2)Manα on domain B is very weak and no specific binding to domain A could be detected. (iii) GzCVNH does not inhibit HIV, in perfect agreement with previous results with CVNH proteins that contain single carbohydrate binding sites.
Materials and Methods
Protein expression and purification
The protein was expressed using a synthetic gene of GzCVNH inserted into the pET-15b(+)vector (Novagen; Madison, WI) and E. coli BL-21(DE3) as the host strain. Using pET-15b(+) introduces three additional N-terminal residues in the final GzCVNH sequence. Uniform 15N and 15N/13C-labeling was achieved utilizing 15NH4Cl (1g/L) and 13C6-D-glucose (2g/L) (Cambridge Isotope Laboratories, Inc; Andover, MA) as sole nitrogen and carbon sources, respectively. The expressed protein was isolated from the periplasmic fraction of E. coli cells by twice heating (70 °C) and cooling (0°C) the cell suspension in PBS buffer (pH 7.4). After removal of insoluble material by centrifugation, the supernatant containing soluble GzCVNH was initially purified by Ni+- affinity chromatography (Amersham Pharmacia Biotech, NJ), using ~ 500 mM imidazole for elution, followed by thrombin cleavage in 50 mM Tris, 100 mM NaCl, 2 mM CaCl2, pH 7.4, and gel filtration chromatography on a Superdex-75 column (HiLoad 2.6 cm × 60 cm, Amersham Pharmacia Biotech, NJ), equilibrated in 20 mM sodium phosphate buffer, 0.01% NaN3, pH 6.0. The G52P mutant of GzCVNH was created using the QuikChange XL II site-directed mutagenesis kit (Stratagene) with two forward/reverse primers: 5'– GCAGCTTTAGCTGGGGCC CGGAAAACTTTAGCGGCAG–3'/5'– CTGCCGCTAAAGTTTTCCGGGCCCCAGCTAAAGC TGC–3'. The protein was expressed and purified as described for wild type GzCVNH. Structural assessment by 1H-15N HSQC spectroscopy revealed that this mutant was not folded correctly.
The quaternary state of GzCVNH was evaluated by native polyacrylamide gel electrophoresis and multi-angle light scattering (data not shown) and the protein was found to be monomeric. The extent of 15N labeling was determined by mass spectrometry and the measured molecular mass was 12,078 Da, compared to an expected mass of 12,093, translating to ~ 90% labeling.
NMR spectroscopy
NMR spectra were recorded at 30°C on Bruker AVANCE 700 and AVANCE 600 spectrometers, equipped with 5-mm, triple resonance, three-axis gradient probes or z-axis gradient cryoprobes. Spectra were processed with NMRPipe16 and analyzed with NMRview.17 Samples contained 1.5 mM protein in 20 mM sodium phosphate buffer, pH 6.0. For chemical shift assignments, a series of heteronuclear, multi-dimensional experiments, routinely used in our laboratory were recorded.18–21 Complete 1H, 15N, and 13C backbone and side chain resonance assignments were obtained using the following heteronuclear 2D and 3D experiments: 1H-15N HSQC, HNCACB, CBCA(CO)NH, HCCH-TOCSY, as well as a 3D simultaneous 13C and 15N NOESY, 22 with NOEs for inter-proton distance constraints extracted from the NOESY spectrum, recorded with a mixing time of 100 ms.
NMR Solution structure determination
All NOESY cross peaks were picked using NMRView17 and inspected/sorted manually for accuracy. NOESY cross peaks were assigned using the ATNOS algorithm23 of CYANA v2.124 in an automated fashion. The input for ATNOS consisted of the amino acid sequence of the protein, chemical shift lists from sequence-specific resonance assignment, and NOEs from 15N and 13C-edited 3D-NOESY spectra. Throughout all calculations, 121 backbone torsion angle constraints derived from TALOS25 were employed. CYANA calculations were performed for seven iterative cycles with an additional cycle for stereospecific assignments for the final structure calculations. From the second cycle onwards, the intermediate protein structures were used as an additional guide for the interpretation of the NOESY spectra. To obtain the best structural quality with the lowest energy target function and no restraints violations, successive runs incorporated optimized input NOE peak lists and chemical shifts assignments. Continued analysis of the NMR data during the process of automated protein structure determination was used for direct feedback between the structure, NOE assignments and experimental NOESY spectra in an iterative fashion. The initial CYANA calculations were used to generate the interproton distance restraints (2391) that were incorporated into further simulated annealing refinement with CNS v1.226, and the final structures were examined using PROCHECK-NMR27 and PSVS28.
The atomic coordinates and NMR constraints have been deposited in the RCSB Protein Data Bank under accession code 2L2F. All structure figures were generated using the program Pymol and Swiss-PdbViewer v3.7.29
NMR titrations
Titration experiments were performed using uniformly 15N-labeled GzCVNH (150 μM) with increasing amounts of dimannose Manα(1-2)Manα in 20 mM sodium phosphate buffer, pH 6.0, 0.01% sodium azide, 90% H2O/10% D2O. A series of 1H-15N HSQC spectra were recorded after the addition of sugar aliquots from a stock solution of 50 mM Manα(1-2)Manα at ligand/protein molar ratios of: 0, 2, 4, 6, 12, 16, 20, 24, 30, and 50.
Free and sugar-bound protein resonances throughout the titration are in fast exchange on the chemical shift scale. The observed chemical shift change during the titration is given by: Δδ = [PL]/[P] (δb − δf), with [P] and [PL] the concentrations of free protein and ligand-protein complex and δb andδf the chemical shifts of protein resonances in the free and fully bound state. The dissociation constant Kd, was obtained from best-fitting the titration curve (chemical shift change Δδ vs. molar ratio M) using KaleidaGraph software and the following equation:
The chemical shift difference was calculated by applying Pitagora’s theorem as following: Δδ = [(ΔδHN) 2 + (ΔδN × 0.17) 2]1/2 , where ΔδH and ΔδN are the observed chemical shift changes for 1H and 15N, respectively. The weighting factor of 0.17 reflects the difference in chemical shift dispersion of 1Hand 15N in folded proteins.30 For determination of dissociation constants, Δδ was plotted as a function of the ligand/protein molar ratio and the data were fitted using the maximum shift and dissociation constant as adjustable parameters.
An accurate concentration of the oligosaccharide was obtained by compositional analysis of the stock solution at the Complex Carbohydrate Research Center at the University of Georgia.
