Abstract
We investigated the effects of growth hormone-releasing hormone (GHRH) antagonists, JV-1-65 and JV-1-63, and bombesin/gastrin-releasing peptide (BN/GRP) antagonist RC-3940-II on DMS-153 human small cell lung carcinoma xenografted into nude mice. Treatment with 10 μg/day JV-1-65 or RC-3940-II decreased tumor volume by 28% (P < 0.05) and 77% (P < 0.01), respectively, after 42 days compared with controls. Combination of JV-1-65 and RC-3940-II induced the greatest inhibition of tumor proliferation (95%; P < 0.01), suggesting a synergism. Western blotting showed that the antitumor effects of these antagonists were associated with inhibition of the expression of the mutant tumor suppressor protein p53 (Tp53). Mutation was detected by sequence analysis of the p53 gene at codon 155: ACC [Thr] → CCC [Pro]. Combination of JV-1-65 and RC-3940-II decreased the levels of mutant p53 protein by 42% (P < 0.01) compared with controls. JV-1-65, JV-1-63, and RC-3940-II, given singly, reduced mutant p53 protein expression by 18-24% (P < 0.05). Serum insulin-like growth factor (IGF)-I levels were diminished in animals receiving GHRH antagonists. mRNA levels for IGF-II, IGF receptor-I, GRP receptor, and EGF receptor in tumors were significantly decreased by combined treatment with JV-1-65 and RC-3940-II. DMS-153 tumors expressed mRNAs for GHRH and GHRH receptor splice variants 1 and 2, suggesting that GHRH could be an autocrine growth factor. Proliferation of DMS-153 cells in vitro was stimulated by GRP and IGF-II and inhibited by JV-1-65. This study indicates that GHRH antagonists and BN/GRP antagonist inhibit the growth of DMS-153 small cell lung carcinoma concomitantly with the expression of mutant Tp53, which might uncouple the signal transduction pathways for cell growth stimulation.
Neoplastic cells are considered to be under the control of specific growth factors and neuropeptides that act by endocrine or autocrine/paracrine mechanisms to stimulate their proliferation and decrease apoptosis. Thus, a strategy for the treatment of cancer could be based on compounds that block or down-regulate components of the stimulatory pathways that support tumor growth (1, 2). Antagonists of growth hormone-releasing hormone (GHRH), which belong to this class of antitumor agents, have been shown to inhibit the proliferation of a broad range of human experimental cancers (1, 2). GHRH antagonists inhibit tumor growth in vivo by suppressing the pituitary GH/hepatic insulin-like growth factor I (IGF-I) axis or by direct actions on cancer cells (1, 2). The direct effects of GHRH antagonists are thought to be mediated by splice variants (SV) of GHRH receptors on the cancer cells and may involve an inhibitory action on the synthesis of tumoral autocrine/paracrine IGF-I and/or IGF-II. The involvement of IGF-I in tumorigenesis is well documented (2-4). IGF-II, the synthesis of which is GH-independent, acts on the same receptor as IGF-I (IGF receptor type I, IGFR-I) and is implicated in malignant transformation, tumor progression, and metastasis (2-5). Human small cell lung cancer (SCLC) and non-SCLC cell lines secrete and respond to IGF-I and -II and express IGF-I and -II genes and IGFR-I (1, 2, 6). GHRH antagonists strongly inhibit growth of H-69 SCLC and H-157 non-SCLC tumors in nude mice and decrease the levels of IGF-I in serum and liver tissue (6). In the H-157 non-SCLC model, the tumoral concentrations of both IGF-I and IGF-II are decreased after treatment with GHRH antagonist (6). However, in H-69 SCLC, the synthesis of tumoral IGF-I or IGF-II is not affected by GHRH antagonists, and the antitumor effects of the GHRH antagonists are apparently exerted through the blocking of the action of local GHRH, which serves as an autocrine growth factor, and not by interfering with the tumoral IGF system (7, 8).
It has also been shown that gastrin-releasing peptide (GRP), a member of the bombesin (BN)-like peptide family, is an autocrine growth factor for SCLC and also stimulates cell proliferation in other neoplasms (2, 9, 10). Several subtypes of receptors for BN/GRP are present in various SCLC lines, and BN-like peptides can promote their proliferation (2, 9-13). We showed that BN/GRP antagonist RC-3095 inhibits growth of H-69 SCLC, but not of H-157 non-SCLC xenografted into nude mice (6). Subsequently, we demonstrated that antagonist RC-3940-II possesses greater antitumor activity on H-69 SCLC than RC-3095 (13). The inhibition of SCLC growth by RC-3095 and RC-3940-II was associated with a decrease in the levels and mRNA expression of epidermal growth factor (EGF) receptors (EGFR) (13).
The tumor suppressor gene p53 is mutated in about half of all human cancers. The p53 protein modulates cellular functions, such as gene transcription, DNA synthesis and repair, cell cycle arrest, and apoptosis. Mutations in the p53 gene can abrogate these functions and may lead to genetic instability and progression to cancer (14). Many studies have shown an abnormal expression of p53 protein by immunohistochemistry in 40-70% of SCLC, suggesting that p53 abnormalities may play a critical role in lung cancer pathogenesis, particularly in SCLC (15). It was also reported that aberrant p53 is associated with a significantly shorter survival time in patients with lung cancer (16). Because the relevance of p53 in the modulation of tumor responsiveness depends on the molecular and biological context, p53 mutations can be a biomarker of carcinogen effect and may help in identifying individuals at increased risk of lung cancer (17, 18).
