Abstract
Leishmania null mutants created by targeted gene replacement are typically complemented with chimeric episomes harboring the replaced gene in order to validate that the observed phenotype is due to the specific gene deletion. However, the current inventory of available episomes for complementation of genetic lesions in Leishmania is unstable in the absence of drug selection, and levels of gene expression cannot be controlled, especially in vivo. To circumvent this impediment, a strategy to re-introduce the targeted gene into the original chromosomal locus to generate “knock-in” parasites within selectable null backgrounds has been developed. A genomic fragment encompassing the ornithine decarboxylase locus and lacking heterologous DNA sequences was transfected into ornithine decarboxylase-deficient L. donovani. The construct randomly integrated into either chromosomal allele by homologous recombination restoring polyamine prototrophy and revealing that LdODC was functionally expressed in the knock-in clones. This strategy offers a mechanism for complementing a genetic lesion amenable to positive selection in a manner that facilitates stable gene expression from its original locus in the absence of continuous drug pressure.
Keywords: Leishmania, genetic manipulations, ornithine decarboxylase
1. Introduction
Protozoan parasites of the Leishmania genus are the etiologic agents of leishmaniasis, a spectrum of devastating and potentially deadly diseases. All Leishmania species are digenetic parasites that exist as both insect vector and mammalian forms. The flagellated, motile, extracellular promastigote proliferates in the midgut of phlebotomine sand fly family members, whereas the nonmotile, intracellular amastigote resides in phagolysosomes of macrophages and other reticuloendothelial cells within the vertebrate host. Because there are no existing vaccines, chemotherapy offers the only avenue to combat the disease. Unfortunately, the current inventory of available drugs to treat leishmaniasis is far from ideal, mainly because of toxicity and therapeutic unresponsiveness. Thus, the identification, characterization, and validation of novel therapeutic targets are urgently needed.
Reverse genetic tools have proven invaluable in validating novel targets in Leishmania (Clayton, 1999). The ability to generate knockout Leishmania by targeted gene replacement using drug resistance cassettes that integrate into the locus of choice by homologous recombination enables an unbiased assessment of gene function in intact parasites (Cruz and Beverley, 1990, Cruz, et al., 1991, Curotto de Lafaille and Wirth, 1992). The ability to complement these genetic lesions using episomal vectors then enables the definitive demonstration that the resultant phenotype can be ascribed to the specific null mutation. The currently episomal vectors available for Leishmania transfections, however, have several shortcomings including the fact that they cannot be stably maintained in the absence of drug pressure. Thus, most leishmanial episomes will eventually be lost if selective pressure is removed. Although the instability of episomal vectors rarely impacts studies with parasites in culture, this drawback poses serious concerns for long-term infectivity studies in animals in which Leishmania are obligatorily passaged for several weeks in the absence of the selective drug. Other disadvantages of episomal vectors are that they lack an active segregation mechanism to ensure equal plasmid distribution between daughter cells during mitosis and usually do no contain endogenous signals for post-transcriptional processing of their encoded mRNAs. Thus, the level of gene expression from an episome cannot be controlled and will fluctuate among individual parasites depending upon the episomal copy number. Furthermore, the episomal vector copy number can only be influenced marginally by the concentration of the selective drug that maintains the plasmid. Thus, expression of the target protein may be low and insufficient for rescue or too high and have deleterious effects on parasite growth. Overall, the need for improved complementation systems to validate targets in Leishmania is considerable.
One biochemical pathway that has received considerable attention as a therapeutic target for the treatment of parasitic diseases, including leishmaniasis, is that for the biosynthesis of polyamines, essential aliphatic cations that are necessary for the survival and growth of all proliferating cells. α-Difluoromethylornithine (DFMO), an irreversible inhibitor of ornithine decarboxylase (ODC), the rate-limiting step in polyamine biosynthesis, has been shown to completely eradicate Trypanosoma brucei brucei infections in mice (Bacchi, et al., 1980) and is an extremely effective agent (>95% cure) against early- and late-stage African sleeping sickness caused by T. b. gambiense, including arsenical-refractory cases that would otherwise prove fatal (Burri and Brun, 2003, Pepin, et al., 1987, Van Nieuwenhove, et al., 1985). Inhibitors of another enzyme crucial for polyamine biosynthesis, S-adenosylmethionine decarboxylase (ADOMETDC), are also effective antitrypanosomal agents (Bitonti, et al., 1990, Bitonti, et al., 1986).