Fluorescent labeling
Purified GzCVNH and CV-N proteins (~500 μM) were incubated with equimolar concentration of the Cy5 dye (GE Healthcare) in 20 mM sodium phosphate buffer, pH 7.4, in the presence of 2 mM TCEP to protect the free cysteines from oxidation. The reaction was carried out for 2 hours at room temperature in the dark. The unreacted fluorophore was removed in two steps. First, the bulk of the free dye was removed by passage over a PD-10 desalting column (GE Healthcare, Uppsala, Sweden) in 20 mM sodium phosphate buffer, pH 7.4. Dye-labeled protein fractions were collected and concentrated to ~150μM using centriprep devices (Millipore). A similar protocol was followed to label a Q50C CV-N mutant, containing a free sulfhydral with Cy5. This protein was used as a control. The labeling efficiency was about 80%, as evaluated by ESI - mass spectrometry. The structural integrity of the labeled proteins was ascertained by recording 1H- 15N HSQC spectra and both proteins were found to be well folded.
Anti-HIV assays
In order to establish whether the GzCVNH protein possesses any anti-HIV activity, HIV infectivity assays were carried out, using CV-N as a control, as described previously.31 For antiviral assays, purified GzCVNH protein was serially diluted in sterile phosphate-buffered saline and 5 μl were added to 500 μl of pre-diluted infectious HIV-1 (produced by transfection of 293T cells with the R9 molecular clone and incubated for 30 min at room temperature). Aliquots of the mixture (125 μL, triplicates) were added to cultures of HeLa-P4 cells (20,000 cells seeded per well the day before in a 48-well format) and after two days, cells were fixed and stained with X-Gal overnight, and counted. Results are expressed as the average number of X-gal-positive cells per well.
Results
GzCVNH sequence
Sequence alignment and comparison of the 108 amino acid containing GzCVNH and the 101 residue CV-N sequences revealed an overall identity of 30% (Figure 1(a)). This is similar to the identity observed between the two sequence repeats of GzCVNH (35%) and their equivalents in CV-N (33%).
Comparing CV-N and GzCVNH, a distinctive sequence difference was noted for the number of cysteine residues and therefore the disulfide bonds in the two proteins. Only one of the cysteine residues present in CV-N, Cys22, is conserved in GzCVNH, while the remaining three Cys residues are replaced by Ser (Cys8-Ser), Ala (Cys58-Ala) and Leu (Cys73-Leu). Furthermore, GzCVNH contains an additional Cys residue at its C-terminus (Cys101) that has no counterpart in CV-N.
Previous biochemical and structural studies showed that CV-N possesses two carbohydrate binding sites, one on domain A (residues 1-38, 90-101) and one on domain B (residues 39-89).32,33 Considering solely sequence, some amino acids involved in sugar binding on domain A of GzCVNH (residues 1-39, 96-108) are the same as in CV-N (Gly2, Asn93), others are conservatively substituted (Leu1-Met, Lys3-Asn and Thr7-Ser) while several are dramatically different (Glu23-Gly and Asp95-Cys; CV-N numbering). Only a slightly smaller difference in sequence is found for residues that make up the carbohydrate binding site on domain B (residues 40-95). Three are identical (Asn42, Asp44, Asn53), two are conservatively substituted (Thr57-Ser), and two are significantly different (Ser52-Glu, Lys74-Asn) (Figure 1(a)).
GzCVNH NMR solution structure
The solution structure of GzCVNH was determined by NMR spectroscopy using uniformly 15N-and 13C,15N-labeled samples and established protocols commonly used in our laboratory. A set of 20 simulated annealing structures was calculated based on 2391 interproton distances and 121 torsion angle constraints. All structures are well defined, satisfy all experimental constraints, display excellent covalent geometry and exhibit atomic r.m.s. deviations of 0.20 ± 0.05 and 0.57 ± 0.04 Å with respect to the mean coordinate positions for the backbone (N, Cα, C’) and all heavy atoms, respectively. About 79% of residues lie in the most favorable region of the Ramachandran plot.34 A summary of all experimental constraints as well as pertinent structural statistics for the refined ensemble of 20 conformers are provided in Table 1.
Table I.
NMR and refinement statistics for GzCVNH
| NMR distance & dihedral constraints | |
| Distance constraints | |
| Total NOE | 2391 |
| Intra residue (|i-j| = 0) | 418 |
| Inter residue | |
| Sequential (|i−j| = 1) | 621 |
| Medium-range (1 <|i−j| < 5) | 383 |
| Long range (|i−j > 5) | 969 |
| Hydrogen bonds | |
| Dihedral angle constraints | |
| φ | 60 |
| ψ | 61 |
| Structure Statistics | |
| Violations (mean and s.d.) | 0 |
| Distance constraints (Å) | 0.006 ± 0.0003 |
| Dihedral angle constraints (°) | 0.4 ±0.02 |
| Max. dihedral angle violation (°) | 2.2 |
| Max. distance constraint violation (Å) | 0.05 |
| Deviations from idealized geometry | |
| Bond lengths (Å) | 0.002 ± 0.0001 |
| Bond angles (°) | 0.3 ± 0.009 |
| Impropers (°) | 0.14 ± 0.006 |
| Energies | |
| Total (kcal/mol) | 88.25 ± 6.2 |
| Bond (kcal/mol) | 2.74 ± 0.3 |
| Angle (kcal/mol) | 36.46 ± 2.2 |
| Improper (kcal/mol) | 2.58 ± 0.2 |
| Van der Waals (kcal/mol) | 37.27 ± 3.2 |
| NOE (kcal/mol) | 6.72 ± 0.7 |
| Dihedral (kcal/mol) | 2.48 ± 0.2 |
| Ramachandran (%)* | |
| Most favorable regions | 79 |
| Allowed regions | 17 |
| Generously allowed regions | 4 |
| Disallowed | 0 |
| Mean r.m.s.d. (Å) ** | |
| Heavy atoms | 0.57±0.04 |
| Backbone atoms (Cα, N, C’) | 0.20 ± 0.05 |
determined by PROCHECK and PSVS
calculated for residues 1-108 (of 108 total for GzCVNH) of each conformer with respect to the minimized mean structure.