Because antagonists of GHRH and BN/GRP antagonists have distinct antiproliferative actions, we investigated whether the administration of GHRH antagonists, JV-1-63 and JV-1-65, singly or in combination with BN/GRP antagonist RC-3940-II could enhance their inhibitory effects on the growth of DMS-153 SCLC in vivo. We also examined effects of GHRH antagonists and BN/GRP antagonists on p53 expression in an attempt to evaluate their mechanism of action.
Materials and Methods
Peptides. The BN/GRP antagonist [Hca6, Leu-13Ψ (CH2N)-Tac14]BN (6-14) (RC-3940-II) (2, 19) was provided by Zentaris (Frankfurt on Main, Germany). The GHRH antagonists JV-1-63 and JV-1-65 were recently synthesized by solid-phase methods (1, 20). The chemical structure of JV-1-63 is [PhAc-Tyr-1, [scapd-Arg-2, Phe(4-Cl)6, Har9, Amp10, Abu15, Nle-27, D-Arg-28, Har29]hGHRH(1-29)NH2, and that of JV-1-65 is [PhAc-Tyr-1, D-Arg-2, Phe(4-Cl)6, Amp9, Tyr(Me)10, Abu15, Nle-27, D-Arg-28, Har29]hGHRH(1-29)NH2, where PhAc is phenylacetyl, Phe(4-Cl) is 4-chlorophenylalanine, Amp is 4-amidinophenylalanine, Abu is α-aminobutyric acid, Nle is norleucine, and Har is homoarginine. For injections, peptides were dissolved in 0.1% dimethyl sulfoxide in sterile aqueous 10% propylene-glycol.
Cell Line and Animals. Human SCLC cell line DMS-153, obtained from American Type Culture Collection, was cultured as described (21). Athymic nude mice were obtained from the National Cancer Institute (Frederick Cancer Research and Development Center, Frederick, MD), and were housed and fed as described (11). Experiments were carried out according to institutional guidelines for animal care.
In Vivo Studies. Tumor xenografts were initiated by s.c. injection of 107 DMS-153 SCLC cells into the male nude mice. The resulting tumors were dissected, minced, and transplanted s.c. by trocar needle into experimental animals. Mice were divided in groups of six to eight animals and received the following treatment for 6 weeks: group 1 (control), vehicle solution; group 2, BN/GRP antagonist RC-3940-II; group 3, GHRH antagonist JV-1-65; group 4, RC-3940-II and JV-1-65; group 5, GHRH antagonist JV-1-63, all at a dose of 10 μg/day per animal s.c. Tumor volumes and body weights were measured twice per week. Tumor volume and tumor volume doubling time between the start and the end of the treatment was calculated as described (11, 13). At the end of treatment, mice were killed, trunk blood was collected for analyses, and various organs were removed and weighed. Tumors were excised, weighed, and stored at -70°C.
Cell Proliferation Assay. DMS-153 cells were seeded at low concentration in 96-well plates, and after 24 h, the culture medium was replaced with medium Waymouth MB containing 3% FBS and the test compounds. After 96 h, cell proliferation was determined by a tetrazolium assay as described (11).
RNA Extraction and RT-PCR. Total RNA was extracted from tumors by using the Tri-Reagent (Sigma) following the manufacturer's instructions. For analysis of GHRH and its receptor SVs, polyadenylated [poly(A)] RNA was isolated as described (22). Amplification of cDNA transcripts was made by using gene-specific primers for human GHRH, GHRH-R, IGF-I, IGF-II, IGFR-I, GRP receptor (GRPR), EGFR, and β-actin as described (7, 8, 11, 13, 22-24). Primers used for SVs of GHRH-R were 5′-CCTACTGCCCTTAGGATGCTGG-3′ (sense) and 5′-CCTTGCTCCTCCAGAGCATGG-3′ (antisense) for the first PCR, and 5′-GCACCTTTGAAGCCAGAGAAGG-3′ (sense) and 5′-CGTGCCAGTGAAGAGCACGG-3′ (antisense) for the second PCR. PCR amplifications of the cDNAs were performed as described (7, 8, 11, 13, 23). PCR products were electrophoresed on a 1.8% agarose gel and stained with ethidium bromide. cDNA Sequencing of p53. Two microliters of cDNA was amplified by RT-PCR in 20 μl of mixture containing 1× PCR buffer, 1.5 mM MgCl2, 200 μM each dNTP, 0.3 μM each primer, and 1 unit per 100 μl AmpliTaq DNA polymerase. PCR consisted of 1 cycle at 94°C for 3 min followed by 30 cycles of 94°C for 45 sec, 60°C for 45 sec, and 72°C for 1 min, and a final extension at 72°C for 7 min. The p53-specific primers were extracted from database (http://www-genome.wi.mit.edu) and were 5′-CAG ACT GCC TTC CGG GTC AC-3′ (sense) and 5′-CCA CAC GCA AAT TTC CTT CCA-3′ (antisense) for exons 2-6 and 5′-TGG AAG GAA ATT TGC GTC GTG TGG-3′ (sense) and 5′-GGC GGG AGG TAG ACT GAC CCC-3′ (antisense) for the exons 6-11 (Invitrogen Life Technologies, Carlsbad, CA). After PCR amplification, PCR products were treated with exonuclease and shrimp alkaline phosphatase (25) and sequenced by using an ABI Prism 3100 Genetic Analyzer (Perkin-Elmer). The sequence was compared with the wild-type p53 sequence, which was extracted from the p53 GenBank database (www.ncbi.nlm.nih.gov; accession no. AF136270, gi4732144).