The polyamine pathway in Leishmania has also been investigated as a promising therapeutic target. The four enzymes of the polyamine biosynthetic pathway of Leishmania are: 1) arginase (ARG) that generates ornithine and urea; 2) ODC that catalyzes the decarboxylation of ornithine to form putrescine; 3) ADOMETDC that produces decarboxylated S-adenosylmethionine for aminopropylation of putrescine; and 4) spermidine synthase (SPDSYN) that transfers the aminopropyl moiety from decarboxylated S-adenosylmethionine to putrescine. The isolation of the ODC (LdODC), ADOMETDC (LdADOMETDC), and SPDSYN (LdSPDSYN) genes from L. donovani and the ARG (LmARG) gene from L. mexicana, as well as their flanking sequences, has enabled a test of polyamine gene function in intact parasites via the creation and phenotypic characterization of Δlmarg, Δldodc, Δldadometdc, and Δldspdsyn knockouts (Jiang, et al., 1999, Roberts, et al., 2001, Roberts, et al., 2002, Roberts, et al., 2004). Each null mutant, as the promastigote, proved to be auxotrophic for polyamines and was only able to proliferate in defined medium supplemented with an appropriate exogenous polyamine. Transfection of episomal copies of the missing genes into the corresponding null mutants complemented the genetic lesions in promastigotes and restored polyamine prototrophy in all cases confirming that the auxotrophic phenotype was caused by the specific gene deletion and not by a secondary, unrelated genetic event. The episomes in each of the complemented “add-back” promastigote strains were stably maintained by appropriate drug selection in these in vitro experiments with Leishmania promastigotes.
The polyamine auxotrophy of Δldodc amastigotes in murine macrophages in cell culture could also be completely reversed by episomal LdODC expression. In these short-term (72 hr) experiments, episomal rescue of the compromised infectivity phenotypes of two independent Δldodc mutants restores the parasite burden in murine macrophages to levels observed for wild type parasite infections (Boitz, et al., 2009). However, the parasite burdens in livers and spleens of mice infected four weeks previously with Δldodc[pLdODC] parasites, although much greater than the parasite loads observed in the organs of mice infected with the two independently derived Δldodc null mutants, were one to two orders of magnitude less than those obtained from mice infected with wild type parasites (Boitz, et al., 2009). Thus, the profoundly compromised virulence phenotype of the Δldodc knockouts is only partially re-established by episomal complementation in these long-term mouse infectivity experiments. Although episome copy number in parasites obtained from these organ harvests could not be directly assessed, the failure to fully complement the virulence defect in vivo can most likely be ascribed to episomal loss in these rodent infections due to the absence of selective pressure.
To circumvent the problem of episomal instability in the rodent model of leishmaniasis in the absence of selective pressure, we developed a strategy for generating an “add-back” strain of Δldodc in which the genotype is stable in the absence of selective pressure.
2. Material and Methods
2.1 Parasites
All genetically manipulated parasites were derived from the wild-type LdBob strain of L. donovani (Goyard, et al., 2003), which was originally obtained from Stephen Beverley (Washington University, St. Louis, MO). LdBob promastigotes and axenic amastigotes were routinely cultivated at 26°C, pH 7.4, and 37°C, pH 5.5, respectively, in the culture media previously reported (Goyard, et al., 2003). Parasite strains were cycled back and forth between the promastigote and axenic amastigote forms several times. Wild-type parasites were routinely cultured in medium with no drug supplementations, while the Δldodc mutants were propagated in 50 μg/ml hygromycin, 20 μg/ml geneticin (G418), and 200 μM putrescine or just 200 μM putrescine alone. The putative knock-in Δldodc::HYG/ODC and Δldodc::NEO/ODC lines were maintained in medium with no putrescine supplementation.