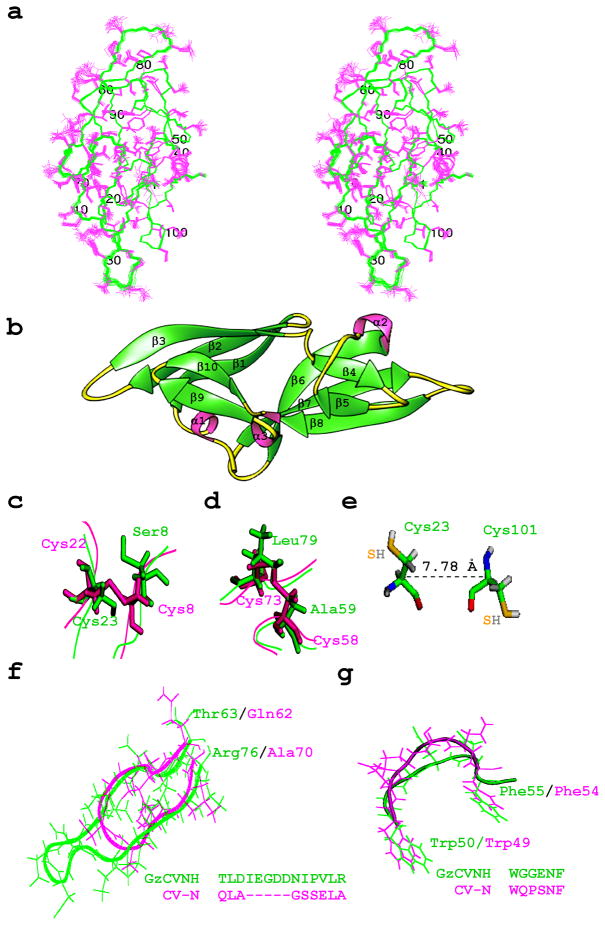
The overall fold of GzCVNH is very similar to the structure of CV-N5, as well as other CVNH family members.6 A superposition of the backbone NMR solution structures of GzCVNH and CV-N is provided in ribbon representation in Figure 1(b) and a stereoview of the refined 20-conformer ensemble is shown in Figure 2(a). Like wild type CV-N, the GzCVNH structure contains two pseudo-symmetric domains and the architecture of both proteins, as well as their secondary structure elements are very similar. The latter are made up from two anti-parallel triple-stranded β-sheets (sheet 1: residues 8–14 (β1), 18-24 (β2) and 30–36 (β3); sheet 2: residues 60–67 (β6), 72–79 (β7) and 87–92 (β8)), two β-hairpins (residues 41-43 (β4) and 48-50 (β5); 97-100 (β9) and 103-106 (β10)) and three 310 helical turns (residues 3-5(α1), 56-58(α2) and 93-95(α3)) (Figure 2(b)). The similarity between the wild type CV-N structure (PDB accession code 2EZM) and the minimized mean structure of GzCVNH is evidenced by a backbone (N, Cα, C’) r.m.s.d. value of 1.04 Å (Table 2). Not surprisingly, in the case of GzCVNH the only conserved cysteine, Cys23 (corresponding to Cys22 of CV-N) is not involved in any disulfide bond since at the position of Cys8, the pairing residue in CV-N, GzCVNH contains a serine. In place of the Cys58-Cys73 disulfide bond in CV-N, two hydrophobic residues, Ala59 and Leu79 are found in GzCVNH (Figure 2(c),(d)). The conserved Cys23 is relatively close to Cys101 and one could have speculated that these two cysteines may possibly form a disulfide bond. However, there is no experimental indication for a disulfide bond and in the final structure the relevant cysteine side chains clearly point in different directions with a Cα-Cα’ distance of 7.8 Å (Figure 2(e)). This distance is incompatible with disulfide bond formation, lying outside the effective range for Cα -Cα ’ distances in native protein disulfide bonds (4.8 to 6.6 Å).35
Figure 2. NMR solution structure of GzCVNH.
(a) Stereoview of the final 20 conformer ensemble. Backbone (N, Cα, C’) atoms are shown in green and all side chain atoms in pink. (b) Ribbon representation of GzCVNH NMR solution structure with β-strands and helical turns labeled. Comparison between CV-N (pink) and GzCVNH (green) for the Cys8-Cys22 (c) and Cys58-Cys73 (d) disulfide regions in CV-N. Cysteine side chains and their counterparts are shown in stick representation and the polypeptide backbone is depicted as a tube. (e) Conformation of the two reduced cysteine residues (Cys23 and Cys101) in GzCVNH. The sulfur atoms are colored in orange. There is no indication that they are able to form a disulfide bond, despite their relative proximity. (f) Cα backbone and side chain traces of the β6, β7 connecting loops in GzCVNH (green) and CV-N (pink), respectively. Note, the loop in GzCVNH is five residues longer than in CV-N. (g) Cα backbone and side chain traces of the hinge-loop region of CV-N (Trp49-Phe54, pink) with the equivalent region inGzCVNH (Trp50-Phe55, green).
Table II.
| CV-N | GzCVNH | CrCVNH | TbCVNH | NcCVNH | |
|---|---|---|---|---|---|
| CV-N | - | 1.04 | 0.93 | 1.01 | 1.08 |
| GzCVNH | 30 | - | 1.14 | 1.35 | 1.42 |
| CrCVNH | 25 | 28 | - | 1.10 | 1.10 |
| TbCVNH | 24 | 37 | 22 | - | 1.22 |
| NcCVNH | 22 | 33 | 19 | 28 | - |
The sequence alignment for CV-N homologs was taken from reference 4 and the gene bank codes are: GzCVNH (EAA74842), CrCVNH (BQ087517), TbCVNH (AAV85993) and NcCVNH (EAA31221). Sequence identity scores were obtained using ClustalW for alignment of the entire sequence of the individual CVNHs.
r.m.s. differences between the minimized mean structure of each CVNH ensemble were calculated for residues 11-14, 17-23, 29-35, 40-42, 46-48, 60-63, 69-74, 81-86, 91-94, and 97-100 of CV-N, 11-14, 18-24, 30-36, 41-43, 47-49, 61-64, 75-80, 87-92, 97-100, and 103-106 of GzCVNH, 12-15, 18-24, 30-36, 41-43, 47-49, 61-64, 70-75, 82-87, 92-95, and 98-101 of CrCVNH, 10-13, 18-24, 30-36, 41-43, 47-49, 61-64, 71-76, 84-89, 93-96, and 99-102 of TbCVNH, and 10-13, 20-26,32-38, 43-45, 49-51, 63-66, 78-83, 90-95, 100-103, and 106-109 of NcCVNH.