Western Blotting Assays. Protein-matched samples were subjected to electrophoresis on SDS/8% PAGE and transferred to nitro-cellulose membranes as described (26). Membranes were incubated with the primary anti-p53 antibody and the secondary horseradish peroxidase-conjugated anti-goat IgG antibody (both from Santa Cruz Biotechnology, 1:1,000). Blots were visualized with the chemiluminescence detection system (Pierce). Bands corresponding to mutant p53 were analyzed by using the Kodak EDAS 290 imaging system with 1D IMAGE ANALYSIS software (Kodak).
RIA for IGF-I. IGF-I levels in serum samples were determined as described (8).
Statistical Analyses. Data are presented as means ± SE and were evaluated by one-way ANOVA and the Student-Newman-Keuls or Bonferroni tests.
Results
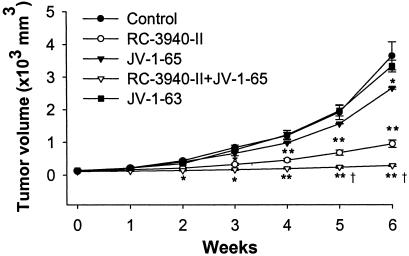
Effect of BN/GRP Antagonist and GHRH Antagonists on the Growth of DMS-153 SCLC Tumor. Nude mice bearing DMS-153 human SCLC tumors were treated with BN/GRP antagonist RC-3940-II, GHRH antagonists JV-1-65 and JV-1-63, or a combination of RC-3940-II and JV-1-65 (Fig. 1 and Table 1). After 42 days of treatment, we observed a 77% inhibition of the tumor volume (P < 0.01) in response to RC-3940-II and a 28% suppression (P < 0.05) after JV-1-65, compared with controls. Combination of RC-3940-II and JV-1-65 caused the greatest inhibition of the tumor volume (95%, P < 0.01 vs. control and P < 0.05 vs. RC-3940-II alone). GHRH antagonist JV-1-63 slightly inhibited tumor volume (10%; not significant), but significantly reduced tumor weight (Fig. 1 and Table 1). The weights of tumors were significantly (P < 0.05) decreased by 73% (P < 0.01) in response to RC-3940-II and by ≈30% after treatment with JV-1-65. The greatest inhibition of tumor weight occurred in animals that received RC-3940-II combined with JV-1-65 (87% reduction; P < 0.01). Combined treatment with both classes of analogs also extended significantly (P < 0.01) tumor doubling time to 31.8 ± 2.1 days from 8.3 ± 0.8 days for the control (Table 1), whereas RC-3940-II alone significantly (P < 0.05) increased tumor doubling time to 14.4 ± 1.0 days vs. control. At the end of the experiment, there were no significant differences in body weights among the groups (Table 1).
Fig. 1.
Tumor volumes in nude mice bearing DMS-153 human SCLC during treatment with the GHRH antagonists JV-1-65, JV-1-63, and BN/GRP antagonist RC-3940-II alone or in combination with JV-1-65, all at a dose of 10 μg/day per animal. Vertical bars indicate SEM. *, P < 0.05 vs. control; **, P < 0.01 vs. control; †, P < 0.05 vs. RC-3940-II.
Table 1. Effect of treatment with BN/GRP antagonist RC-3940-II and GHRH antagonists JV-1-63 and JV-1-65 on tumor volume and weight and tumor volume doubling time of human SCLC cell line DMS-153 xenografted into nude mice.
| Tumor volume, mm3
|
|||||
|---|---|---|---|---|---|
| Groups | Body weight, g | Tumor weight, g | Initial | Final | Tumor volume doubling time, days |
| Control | 33.0 ± 1.2 | 2.31 ± 0.25 | 112 ± 13 | 3,641 ± 431 | 8.3 ± 0.8 |
| RC-3940-II | 31.7 ± 1.3 | 0.63 ± 0.02** | 127 ± 10 | 949 ± 14** | 14.4 ± 1.0* |
| JV-1-65 | 33.4 ± 3.6 | 1.60 ± 0.19* | 132 ± 24 | 2,650 ± 30* | 9.7 ± 0.9 |
| RC-3940-II+JV-1-65 | 31.3 ± 4.8 | 0.31 ± 0.02**,† | 115 ± 13 | 286 ± 24**† | 31.8 ± 2.1**† |
| JV-1-63 | 31.7 ± 2.5 | 1.58 ± 0.11* | 138 ± 20 | 3,329 ± 180 | 9.1 ± 0.8 |
Values are mean ± SE. *, P < 0.05 vs. control. **, P < 0.01 vs. control. †, P < 0.05 vs. RC-3940-II.
Serum IGF-I Measurements. There were no significant differences in serum IGF-I levels after treatment with RC-3940-II (167.6 ± 9.9 ng/ml), compared with the controls (169.0 ± 9.9 ng/ml). However, groups of mice that received the GHRH antagonists showed significant decreases in the serum IGF-I levels that were as follows: JV-1-65 (117.9 ± 17.2 ng/ml), RC-3940-II+JV-1-65 (114.5 ± 21.9 ng/ml), and JV-1-63 (118.4 ± 12.5 ng/ml) (P < 0.05 in all cases).