2.2. Transfections
A ~15 kb genomic fragment encompassing the 2,121 bp LdODC open reading frame and extensive flanking regions was derived from a cosmid clone (Jiang, et al., 1999) by a HindIII-EcoRI restriction digest. The genomic fragment was gel purified and used to transfect Δldodc promastigotes according to standard procedures (Jiang, et al., 1999). Transfected parasites were plated on semisolid agar plates lacking putrescine to select for integration of the LdODC containing fragment. Four colonies were obtained per μg of transfected 15 kb DNA fragment. Transfections were also performed with a polymerase chain reaction- generated fragment that encompassed the entire LdODC open reading frame and ~500 bp of each of the two LdODC flanking regions. However, no colonies that grew in putrescine-deficient medium were obtained after transfection of the ~3 kb fragment, and this strategy was not pursued further since the transfections with the 15 kb genomic fragment were successful. These observations are consistent with the hypothesis that transfections with DNA molecules containing longer homologous flanking sequences have a higher rate of homologous recombination.
2.3. Growth curves
Parasites were seeded at 1 × 105 parasites/ml in normal growth medium with no additions, 200 μM putrescine, 50 μg/ml hygromycin, or 20 μg/ml neomycin. After five days of incubation at 28°C, parasites were enumerated on a hemacytometer.
2.4. Southern and western blot
Genomic DNA from wild type, Δldodc and knock-in parasites was prepared for Southern blot analysis using the DNAeasy kit (Qiagen Inc., Valencia, CA) according to the manufacturer's protocol. DNA was digested with SalI and probed with ODC, HYG or NEO coding regions using highly stringent hybridization conditions. Parasite lysates from exponentially growing cell populations were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Laemmli, 1970) and blotted onto either nitrocellulose or Nytran membranes (Schleicher and Schuell, Keene, NH), and Western blot analysis was performed according to standardized procedures (Towbin, et al., 1979). The membranes were probed with polyclonal antibodies raised to the L. donovani ODC and SPDSYN (Jiang, et al., 1999, Roberts, et al., 2001).
3. Results and Discussion
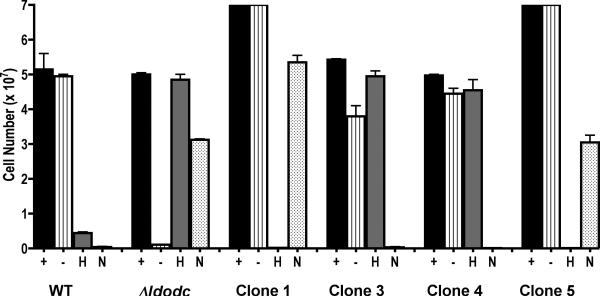
Our approach involves the re-introduction of a “markerless” LdODC gene copy into a Δldodc strain at the original LdODC genomic locus that had been previously genetically transformed by homologous integration of two drug resistance cassettes. The Δldodc parasite strain was originally created by double targeted gene replacement using the hygromycin and neomycin drug resistance markers (HYG and NEO) and is designated according to the generally accepted genetic nomenclature for Leishmania as Δldodc::HYG/Δldodc::NEO (Clayton, et al., 1998). The Δldodc::HYG/Δldodc::NEO strain, which requires exogenous putrescine supplementation for growth, was transfected with a linearized ~15 kb genomic fragment encompassing the LdODC open reading frame and extensive flanking regions. This ~15 kb HindIII-EcoRI fragment was derived from a cosmid clone (Jiang, et al., 1999), contained only L. donovani genomic DNA and lacked drug resistance markers. The transfected Δldodc parasites were plated on semi-solid agar plates lacking putrescine, G418, and hygromycin to select for clonal cell lines in which the linearized LdODC genomic fragment had integrated into either the Δldodc::HYG or the Δldodc::NEO allele by homologous combination. These knock-in clones, therefore, were conjectured to have randomly replaced one of the two drug resistance markers in the knockout line as a consequence of integration of the LdODC genomic fragment. Five colonies were picked from the semi-solid agarose and expanded in growth medium lacking putrescine, G418, and hygromycin, and the growth phenotypes of four of these clones were subsequently characterized. All four clones exhibited polyamine prototrophy as exemplified by their ability to proliferate normally in the absence of putrescine supplementation, whereas the Δldodc progenitor was auxotrophic for polyamines (Fig. 1). Moreover, two of the clones, clone 1 and clone 5, were now sensitive to 50 μg/ml hygromycin and two, clones 3 and 4, were sensitive to 20 μg/ml G418. Conversely, clones 1 and 5 retained their resistance to G418, whereas clones 3 and 4 maintained their refractoriness to hygromycin. As expected, wild type promastigotes were sensitive to both 20 μg/ml G418 and 50 μg/ml hygromycin, while the Δldodc::HYG/Δldodc::NEO (Δldodc) null mutant was refractory to both drugs (Fig. 1). Thus, the growth phenotypes of the four clones were consistent with the loss of a single drug resistance allele and the concomitant gain of LdODC function.