When comparing GzCVNH with other CVNH members, the closest CVNH relative is CrCVNH with a backbone (N, Cα, C’) r.m.s.d of 1.14 Å (Table 2). In CrCVNH, the two disulfide bonds found in CV-N (Cys8-Cys22 and Cys58-Cys73 are structurally conserved (Cys9-Cys23 and Cys59-Cys74)6, with an additional disulfide bond between Cys3 and Cys102. The other two CVNH proteins, TbCVNH and NcCVNH, contain no disulfide bridges.6 Therefore, it appears that the presence or absence of disulfide bonds does not determine which members of the CVNH family are structurally most similar.
Another distinct feature in the GzCVNH structure is the presence of an extended loop in the Thr63-Arg76 region (equivalent to the Gln62-Ala70 loop in CV-N (Figure 1(b)) that is created by the insertion of five extra amino acids (Figure 2(f)). In addition, a different local conformation exists in the loop region around Trp50-Phe55 in GzCVNH, corresponding to Trp49-Phe54 in CV-N. In this stretch of sequence, three out of the six amino acids are different, causing a change in the overall structure, even if the two aromatic side chains, Trp and Phe, occupy essentially identical positions in space. A superposition of this region for the two proteins, including side chains, is depicted in Figure 2(g). Interestingly, the pivotal Pro51 in the hinge-loop region of CV-N, associated with domain swapping, is Gly52 in GzCVNH.
Glycoarray screening of GzCVNH
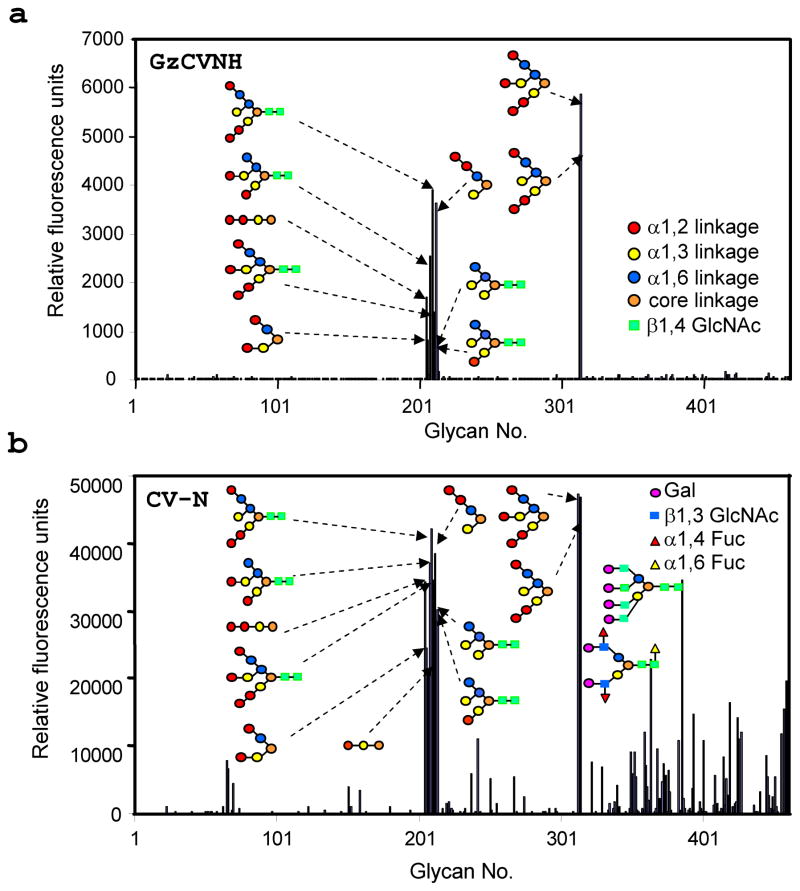
To gain information about the sugar-binding specificity of GzCVNH, a series of glycan array screens was carried out. Over 400 glycans were tested, covering a variety of hybrid and N-linked carbohydrates (http://www.functionalglycomics.org/static/index.shtml). Direct fluorescent labeling with Cy5 was used for detection and fluorescent labeled CV-N was used as a control in these experiments. The results of two representative experiments are provided in Figures 3(a) and (b). GzCVNH like CV-N recognized mannose-containing structures, with GzCVNH exhibiting slightly more restricted specificity (Figure 3(a)). Both proteins interacted with several high-mannose N-glycans and branched n-pentyl glycosides (nano-, octa-, penta-mannosides) that lack the chitobiose moiety at the reducing end of the oligosaccharide structure, as well as the linear tri- and dimannose. In addition, some glycans containing Galβ1-4GlcNAc and Galβ1-3(Fucα1-4)GlcNAc structural motifs, were found within the carbohydrate set that interacted with CV-N (Figure 3(b)), but these appear not to bind to GzCVNH.
Figure 3. Glycan array screening of GzCVNH (a) and CV-N (b).
Six replicates were performed, the highest and lowest points were discarded, and the displayed results represent the relative fluorescence unit (RFU) average of the four remaining determinations. A list of all glycans present on the arrays can be found at: http://www.functionalglycomics.org/static/index.shtml. Interacting sugars are depicted schematically and the individual sugar units in the structures of the oligosaccharides are color coded according to their linkage pattern.