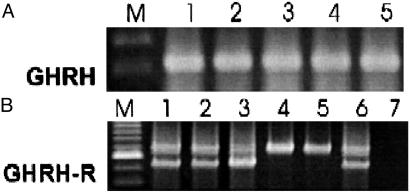
Expression of mRNA for GHRH and GHRH Receptors in DMS-153 SCLC. RT-PCR analyses for GHRH and GHRH receptors were performed on control tumors and those treated with GHRH and BN/GRP antagonists. Fig. 2A shows one representative tumor from each group. We found a PCR product of 322 bp, specific for GHRH in all of the DMS-153 SCLC tumor samples examined. No differences were found in the level of expression of GHRH mRNA in the treated groups compared with the control tumors (data not shown). The analyses for SV receptors of GHRH were also performed on individual RNA samples from each treatment group. PCR products of 720 bp and 566 bp, corresponding to SV1 and SV2 isoforms, were detected by nested PCR in DMS-153 tumors from the control and treated groups (Fig. 2B). No PCR products were amplified from the negative controls, ruling out the possibility of genomic DNA contamination. Amplification with β-actin-specific primers produced a product of 459 bp from all samples, confirming that there was no degradation of the RNA preparations (not shown).
Fig. 2.
Expression of mRNA for GHRH (A) and GHRH-SV receptors (B) in DMS-153 SCLC tumors. Poly(A) RNAs from individual tumor samples were subjected to RT-PCR analysis, electrophoresed on agarose gel, and stained with ethidium bromide. (A) RT-PCR analysis shows a product of 322 bp for GHRH. (B) PCR products of 720 bp for SV1 and 566 bp for SV2 were detected. Lane M, 100-bp DNA molecular marker; lane 1, untreated tumor; lane 2, tumor treated with RC-3940-II; lane 3, tumor treated with JV-1-65; lane 4, tumor treated with RC-3940-II and JV-1-65; lane 5, tumor treated with JV-1-63; lane 6, positive control from LNCaP human prostate cancer cells; lane 7, negative control.
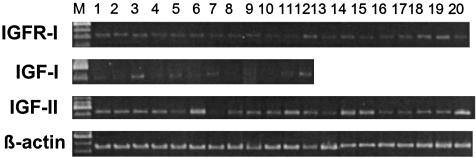
Expression of mRNA for IGF-I, IGF-II, and IGFR-I in DMS-153 SCLC. RT-PCR analyses demonstrated that DMS-153 tumors of control animals expressed mRNA for IGF-I (not shown) as well as for IGF-II and IGF receptors type I (Fig. 3). Amplification of cDNAs from tumor samples of control and treated animals yielded products of 538 bp and 447 bp corresponding to IGF-II and IGFR-I, respectively. Densitometric analyses of the RT-PCR products and normalization of levels according to β-actin values were performed to determine the effect of treatment with various antagonists on the expression of mRNAs for IGF-II, and IGFR-I (Table 2). In the case of IGF-I, this effect could not be established because of high intragroup variations in the levels of IGF-I mRNA. A tendency for the suppression of mRNA levels for IGF-II was noted after treatment with either the GHRH antagonists or BN/GRP antagonist alone, but this effect was only significant in the case of JV-1-63 (P < 0.01). JV-1-65 potentiated the effect of RC-3940-II and the greatest decrease in the mRNA levels for IGF-II of 73% (P < 0.01) occurred in tumors treated with a combination of RC-3940-II and JV-1-65 (Table 2). The expression of mRNA for IGFR-I was significantly suppressed (P < 0.05) after treatment with JV-1-65 combined with RC-3940-II (Table 2).
Fig. 3.
RT-PCR analysis of IGF-II and IGFR-I in samples of DMS-153 SCLC tumors. PCR products were of the expected sizes of 538 bp for IGF-II, 447 bp for IGFR-I, and 459 bp for β-actin. Lane M, 100-bp DNA molecular weight marker; lanes 1-5, untreated animals; lanes 6-9, animals treated with RC-3949-II; lanes 10-13, animals treated with JV-1-65; lanes 14-16, animals treated with RC-3940-II + JV-1-65; lanes 17-20, animals treated with JV-1-63.
Table 2. Effect of GHRH antagonists JV-1-65 and JV-1-63 and BN/GRP antagonist RC-3940-II on the mRNA expression for IGF-II, IGFR -I and -II, GRPR, and EGFR in DMS-153 SCLC.
| mRNA, % of control
|
|||||
|---|---|---|---|---|---|
| Control | RC-3940-II | JV-1-65 | RC-3940-II + JV-1-65 | JV-1-63 | |
| IGF-II | 100.0 ± 1.5 | 61.0 ± 19.5 | 78.0 ± 44.3 | 27.0 ± 2.0** | 42.7 ± 18.5** |
| IGFR-I | 100.0 ± 4.2 | 81.2 ± 9.2 | 62.2 ± 12.6 | 51.2 ± 16.1* | 117.7 ± 11.9 |
| GRPR | 100.0 ± 10.0 | 64.2 ± 2.0** | NI | 74.9 ± 3.8* | NI |
| EGFR | 100.0 ± 7.3 | 93.1 ± 6.5 | NI | 48.3 ± 4.8** | NI |
Values are mean ± SE. Results were quantified by densitometric analysis of three to six tumor samples from each group. Data were normalized to β-actin values and presented as percentage of control. NI, not investigated. *, P < 0.05 vs. control. **, P < 0.01 vs. control.
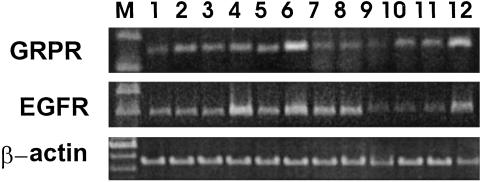
Expression of mRNA for EGFR and BN/GRPR in DMS-153 Tumors. The expression of mRNA for EGFR and GRPR in DMS-153 SCLC was also investigated (Fig. 4). Densitometric analyses revealed a 51.7% (P < 0.01) decrease in the EGFR mRNA levels after combined treatment with RC-3940-II and JV-1-65; mRNA levels for GRPR were significantly decreased by 35.8% (P < 0.01) and 25.1% (P < 0.05), respectively, after treatment with the BN/GRP antagonist alone or in combination with GHRH antagonist (Fig. 4 and Table 2).