Figure 1.
Growth phenotypes of wild type, Δldodc, and knock-in parasites. L. donovani wild type (WT), Δldodc::NEO/Δldodc::HYG (Δldodc), and four independent knock-in clones (clones 1, 3, 4, and 5) were seeded at 1 × 105 parasites/ml in normal growth medium with either 200 μM putrescine (+), no additions (-), 50 μg/ml hygromycin (H), or 20 μg/ml neomycin (N). After five days of incubation at 28 °C, parasites were enumerated on a hemacytometer. The results are those of a single typical experiment that was performed in duplicate. Quantitation of parasite numbers employing Alamar Blue® as the growth indicator provided essentially identical results (data not shown).
To assess the genotypic alterations in the four putative knock-in lines that were created by transfection with the ~15 kb LdODC cosmid fragment; Southern blots were performed (Fig. 2A). This Southern analysis revealed that all four clones isolated from the transfection with the ~15 kb genomic fragment had assimilated an LdODC copy into the genome. Furthermore, these integrations occurred within the original LdODC genomic locus as revealed by the loss of the HYG allele in clones 1 and 5 and the lack of the NEO allele in clones 3 and 4 (Fig. 2A). The Δldodc::HYG/ODC lines (clones 3 and 4) were genetically indistinguishable from the heterozygous line from which the Δldodc null mutant was originally derived (Boitz, et al., 2009). However, our laboratory has never generated a neomycin resistant heterozygous strain, thus the two Δldodc::NEO/ODC clones represent a novel genotype that could not have resulted from accidental contamination of the Δldodc by its parental heterozygous progenitor (Boitz, et al., 2009).
Figure 2.
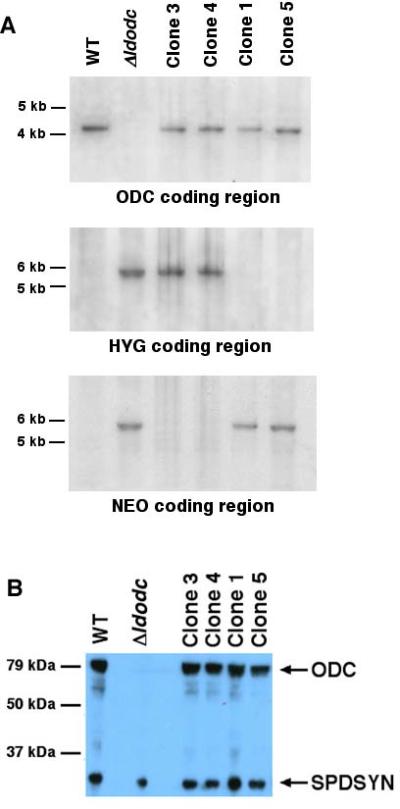
Southern and western blot analyses of knock-in parasites. (A) Genomic DNA from wild type (WT), Δldodc::NEO/Δldodc::HYG (Δldodc), and the four knock-in clones (clones 1, 3, 4, and 5) was digested with Sal I and hybridized separately to either LdODC, HYG, or NEO as indicated. Molecular weight markers are shown to the left. (B) Cell lysates from wild type (WT), Δldodc::NEO/Δldodc::HYG (Δldodc), and the four knock-in clones were fractionated by SDS polyacrylamide gel electrophoresis and probed with previously characterized rabbit polyclonal antibodies to LdODC and LdSPDSYN (as loading control). Molecular weight markers are indicated to the left.
Western blotting was then carried out to assess the levels of endogenous LdODC protein expression in the knock-in clones. This western blot analysis, employed monospecific LdODC antisera (Jiang, et al., 1999) and demonstrated that the levels of LdODC protein in cell lysates prepared from the four knock-in clones were comparable to that of wild type parasites, while no LdODC protein, as anticipated, could be detected in Δldodc cell extracts. Although a gene dosage effect might be expected to produce less protein in the knock-in parasite line, equivalent LdODC expression levels were observed in the diploid wild type and knock-in clones, each containing a single LdODC chromosomal integration. The genotypic and phenotypic analysis of the knock-in clones established that the transfection of the linearized ~15 kb LdODC construct into the Δldodc line enabled stable integration and expression of LdODC from its original locus. Furthermore, all four clones stably maintained their genotypes and phenotypes when continuously cultured in growth medium lacking putrescine, G418, and hygromycin for over three months (data not shown).