NMR Titration
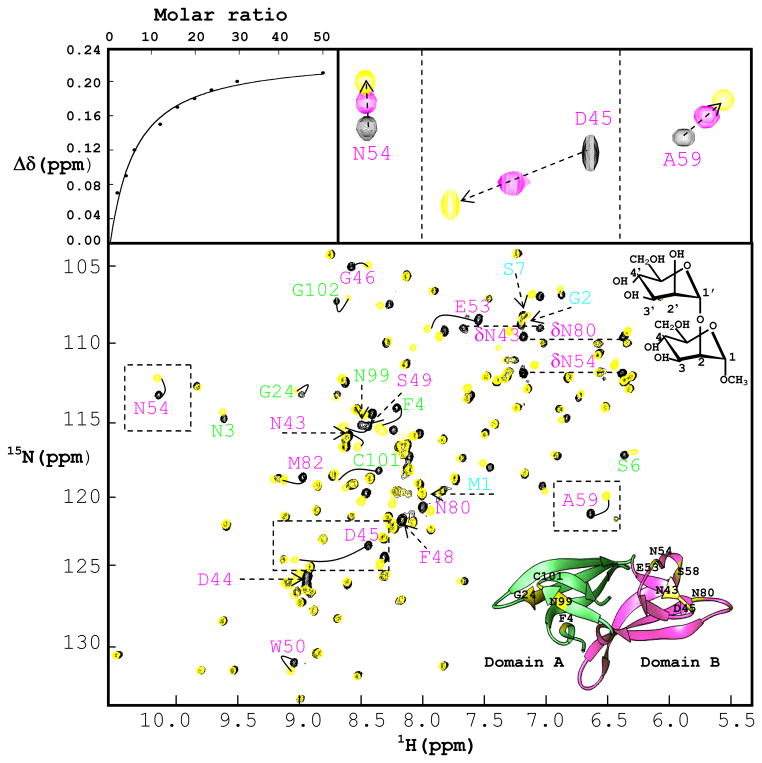
Subsequent to the array screening, direct NMR titration experiments were carried out with Manα(1-2)Manα, the smallest epitope of the branched high mannoses that was identified in the array screening. Carbohydrate binding sites on GzCVNH were mapped by following chemical shift perturbations in the 1H-15N HSQC spectrum of 15N uniformly labeled protein as a function of ligand addition. Titrations were carried out at 150 μM protein concentration up to a final concentration of 7.5 mM Manα(1-2)Manα (~ 50 fold molar excess), ensuring that low micromolar interactions could be detected. A superposition of the 1H-15N HSQC spectra of 150μM GzCVNH in the absence (black contours) and presence of 50 molar equivalents of Manα(1-2)Manα (yellow contours) is displayed in Figure 4, with an expanded view for three selected regions shown in the top right panels. The binding isotherm derived from the chemical shift changes is depicted in the top left panel. The location of residues whose amide resonances were perturbed, Asn43, Asp44, Asp45, Gly46, Glu53, Asn54, Phe55, Ala59, Asn80, Met82, as well as the Asn43, Asn54, Asn 80 side chains, clearly delineate the binding site in domain B (Figure 4(b)). The degree of perturbation is largest for Asp45, located on the first strand of the β4-β5 hairpin in domain B. The amide resonance of Asp45 is in intermediate exchange, while all other resonances exhibit fast exchange on the chemical shift scale.
Figure 4. Interaction of GzCVNH and CV-N with Manα(1-2)Manα.
Superposition of 1H-15N HSQC NMR spectra of GzCVNH in the absence (black) and presence of 50 molar equivalents of Manα(1-2)Manα (yellow). The identity of the perturbed amide NH resonances in the spectra clearly demonstrates the presence of a sugar binding site in domain B (all resonances arising from residues in domain B and A are labeled in pink and green, respectively). The top left panel displays the titration curve for Ala 59 (chemical shift difference versus ligand/protein molar ratio) with the chemical shift difference defined as: Δδ = [(ΔδHN)2 + (ΔδN × 0.17)2]1/2, where ΔδHN and ΔδN are the observed chemical shift changes for 1H and 15N, respectively. The top right panel displays expanded regions for three selected resonances and 3 titration points (0, 6 and 50 sugar/protein equivalents; these resonances are boxed in the 1H-15N HSQC spectra). A ribbon representation of the GzCVNH structure is shown at the bottom right in the superimposed spectra. Domains A and B are colored in green and pink, respectively, and the positions of amino acids involved in mannose binding are indicated in yellow.
The equilibrium dissociation constant (Kd) for Manα(1-2)Manα binding was determined from the titration shifts (chemical shift Δδ (ppm) versus the ratio of ligand to protein concentration) using several resonances. Analysis of the binding isotherm for Manα(1-2)Manα binding to the site on domain B yields a Kd value of 768 ± 60 μM. This value is very similar to the one determined for Manα(1-2)Manα binding to site A on CV-N (757 ± 80 μM).11 In order to probe whether any dimannose binding to domain A of GzCVNH could be detected, an even larger excess of Manα(1-2)Manα was used. Only at very high sugar/protein molar ratios (20-50 fold molar excess), some resonances of domain A residues were affected. However, the magnitude of chemical shifts changes was very small, suggesting that the affinity for Manα(1-2)Manα is extremely low. Resonances that were affected at the highest sugar concentration belonged to amino acids Phe4, Asn99 and Cys101. These residues occupy equivalent locations to residues in the sugar binding site in domain A of CV-N.
Discussion
The high-resolution solution NMR structure of GzCVNH reported here allows for a detailed comparison with the structure of the original, class-defining member of the CVNH family, CV-N. Despite a significant degree of sequence conservation and high structural similarity between GzCVNH and CV-N, several notable differences exist. The four cysteine residues in CV-N that form two disulfide bridges, and which were initially thought to be important for structural integrity and activity of CV-N,36 are not present. This finding complements results for other CVNH homologues that also contain varying numbers of cysteines.6 On the other hand , the lower thermal stability (Tm=48°C; data not shown) of GzCVNH, compared to wild type CV-N (Tm=61.3°C), may be partially caused by the absence of the disulfide bonds.
It is interesting to note that unlike CV-N which can exist either as a monomer5 or domain-swapped dimer,37,38 domain-swapped dimer structures for any CVNH member, including GzCVNH, have not been observed to date. In solution, the domain-swapped dimeric form of wild type CV-N is a trapped kinetic intermediate,38 while in the crystal, wild type CV-N is always found as a domain-swapped dimer.37,39 Amino acid composition of the hinge-loop region (Trp49-Phe54) is one of the determinants in domain swapping, and significant sequence variability is observed in the equivalent region of GzCVNH (Trp50-Phe55), with Gly52 taking the place of Pro51. Interestingly, the equivalent P51G substitution in CV-N does not prevent domain swapping and this mutant of CV-N still is capable of domain swapping, although it renders both monomeric and dimeric forms significantly more stable than wild type.38 The reverse mutation in GzCVNH, namely changing Gly52 to Pro interfered with correct folding of the protein, preventing the structural characterization of this mutant (data not shown). This result indicates the importance of the precise sequence context for structural stability, even for closely related homologous proteins.
The glycan array screening data revealed similar carbohydrate binding profiles for GzCVNH and CV-N, with Manα(1-2)Manα linked sugars as the recognition unit in a variety of branched oligomannoses. However, no substantive anti-HIV activity was detected for GzCVNH, up to a concentration of ~1 μM, when tested in single-cycle HIV-1 infection assays using Hela-P4 cells (data not shown). This can be rationalized by the fact that GzCVNH contains only a single, weak sugar binding site on domain B. When comparing GzCVNH with other CVNH members, only CrCVNH was found to exhibit two carbohydrate binding sites, one on domain A and one on domain B. In TbCVNH and NcCVNH only one binding site is present, on domain A and domain B, respectively. Some, but very little, anti-HIV activity was detected for CrCVNH; albeit the fact that a ~1000-fold higher protein concentration was necessary to detect any activity, supports the notion that the correct disposition of sugar binding sites as well as optimal affinities for the dimannose and trimannose units on the D1 and D3 arms of high mannose sugars are necessary for anti-HIV activity.6 We and others10 previously demonstrated that multi-site and multi-valent interactions between the protein and the high-mannose glycans on gp120 are necessary of CV-N’s HIV-inactivating activity,12 therefore a clear rational for the observed lack of anti-HIV activity for GzCVNH exists.