Fig. 4.
Expression of mRNA for GRPR and EGFR in samples of DMS-153 SCLC. PCRs yielded products the size of 159 bp for GRPR, 400 bp for EGFR, and 459 bp for β-actin. Lane M, 100-bp DNA molecular mass marker; lanes 1-5, untreated animals; lanes 6-9, animals treated with RC-3940-II; lanes 10-12, animals treated with RC-3940-II + JV-1-65.
cDNA Sequencing Analysis. We analyzed p53 gene in DMS-153 SCLC by RT-PCR and automated sequencing and compared it with wild-type p53 sequence, which was extracted from the p53 database (www.ncbi.nlm.nih.gov) and supplemented from published reports. We sequenced the entire coding region of the p53 gene, from exons 2 to11 by using two overlapping p53-specific primer pairs (see Materials and Methods). We determined that, in DMS-153 SCLC, there was a mutation in exon-5, codon-155: ACC [Thr] → CCC [Pro].
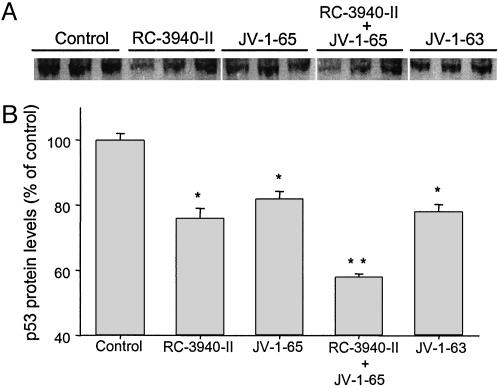
Effect of GHRH and BN/GRP Antagonists on Mutant p53 Tumor Suppressor Protein Expression in DMS-153 Human SCLC. We also investigated the expression of mutant p53 tumor suppressor protein by Western blotting assays with specific antibody in DMS-153 tumors from animals treated with GHRH and BN/GRP antagonists. Fig. 5A shows three representative tumors from each group. All of the tumor samples examined presented a band of 53 kDa corresponding to p53 protein (Fig. 5A), shown to be a mutant on the basis of cDNA sequencing analysis. Densitometric analyses of the band representing this mutant p53, after normalization to β-actin levels, demonstrated that RC-3940-II decreased the mutant p53 protein expression by 24% (P < 0.05) (Fig. 5B). Similarly, GHRH antagonists JV-1-65 and JV-1-63 alone also significantly reduced mutant p53 expression by 18% and 22%, respectively (both P < 0.05) (Fig. 5B). Combination of RC-3940-II and JV-1-65 caused the highest decrease of 42% (P < 0.01) in the mutant p53 level.
Fig. 5.
Expression of mutant p53 in DMS-153 SCLC tumors. Protein matched samples of tumor tissues from mice untreated or treated with JV-1-65, JV-1-63, and RC-3940-II alone or combined with JV-1-65 were submitted to Western blotting assays. Whole tissue homogenate was examined for p53 by using specific antibody (1:1,000) and immunoblot analysis. (A) Three representative tumors from each group are shown. (B) Percentages of protein measurements of mutant p53 are presented as means ± SEM of six to eight tumor samples from each group. Values were normalized with the expression of β-actin. *, P < 0.05 vs. control; **, P < 0.01 vs. control.
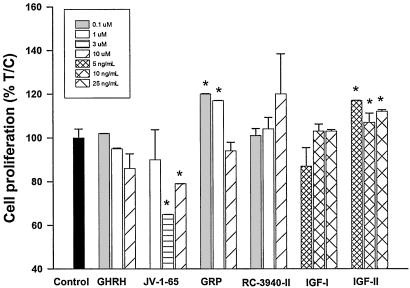
Effects of GHRH, GRP, Their Antagonists, and IGF-I and IGF-II on the Proliferation of DMS-153 SCLC in Vitro. DMS-153 SCLC cells cultured in vitro were exposed to various concentrations of GHRH(1-29)NH2, GHRH antagonist JV-1-65, GRP (14-27), BN/GRP antagonist RC-3940-II, IGF-I, and IGF-II, and the effects on the cell proliferation were determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay. GHRH antagonist JV-1-65 at 3 and 10 μM (P < 0.05) inhibited cell proliferation by 20-35%, compared with the control, whereas GHRH did not affect cell growth at concentrations between 0.1 and 10 μM (Fig. 6). GRP at 0.1 and 1 μM significantly (P < 0.05) increased the proliferation of DMS-153 SCLC cells by 20% and 17%, respectively, compared with controls. Growth rate of cells treated with BN/GRP antagonist RC-3940-II at 0.1-10 μM showed no significant changes. Treatment of cells with exogenous IGF-II at different concentrations significantly (P < 0.05) stimulated the proliferation by 12-17%, but IGF-I had no effect (Fig. 6).
Fig. 6.
Effects of GHRH, GRP, their antagonists, and IGF-I and IGF-II on cell proliferation of DMS-153 human SCLC line in vitro. Cultured cells were treated for 96 h with GHRH, GHRH antagonist JV-1-65, GRP, BN/GRP antagonist RC-3940-II, IGF-I, and IGF-II at different concentrations. Cell growth was determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) test. Data are expressed as percentage T/C values, where T represents absorbance of treated cells and C represents the absorbance of control cells. The measured absorbance is proportional to the number of living cells.