Recently, the targeted chromosomal integration strategy has been exploited to generate an “add-back” strain from a conventional gene deletion mutant in L. infantum (Denise, et al., 2006). In this case, the gene encoding the L. infantum CPA cysteine (LiCPA) peptidase was re-introduced into the CPA chromosomal locus of two ΔLicpa strains using a construct containing the CPA gene. However, unlike the linear 15 kb genomic fragment employed in the LdODC integration, which contains exclusively autologous DNA, the 3’ sequence of the L. infantum LiCPA re-expression construct contained heterologous DNA encoding the Bacillus cereus blasticidin-resistance marker and sequences from the L. major dihydrofolate reductase-thymidylate synthase locus (Denise, et al., 2006). Although the integrated LiCPA gene was expressed in the Δlicpa::CPA transfectants, there was no rescue of the virulence defect observed as a consequence of the parental CPA-deficient mutants in hamsters. The failure of the homologous chromosomal integration to restore ΔLicpa virulence was postulated by the authors to the interference of the exogenous 3’ flanking sequences with LiCPA gene expression in vivo (Denise, et al., 2006).
Although we have not yet evaluated the capacity of LdODC knock-in strains to restore virulence of Δldodc parasites to wild type levels in animals, the homologous integrations at the LdODC locus, unlike those at the LiCPA locus (Denise, et al., 2006), did not involve selection for drug resistance traits or integration of heterologous DNA sequences that could interfere with endogenous gene expression. Thus, the LdODC knock-in integrations were markerless and effectively recreated the genotype of the LdODC/ldodc heterozygote whose phenotype was identical to the wild type parent. This markerless integration complementation strategy can be effectively applied to any null mutant that is amenable to positive selection, e.g., restoration of putrescine prototrophy, and offers an avenue for stable complementation of a genetic defect in the absence of continuous drug selection.
Acknowledgements
This work was supported by grant R01 AI041622 from the National Institute of Allergy and Infectious Disease.
Abbreviations
- DFMO
α-difluoromethylornithine
- ODC
ornithine decarboxylase
- ADOMETDC
S-adenosylmethionine decarboxylase
- SPDSYN
spermidine synthase
- LdODC
Leishmania donovani ODC
- G418
Geneticin
- LiCPA
CPA cysteine protease
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bacchi CJ, Nathan HC, Hutner SH, McCann PP, Sjoerdsma A. Polyamine metabolism: a potential therapeutic target in trypanosomes. Science. 1980;210:332–334. doi: 10.1126/science.6775372. [DOI] [PubMed] [Google Scholar]
- Bitonti AJ, Byers TL, Bush TL, Casara PJ, Bacchi CJ, Clarkson AB, Jr., McCann PP, Sjoerdsma A. Cure of Trypanosoma brucei brucei and Trypanosoma brucei rhodesiense infections in mice with an irreversible inhibitor of S-adenosylmethionine decarboxylase. Antimicrob Agents Chemother. 1990;34:1485–1490. doi: 10.1128/aac.34.8.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitonti AJ, Dumont JA, McCann PP. Characterization of Trypanosoma brucei brucei S-adenosyl-L-methionine decarboxylase and its inhibition by Berenil, pentamidine and methylglyoxal bis(guanylhydrazone). Biochem J. 1986;237:685–689. doi: 10.1042/bj2370685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boitz JM, Yates PA, Kline C, Gaur U, Wilson ME, Ullman B, Roberts SC. Leishmania donovani ornithine decarboxylase is indispensable for parasite survival in the mammalian host. Infect Immun. 2009;77:756–763. doi: 10.1128/IAI.01236-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burri C, Brun R. Eflornithine for the treatment of human African trypanosomiasis. Parasitol Res. 2003;90(Supp 1):S49–52. doi: 10.1007/s00436-002-0766-5. [DOI] [PubMed] [Google Scholar]