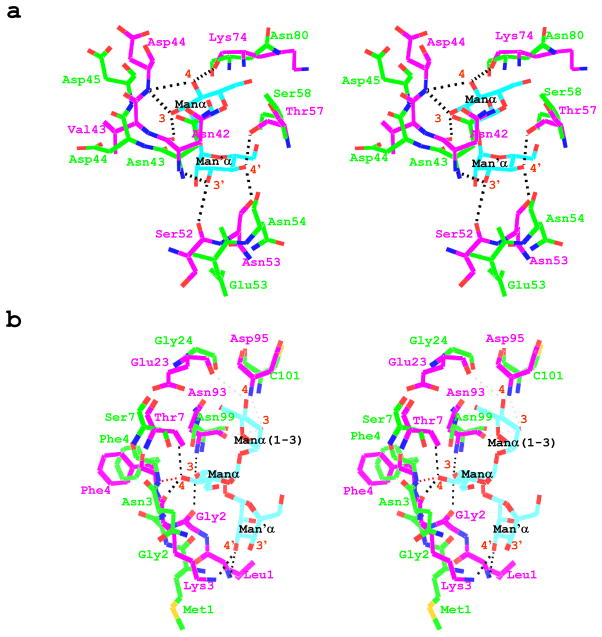
Comparison of the sequences and structures of GzCVNH and CV-N for the sugar binding regions revealed that most of residues involved in carbohydrate binding in CV-N are not the same in GzCVNH. In domain A, two more side chains are different than in domain B. Inspection of the crystal structure of a mutant CV-N protein, in which the carbohydrate binding site in domain A was abolished, complexed with a dimannose ligand, P51G-m4-CVN: Manα(1-2)Manα (PDB accession code 2RDK), revealed that in the binding site on domain B, four of the sugar hydroxyl groups simultaneously function as donor and acceptor groups, resulting in eight hydrogen bonds.10,40 These H-bonds are equally distributed between 3, 4, 3’ and 4’-hydroxyl groups, each forming a bifurcated hydrogen bond. These hydroxyls act as donors to the main chain oxygen atoms of Asn42, Ser52, Asn53 and Lys74, and hydrogen bond acceptors for the main chain nitrogen atoms of the Asn42, Asp44, and the side chain Oγ of Thr57, respectively (Figure 5(a)). In the case of GzCVNH, three of the corresponding residues in domain B are conserved (Asn42/Asn43, Asp44/Asp45 and Asn53/Asn54), one is conservatively substituted (Thr57/Ser60), while two are quite different (Ser52/Glu53, Lys74/Asn80) (Figure 4(d)). This partial sequence conservation in equivalent structural positions explains why GzCVNH is able to bind to Manα(1-2)Manα linked sugars in domain B. Since seven of the H-bonds between Manα(1-2)Manα and CV-N involve main-chain atoms and 3’ and 4’-hydroxyl groups on the sugars are important for selectivity and high-affinity binding,41 one could argue that a similar H-bonding network could be formed in the interaction of dimannose or oligomannose with GzCVNH in domain B. However, subtle structural changes may modulate these interactions, causing a substantial drop in affinity.
Figure 5. Superposition of the CV-N and GzCVNH structures of the carbohydrate binding regions in domains A and B.
(a) Stereo view representation of the superposition of GzCVNH solution NMR and the P51G-m4-CVN:dimannose complex (PDB accession code 2RDK) crystal structure, depicting the H-bonding network in domain B. Hydrogen bonds are indicated by dashed lines. (b) Stereo view representation of the superposition of the GzCVNH solution NMR structure and the wild type CV-N:Man-9 complex (PDB accession code 3GXZ) crystal structure, illustrating residues involved in protein-sugar interaction in domain A. Hydrogen bonds in the CV-N complex are indicated by dashed lines in black and a potential hydrogen bond at the N-terminus of GzCVNH is indicated by a dashed red line. The hydrogen atoms of the GzCVNH NMR solution structure were removed for clarity.
The extremely weak and virtually absent second carbohydrate binding site on domain A of GzCVNH, that is only detectable in the presence of a massive excess of sugar, can be explained by the more pronounced structural differences associated with the amino acid composition in the binding site on domain A. From the crystal structures of wild type CV-N bound to Man-942 (PDB accession code 3GXZ) residues of domain A that form H-bonds with the trimannose on the D1 arm of Man-9 can be delineated. These are Leu1, Gly2, Lys3, Thr7, Glu23, Asn93 and Asp95. Only two of these residues are conserved in GzCVNH (Gly2/Gly2 and Asn93/Asn99), while two are conservatively substituted (Lys3/Asn3, Thr7/Ser7), and three are very different (Leu1/Met1, Glu23/Gly24, Asp95/Cys101). In this structure, three of the terminal Manα(1- 2)Manα hydroxyl groups are involved in six hydrogen bonds.42 These H-bonds involve the 3, 4, and 3’-hydroxyl groups (Figure 5(b), dashed black lines), with the 3 and 4 hydroxyls acting as donors to the main chain oxygen atoms of Gly2 and Lys3, and hydrogen bond acceptors for the main chain nitrogen atoms of Asn93, and the side chains Oγ of Thr7. A bifurcated hydrogen bond is formed between the 3’ hydroxyl of the sugar and the main chain nitrogen of Leu1 and the nitrogen side chain Nζ of Lys3. Some weak hydrogen bonds (distance ~ 3.2 Å) may also be present between the 3 and 4 hydroxyls of the Manα(1-3) unit of the D1 arm and residues Glu23, Asn93, and Asn95 (Figure 5(b), dashed gray lines). A superposition of the sugar binding region on domain A reveals clear differences in the spatial disposition of the N-terminal residues (Met1-Asn3) in GzCVNH, when compared with the Man-9/wild type CV-N complex crystal structure. No chemical shift changes were observed for the Met1, Gly2 and Ser7 in the NMR titration experiments with Manα(1-2)Manα (Figure 4 bottom panel, cyan), even when a large excess of sugar was present, and only very small chemical shift changes were observed for Phe4, Asn99 and C101. Since it is impossible to ascertain whether these changes are saturable, one cannot determine with certainty that a binding site is present. Such small changes could also be caused by non-specific binding. A modeled structural depiction of a possible hydrogen bonding network for GzCVNH suggests that neither of the main chain amides of Met1, Gly2 or the side chain of Asn3, can engage in H-bonding. Only the main chain nitrogen of Phe4 is potentially capable to hydrogen-bond with the 4 hydroxyl of Manα(1-2)Manα (Figure 5(b), red dashed line). It therefore seems reasonable to conclude that the structural difference at the protein’s N-terminus is responsible for the extremely weak binding of Manα(1-2)Manα to domain A of GzCVNH.