Discussion
The present study reports inhibitory effects of GHRH antagonists JV-1-65 and JV-1-63 and BN/GRP antagonist RC-3940-II on the growth of the DMS-153 human SCLC cell line xenografted into nude mice. We also evaluated the suppressive effects of these antagonists on tumoral IGF-II, the IGF-I receptor, and receptors for EGF and BN. Moreover, we attempted to shed some light on the signal transduction pathways that might be involved in the tumor growth processes by investigating the expression of p53 tumor suppressor gene.
Combined treatment with BN/GRP antagonist RC-3940-II and the GHRH antagonist JV-1-65 resulted in the greatest inhibition of growth of DMS-153 SCLC. Treatment with RC-3940-II alone also caused a significant but smaller decrease in tumor growth. Similarly, both GHRH antagonists exhibited tendencies to inhibit the tumor growth, but this effect was statistically significant only in the case of JV-1-65.
We have previously documented that the mechanism of antitumor action of GHRH antagonists involves the blocking of endocrine, paracrine, or autocrine stimulatory pathways of proliferation (1, 2, 8). RT-PCR analysis showed that DMS-153 tumors express the mRNAs corresponding to GHRH peptide as well as the SV1 and SV2 isoforms of GHRH receptors. The GHRH antagonist JV-1-65 inhibited the proliferation of DMS-153 cells in vitro. Thus, GHRH peptide and its tumoral receptors might form an autocrine stimulatory loop in the DMS-153 cell line. The inhibition of tumor growth after treatment with GHRH antagonists could be due in part to their blockade of the actions of locally produced GHRH. GHRH peptide has been previously reported to serve as an autocrine growth factor for various human cancer cell lines including SCLC and non-SCLC (2, 8, 22). The effects of GHRH on the proliferation of various cancers are thought to be mediated by the tumoral isoforms of GHRH receptors, which are generated by alternative splicing from the GHRH receptor gene and are different from the GHRH receptor found in the pituitary (23, 24). The putative protein sequence encoded by SV1 mRNA is consistent with a functional G protein-coupled receptor, whereas the mRNA for SV2 encodes a truncated receptor form that might not be functional. Because DMS-153 tumor samples in the present study were found to express mRNA for SV1, this tumoral isoform of GHRH receptor could mediate the direct effect of GHRH antagonists on DMS-153 cells.
RIAs demonstrated a significant reduction of serum IGF-I levels in the animals treated with GHRH antagonists JV-1-65 and JV-1-63. Thus, the reduction of serum IGF-I could partially account for the inhibition of tumor growth induced by GHRH antagonists in vivo. In addition, RT-PCR analyses demonstrated that GHRH antagonists tend to decrease the expression of mRNAs for tumoral IGF-II and IGFR-I. BN/GRP antagonist shows a similar tendency when given alone, and a strong additive inhibitory effect was observed in the group that received combined therapy with JV-1-65 and RC-3940-II. In this group, the growth of tumors was virtually arrested (95% inhibition of tumor volume), and the suppression of the expression of mRNAs for tumoral IGF-II and IGFR-I approached 73% and 49%, respectively. These effects could be relevant for tumor inhibition, because we showed that IGF-II stimulates growth of DMS-153 cell line.
Most SCLC cell lines, including DMS 153 SCLC (25), and many other tumors produce BN-like peptides, which act as autocrine growth factors (2, 10, 12, 27). These findings led to the development of BN/GRP antagonists for cancer therapy (2). BN and GRP up-regulate the binding of EGF and BN mediates its cell growth stimulus, at least partially, through the amplification of the EGFR tyrosine kinase system (2, 27). BN/GRP antagonists, such as RC-3940-II, inhibit tumor growth and cause a marked down-regulation of EGFRs (2, 13, 28). In the present study, we found that the BN/GRP antagonist, given alone or in combination with a GHRH antagonist, decreased the mRNA levels for GRPR and EGFR in DMS-153 tumor tissue in accordance with the assumed mechanisms of action. The growth of DMS-153 cells in vitro was also significantly stimulated by exogenous GRP, although the antagonist RC-3940-II did not inhibit the proliferation in vitro at concentrations up to 10 μM.
Mutations of p53 gene are reported to be among the most common genetic changes found in malignant tumors (29). It has been demonstrated that chemotherapy with DNA-damaging agents and radiotherapy are influenced by the p53 status of tumors (30). Based on the role of p53 in the control of apoptosis after DNA damage, the p53 gene has been implicated as a major determinant of tumor responsiveness to cytotoxic therapies. Wild-type p53 protein can suppress tumorigenesis and promote apoptosis, being a transcriptional factor (17, 18). Mutated p53 proteins in cancer cells have been linked to a number of pathophysiological processes and have been shown to exert effects that are opposite to those of wild-type p53. Thus, gene transfection and expression of mutant p53 proteins in cancer lines that were originally p53-null confers increased resistance to apoptosis (31) and stimulates the transcription of IGFR-I gene, in contrast to wild-type p53, which inhibits it (32). The presence of mutant p53 protein in tumors suggests that the cells acquire a selective growth advantage (18). It has also been shown that a mutation of the p53 gene, frequently occurring in hepatocellular carcinomas, markedly increases the production of IGF-II, which has an antiapoptotic effect, and thus could contribute to the selection of transformed hepatocytes with an increased resistance to apoptosis (33). The majority of p53 mutations are clustered into the central conserved DNA binding domain, and within this region, a number of hot spot p53 mutations were observed (18, 34, 35). In our study, we found a mutation on the so-called hot spot region of exons 5-8, specifically in exon-5, codon-155: ACC [Thr] → CCC [Pro]. A search of the database on p53 mutations maintained by International Agency for Research on Cancer (IARC database, version R7, accessible at www.iarc.fr/p53) (36) showed a deposit of 75 entries of mutations at codon 155, occurring in various human cancers. Fourteen of the mutations were of the ACC [Thr] → CCC [Pro] type, and 5 of 14 cases were observed in lung cancers (36).