- Clayton C, Adams M, Almeida R, Baltz T, Barrett M, Bastien P, Belli S, Beverley S, Biteau N, Blackwell J, Blaineau C, Boshart M, Bringaud F, Cross G, Cruz A, Degrave W, Donelson J, El-Sayed N, Fu G, Ersfeld K, Gibson W, Gull K, Ivens A, Kelly J, Vanhamme L, et al. Genetic nomenclature for Trypanosoma and Leishmania. Mol Biochem Parasitol. 1998;97:221–224. doi: 10.1016/s0166-6851(98)00115-7. [DOI] [PubMed] [Google Scholar]
- Clayton CE. Genetic manipulation of kinetoplastida. Parasitol Today. 1999;15:372–378. doi: 10.1016/s0169-4758(99)01498-2. [DOI] [PubMed] [Google Scholar]
- Cruz A, Beverley SM. Gene replacement in parasitic protozoa. Nature. 1990;348:171–173. doi: 10.1038/348171a0. [DOI] [PubMed] [Google Scholar]
- Cruz A, Coburn CM, Beverley SM. Double targeted gene replacement for creating null mutants. Proc Natl Acad Sci U S A. 1991;88:7170–7174. doi: 10.1073/pnas.88.16.7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curotto de Lafaille MA, Wirth DF. Creation of Null/+ mutants of the alpha-tubulin gene in Leishmania enriettii by gene cluster deletion. J Biol Chem. 1992;267:23839–23846. [PubMed] [Google Scholar]
- Denise H, Poot J, Jimenez M, Ambit A, Herrmann DC, Vermeulen AN, Coombs GH, Mottram JC. Studies on the CPA cysteine peptidase in the Leishmania infantum genome strain JPCM5. BMC Mol Biol. 2006;7:42. doi: 10.1186/1471-2199-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyard S, Segawa H, Gordon J, Showalter M, Duncan R, Turco SJ, Beverley SM. An in vitro system for developmental and genetic studies of Leishmania donovani phosphoglycans. Mol Biochem Parasitol. 2003;130:31–42. doi: 10.1016/s0166-6851(03)00142-7. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Roberts SC, Jardim A, Carter NS, Shih S, Ariyanayagam M, Fairlamb AH, Ullman B. Ornithine decarboxylase gene deletion mutants of Leishmania donovani. J Biol Chem. 1999;274:3781–3788. doi: 10.1074/jbc.274.6.3781. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Pepin J, Milord F, Guern C, Schechter PJ. Difluoromethylornithine for arseno-resistant Trypanosoma brucei gambiense sleeping sickness. Lancet. 1987;2:1431–1433. doi: 10.1016/s0140-6736(87)91131-7. [DOI] [PubMed] [Google Scholar]
- Roberts SC, Jiang Y, Gasteier J, Frydman B, Marton LJ, Heby O, Ullman B. Leishmania donovani polyamine biosynthetic enzyme overproducers as tools to investigate the mode of action of cytotoxic polyamine analogs. Antimicrob Agents Chemother. 2007;51:438–445. doi: 10.1128/AAC.01193-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SC, Jiang Y, Jardim A, Carter NS, Heby O, Ullman B. Genetic analysis of spermidine synthase from Leishmania donovani. Mol Biochem Parasitol. 2001;115:217–226. doi: 10.1016/s0166-6851(01)00293-6. [DOI] [PubMed] [Google Scholar]
- Roberts SC, Scott J, Gasteier JE, Jiang Y, Brooks B, Jardim A, Carter NS, Heby O, Ullman B. S-adenosylmethionine decarboxylase from Leishmania donovani. Molecular, genetic, and biochemical characterization of null mutants and overproducers. J Biol Chem. 2002;277:5902–5909. doi: 10.1074/jbc.M110118200. [DOI] [PubMed] [Google Scholar]
- Roberts SC, Tancer MJ, Polinsky MR, Gibson KM, Heby O, Ullman B. Arginase plays a pivotal role in polyamine precursor metabolism in Leishmania. Characterization of gene deletion mutants. J Biol Chem. 2004;279:23668–23678. doi: 10.1074/jbc.M402042200. [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nieuwenhove S, Schechter PJ, Declercq J, Bone G, Burke J, Sjoerdsma A. Treatment of gambiense sleeping sickness in the Sudan with oral DFMO (DL-alpha-difluoromethylornithine), an inhibitor of ornithine decarboxylase; first field trial. Trans R Soc Trop Med Hyg. 1985;79:692–698. doi: 10.1016/0035-9203(85)90195-6. [DOI] [PubMed] [Google Scholar]