In summary, although structurally similar to CV-N, GzCVNH, like several other CVNH family members with single carbohydrate binding sites that interact weakly with mannose sugars, does not possess any anti-HIV activity. This finding lends further support to our previous assertion that multi-site and multi-valent interactions are necessary for potent HIV-inactivation.
Acknowledgments
We thank Annie Aniana for useful discussions, In-Ja Byeon for expert help with NMR experiments and Chris Aiken and Jeremy Rose for carrying out HIV activity assays. The Protein-Glycan Interaction Core (Core H) of the Consortium for Functional Glycomics (supported by NIH grant GM62116) is gratefully acknowledged for glycan array analyses. This work was supported by National Institutes of Health Grant GM080642 (to A.M.G)
References
- 1.Sharon N, Lis H. Lectins: cell-agglutinating and sugar-specific proteins. Science. 1972;177(53):949–959. doi: 10.1126/science.177.4053.949. [DOI] [PubMed] [Google Scholar]
- 2.Sharon N. Bacterial lectins, cell-cell recognition and infectious disease. FEBS Lett. 1987;217(2):145–157. doi: 10.1016/0014-5793(87)80654-3. [DOI] [PubMed] [Google Scholar]
- 3.Swamy BM, Bhat AG, Hegde GV, Naik RS, Kulkarni S, Inamdar SR. Immunolocalization and functional role of Sclerotium rolfsii lectin in development of fungus by interaction with its endogenous receptor. Glycobiology. 2004;14(11):951–957. doi: 10.1093/glycob/cwh130. [DOI] [PubMed] [Google Scholar]
- 4.Percudani R, Montanini B, Ottonello S. The anti-HIV cyanovirin-N domain is evolutionarily conserved and occurs as a protein module in eukaryotes. Proteins. 2005;60(4):670–678. doi: 10.1002/prot.20543. [DOI] [PubMed] [Google Scholar]
- 5.Bewley CA, Gustafson KR, Boyd MR, Covell DG, Bax A, Clore GM, Gronenborn AM. Solution structure of cyanovirin-N, a potent HIV-inactivating protein. Nat Struct Biol. 1998;5(7):571–578. doi: 10.1038/828. [DOI] [PubMed] [Google Scholar]
- 6.Koharudin LM, Viscomi AR, Jee JG, Ottonello S, Gronenborn AM. The evolutionarily conserved family of cyanovirin-N homologs: structures and carbohydrate specificity. Structure. 2008;16(4):570–584. doi: 10.1016/j.str.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 7.Boyd MR, Gustafson KR, McMahon JB, Shoemaker RH, O'Keefe BR, Mori T, Gulakowski RJ, Wu L, Rivera MI, Laurencot CM, Currens MJ, Cardellina JH, 2nd, Buckheit RW, Jr, Nara PL, Pannell LK, Sowder RC, 2nd, Henderson LE. Discovery of cyanovirin-N, a novel human immunodeficiency virus-inactivating protein that binds viral surface envelope glycoprotein gp120: potential applications to microbicide development. Antimicrob Agents Chemother. 1997;41(7):1521–1530. doi: 10.1128/aac.41.7.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Keefe BR, Smee DF, Turpin JA, Saucedo CJ, Gustafson KR, Mori T, Blakeslee D, Buckheit R, Boyd MR. Potent anti-influenza activity of cyanovirin-N and interactions with viral hemagglutinin. Antimicrob Agents Chemother. 2003;47(8):2518–2525. doi: 10.1128/AAC.47.8.2518-2525.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barrientos LG, Matei E, Lasala F, Delgado R, Gronenborn AM. Dissecting carbohydrate-Cyanovirin-N binding by structure-guided mutagenesis: functional implications for viral entry inhibition. Protein Eng Des Sel. 2006;19(12):525–535. doi: 10.1093/protein/gzl040. [DOI] [PubMed] [Google Scholar]
- 10.Fromme R, Katiliene Z, Giomarelli B, Bogani F, Mc Mahon J, Mori T, Fromme P, Ghirlanda G. A monovalent mutant of cyanovirin-N provides insight into the role of multiple interactions with gp120 for antiviral activity. Biochemistry. 2007;46(32):9199–9207. doi: 10.1021/bi700666m. [DOI] [PubMed] [Google Scholar]
- 11.Matei E, Furey W, Gronenborn AM. Solution and crystal structures of a sugar binding site mutant of cyanovirin-N: no evidence of domain swapping. Structure. 2008;16(8):1183–1194. doi: 10.1016/j.str.2008.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matei E, Zheng A, Furey W, Rose J, Aiken C, Gronenborn AM. Anti-HIV activity of defective cyanovirin-N mutants is restored by dimerization. J Biol Chem. 285(17):13057–13065. doi: 10.1074/jbc.M109.094938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bai GH, Chen LF, Shaner GE. In: Breeding for resistance to Fusarium head blight of wheat in China. Leonard KJ, Bushnell WR, editors. APS Press; St Paul, MN: 2003. pp. 296–317. [Google Scholar]
- 14.Bennett JW, Klich M. Mycotoxins. Clin Microbiol Rev. 2003;16(3):497–516. doi: 10.1128/CMR.16.3.497-516.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gale LR, Bryant JD, Calvo S, Giese H, Katan T, O'Donnell K, Suga H, Taga M, Usgaard TR, Ward TJ, Kistler HC. Chromosome complement of the fungal plant pathogen Fusarium graminearum based on genetic and physical mapping and cytological observations. Genetics. 2005;171(3):985–1001. doi: 10.1534/genetics.105.044842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 17.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 18.Bax A, Grzesiek S. Methodological Advances in Protein NMR. Acc Chemical Research. 1993;26:131–138. [Google Scholar]
- 19.Fesik SW, Zuiderweg ER. Heteronuclear three-dimensional NMR spectroscopy of isotopically labelled biological macromolecules. Q Rev Biophys. 1990;23(2):97–131. doi: 10.1017/s0033583500005515. [DOI] [PubMed] [Google Scholar]