Sequencing yields detailed molecular information on the p53 status and allows a better understanding of how the different mutated p53 versions interact with downstream molecules of the signaling pathway. The Thr155Val mutation in p53 has severe structural and functional consequences, including the appearance of a mutant protein conformation, loss of the normal phosphorylation pathway dependent on Thr-155, reduced binding to Mdm2, altered trafficking, and increased accumulation (37). All of the anomalies caused by the nullification of phosphorylation at position 155 are also present in the case of Thr155Pro mutant, and the conformational consequences of a Thr to Pro mutation could be even more drastic than those observed in the more conservative Thr to Val change.
The reduction in the levels of mutant P-53 protein shown in our study may be indicative of a response to therapy with antagonists of GHRH and BN/GRP antagonists. DMS-153 tumors from animals treated with BN/GRP antagonist RC-3940-II alone or with the combination of RC-3940-II and JV-1-65 showed the greatest decreases in the expression of mutant p53 protein. GHRH antagonists, JV-1-65 or JV-1-63 alone, likewise caused a significant but somewhat smaller reduction in mutant p53 expression. On the basis of these results, it is possible that the inhibition of tumor growth observed in vivo can be partially explained by the interference with mutant p53 protein expression.
Consequently, the suppression of growth of DMS-153 SCLC tumors by GHRH antagonists and BN/GRP antagonists might be mediated in part by restraining of the defective p53 function. Furthermore, because the expression of wild-type, proapoptotic p53 protein is apparently not required for tumor inhibition by GHRH and BN/GRP antagonists, these classes of compounds could offer a distinct advantage for the treatment of cancers with mutant p53 status, where the conventional anticancer drugs, including DNA cross linking agents, antimetabolites, and topo-isomerase I and II inhibitors, are less effective (38). Our findings demonstrate the ability of GHRH antagonists and BN/GRP antagonists to suppress the production of defective p53, which might contribute to their efficacy as antitumor agents. Our work supports the merit of therapeutic trials with GHRH antagonists and BN/GRP antagonists in the management of lung cancers.
Acknowledgments
This work was supported by the Medical Research Service of the Department of Veterans Affairs and a grant from Zentaris AG to Tulane University (all to A.V.S.). C.A.K. is a recipient of fellowship from Aché Laboratórios Farmacêuticos S/A (Sao Paulo, Brazil).
Abbreviations: BN, bombesin; GRP, gastrin-releasing peptide; GRPR, GRP receptor; EGF, epidermal growth factor; EGFR, EGF receptor; GHRH, growth hormone-releasing hormone; IGF, insulin-like growth factor; IGFR, IGF receptor; SCLC, small cell lung carcinoma; SV, splice variant.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY429684).
References
- 1.Schally, A. V. & Varga, J. L. (1999) Trends Endocrinol. Metab. 10, 383-391. [DOI] [PubMed] [Google Scholar]
- 2.Schally, A. V., Comaru-Schally, A. M., Nagy, A., Kovacs, M., Szepeshazi, K., Plonowski, A., Varga, J. L. & Halmos, G. (2001) Front. Neuroendocrinol. 22, 248-291. [DOI] [PubMed] [Google Scholar]
- 3.Werner, H. & LeRoith, D. (1996) Adv. Cancer Res. 68, 183-223. [DOI] [PubMed] [Google Scholar]
- 4.Khandwala, H. M., McCutcheon, I. E., Flyvbjerg, A. & Friend, K. E. (2000) Endocrine Rev. 21, 215-244. [DOI] [PubMed] [Google Scholar]
- 5.Moorehead, R. A., Sanchez, O. H., Baldwin, R. M. & Khokha, R. (2003) Oncogene 22, 853-857. [DOI] [PubMed] [Google Scholar]
- 6.Pinski, J., Schally, A. V., Jungwirth, A., Groot, K., Halmos, G., Armatis, P., Zarandi, M. & Vadillo-Buenfil, M. (1996) Int. J. Oncol. 9, 1099-1105. [DOI] [PubMed] [Google Scholar]
- 7.Kiaris, H., Schally, A. V. & Varga, J. L. (2000) Cancer Lett. 161, 149-155. [DOI] [PubMed] [Google Scholar]
- 8.Kiaris, H., Schally, A. V., Varga, J. L., Groot, K. & Armatis, P. (1999) Proc. Natl. Acad. Sci. USA 96, 14894-14898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siegfried, J. M., DeMichele, M. A., Hunt, J. D., Davis, A. G., Yohra, K. P. & Pilewski, J. M. (1997) Am. J. Respir. Crit. Care Med. 156, 358-366. [DOI] [PubMed] [Google Scholar]