- 20.Grzesiek S, Vuister GW, Bax A. A simple and sensitive experiment for measurement of JCC couplings between backbone carbonyl and methyl carbons in isotopically enriched proteins. J Biomol NMR. 1993;3(4):487–493. doi: 10.1007/BF00176014. [DOI] [PubMed] [Google Scholar]
- 21.Logan TM, Olejniczak ET, Xu RX, Fesik SW. A general method for assigning NMR spectra of denatured proteins using 3D HC(CO)NH-TOCSY triple resonance experiments. J Biomol NMR. 1993;3(2):225–231. doi: 10.1007/BF00178264. [DOI] [PubMed] [Google Scholar]
- 22.Sattler M, Maurer M, Schleucher J, Griesinger C. A Simultaneous 15N, 1H-HSQC and 13C, 1H-HSQC with sensitivity enhancement and a heteronuclear gradient-echo. J Biomol NMR. 1995;5:97–102. doi: 10.1007/BF00227475. [DOI] [PubMed] [Google Scholar]
- 23.Herrmann T, Guntert P, Wüthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol. 2002;319(1):209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 24.Guntert P, Mumenthaler C, Wuthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol. 1997;273(1):283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 25.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13(3):289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 26.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 27.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK - a program to check the stereochemical quality of protein structures. J Appl Crystallog. 1993;26:83–291. [Google Scholar]
- 28.Bhattacharya A, Wunderlich Z, Monleon D, Tejero R, Montelione GT. Assessing model accuracy using the homology modeling automatically software. Proteins. 2007;66:778–795. doi: 10.1002/prot.21466. [DOI] [PubMed] [Google Scholar]
- 29.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18(15):2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 30.Wuthrich K. NMR of Proteins and Nucleic Acids. Wiley; New York: 1986. [Google Scholar]
- 31.Yang R, Aiken C. A mutation in alpha helix 3 of CA renders humanimmunodeficiency virus type 1 cyclosporin A resistant and dependent: rescue by a second-site substitution in a distal region of CA. " J Virol. 2007;81(8):3749–3756. doi: 10.1128/JVI.02634-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bewley CA, Otero-Quintero S. The potent anti-HIV protein cyanovirin-N contains two novel carbohydrate binding sites that selectively bind to Man(8) D1D3 and Man(9) with nanomolar affinity: implications for binding to the HIV envelope protein gp120. J Am Chem Soc. 2001;123(17):3892–3902. doi: 10.1021/ja004040e. [DOI] [PubMed] [Google Scholar]
- 33.Shenoy SR, Barrientos LG, Ratner DM, O'Keefe BR, Seeberger PH, Gronenborn AM, Boyd MR. Multisite and multivalent binding between cyanovirin-N and branched oligomannosides: calorimetric and NMR characterization. Chem Biol. 2002;9(10):1109–1118. doi: 10.1016/s1074-5521(02)00237-5. [DOI] [PubMed] [Google Scholar]
- 34.Ramachandran GN, Sasisekharan V. Conformation of polypeptides and proteins. Adv Protein Chem. 1968;23:283–438. doi: 10.1016/s0065-3233(08)60402-7. [DOI] [PubMed] [Google Scholar]
- 35.Thoronton JM. Disulphide Bridges in Globular Proteins. J Mol Biol. 1981;151:261–287. doi: 10.1016/0022-2836(81)90515-5. [DOI] [PubMed] [Google Scholar]
- 36.Gustafson KR, Sowder RC, 2nd, Henderson LE, Cardellina JH, 2nd, McMahon JB, Rajamani U, Pannell LK, Boyd MR. Isolation, primary sequence determination, and disulfide bond structure of cyanovirin-N, an anti-HIV (human immunodeficiency virus) protein from the cyanobacterium Nostoc ellipsosporum. Biochem Biophys Res Commun. 1997;238(1):223–228. doi: 10.1006/bbrc.1997.7203. [DOI] [PubMed] [Google Scholar]
- 37.Yang F, Bewley CA, Louis JM, Gustafson KR, Boyd MR, Gronenborn AM, Clore GM, Wlodawer A. Crystal structure of cyanovirin-N, a potent HIV-inactivating protein, shows unexpected domain swapping. J Mol Biol. 1999;288(3):403–412. doi: 10.1006/jmbi.1999.2693. [DOI] [PubMed] [Google Scholar]
- 38.Barrientos LG, Louis JM, Botos I, Mori T, Han Z, O'Keefe BR, Boyd MR, Wlodawer A, Gronenborn AM. The domain-swapped dimer of cyanovirin-N is in a metastable folded state: reconciliation of X-ray and NMR structures. Structure. 2002;10(5):673–686. doi: 10.1016/s0969-2126(02)00758-x. [DOI] [PubMed] [Google Scholar]
- 39.Botos I, Mori T, Cartner LK, Boyd MR, Wlodawer A. Domain-swapped structure of a mutant of cyanovirin-N. Biochem Biophys Res Commun. 2002;294(1):184–190. doi: 10.1016/S0006-291X(02)00455-2. [DOI] [PubMed] [Google Scholar]
- 40.Fromme R, Katiliene Z, Fromme P, Ghirlanda G. Conformational gating of dimannose binding to the antiviral protein cyanovirin revealed from the crystal structure at 1.35 A resolution. Protein Sci. 2008;17(5):939–944. doi: 10.1110/ps.083472808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sandstrom C, Hakkarainen B, Matei E, Glinchert A, Lahmann M, Oscarson S, Kenne L, Gronenborn AM. Atomic mapping of the sugar interactions in one-site and two-site mutants of cyanovirin-N by NMR spectroscopy. Biochemistry. 2008;47(12):3625–3635. doi: 10.1021/bi702200m. [DOI] [PubMed] [Google Scholar]
- 42.Botos I, O'Keefe BR, Shenoy SR, Cartner LK, Ratner DM, Seeberger PH, Boyd MR, Wlodawer A. Structures of the complexes of a potent anti-HIV protein cyanovirin-N and high mannose oligosaccharides. J Biol Chem. 2002;277(37):34336–34342. doi: 10.1074/jbc.M205909200. [DOI] [PubMed] [Google Scholar]