- 10.Cuttita, F., Carney, D. N., Mulshine, J., Moody, T. W., Fedorko, J., Fischler, A. & Minna, J. D. (1985) Nature 316, 823-826. [DOI] [PubMed] [Google Scholar]
- 11.Kiaris, H., Schally, A. V., Nagy, A., Sun, B., Armatis, P. & Szepeshazi, K (1999) Br. J. Cancer 81, 966-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spindel, E. R., Giladi, E., Segerson, T. P. & Nagalla, S. (1993) Rec. Progr. Horm. Res. 48, 365-391. [DOI] [PubMed] [Google Scholar]
- 13.Koppan, M., Halmos, G., Arencibia, J. M., Lamharzi, N. & Schally, A. V. (1998) Cancer 83, 1335-1343. [DOI] [PubMed] [Google Scholar]
- 14.Hussain, S. P., Hofseth, L. J. & Hans, C. C. (2001) Lung Cancer 34, S7-S15. [DOI] [PubMed] [Google Scholar]
- 15.Sekido, Y., Fong, K. M. & Minna, J. D. (1998) Biochim. Biophys. Acta 1378, F21-F59. [DOI] [PubMed] [Google Scholar]
- 16.Ahrendt, S. A., Hu, Y., Buta, M., McDermott, P., Benoit, N., Yang, S. C., Wu, L. & Sidransky, D. (2003) Natl. Cancer Inst. 95, 961-970. [DOI] [PubMed] [Google Scholar]
- 17.Vogelstein, B. & Kinzler, K. W. (1992) Cell 70, 523-526. [DOI] [PubMed] [Google Scholar]
- 18.Greenblatt, M. S., Bennett, W. P., Hollstein, M. & Harris, C. C. (1994) Cancer Res. 54, 4855-4878. [PubMed] [Google Scholar]
- 19.Reile, H., Cai, R.-Z., Armatis, P. & Schally, A. V. (1995) Int. J. Oncol. 7, 749-754. [DOI] [PubMed] [Google Scholar]
- 20.Varga, J. L., Schally, A. V., Csernus, V. J., Zarandi, M., Halmos, G. & Rekasi, Z. (1999) Proc. Natl. Acad. Sci. USA 96, 692-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sorenson, G. D., Pettengill, O. S., Cate, C. C., Ghatei, M. A., Molyneux, K. E., Gosselin, E. J. & Bloom, S. R. (1983) Life Sci. 33, 1939-1944. [DOI] [PubMed] [Google Scholar]
- 22.Busto, R., Schally, A. V., Varga, J. L., Garcia-Fernandez, M. O., Groot, K., Armatis, P. & Szepeshazi, K. (2002) Proc. Natl. Acad. Sci. USA 99, 11866-11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rekasi, Z., Czompoly, T., Schally, A. V. & Halmos, G. (2000) Proc. Natl. Acad. Sci. USA 97, 10561-10566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halmos, G., Schally, A. V., Varga, J. L., Plonowski, A., Rekasi, Z. & Czompoly, T. (2000) Proc. Natl. Acad. Sci. USA 97, 10555-10560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Werle, E., Schneider, C., Renner, M., Volker, M. & Fiehn, W. (1994) Nucleic Acids Res. 22, 4354-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanashiro, C. A. & Khalil, R. A. (2001) Am. J. Physiol. 280, C34-C45. [DOI] [PubMed] [Google Scholar]
- 27.Liebow, C., Crean, D. H., Lee, M. T., Kamer, A. R., Mang, T. S. & Schally, A. V. (1994) Proc. Natl. Acad. Sci. USA 91, 3804-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plonowski, A., Schally, A. V., Varga, J. L., Rekasi, Z., Hebert, F., Halmos, G. & Groot, K. (2000) Prostate 44, 172-180. [DOI] [PubMed] [Google Scholar]
- 29.Hollstein, M., Sidransky, D., Vogelstein, B. & Harris, C. C. (1991) Science 253, 49-53. [DOI] [PubMed] [Google Scholar]
- 30.Natsume, T., Kobayashi, M. & Fujimoto, S. (2001) Cancer 92, 386-394. [DOI] [PubMed] [Google Scholar]
- 31.Ohnishi, K., Ota, I., Takahashi, A., Yane, K., Matsumoto, H. & Ohnishi, T. (2002) Apoptosis 7, 367-372. [DOI] [PubMed] [Google Scholar]
- 32.Werner, H., Karnichi, E., Rauscher, F. J. & LeRoith, D. (1996) Proc. Natl. Acad. Sci. USA 93, 8318-8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, Y. I., Lee, S., Das, G. C., Park, U. S., Park, S. M. & Lee, Y. I. (2000) Oncogene 19, 3717-3726. [DOI] [PubMed] [Google Scholar]
- 34.Cho, Y., Gorina, S., Jeffrey, P. D. & Pavletich, N. P. (1994) Science 265, 346-356. [DOI] [PubMed] [Google Scholar]
- 35.Hollstein, M., Shomer, B., Greenblatt, M., Soussi, T., Hovig, E., Montesano, R. & Harris, C. C. (1996) Nucleic Acids Res. 24, 141-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olivier, M., Eales, R., Hollstein, M., Khan, M. A., Harris, C. C. & Hainaut, P. (2002) Hum. Mutat. 19, 607-614. [DOI] [PubMed] [Google Scholar]
- 37.Bech-Otschir, D., Kraft, R., Huang, X., Henklein, P., Kapelari, B., Pollmann, C. & Dubiel, W. (2001) EMBO J. 20, 1630-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Connor, P. M., Jackman, J., Bae, I., Myers, T. G., Fan, S., Mutoh, M., Scudiero, D. A., Monks, A., Sausville, E. A., Weinstein, J. N., et al. (1997) Cancer Res. 57, 4285-4300. [PubMed] [Google Scholar]