Abstract
A multitude of results from genome-wide association studies have been published in recent years in relation to different human diseases and phenotypic traits. However, the identified polymorphisms explain just a small fraction of the variability of the traits and they are poor predictors of occurrence of disease. Although part of the missing variability may be found in still to be identified rare genetic variants, the present work proposes that a major part of the problem is due to our conceptual limitations regarding functional loci and its variants. Functional variants are currently defined in absolute positional terms; they are just sequence variations in fixed positions along the DNA molecule. In the present study is postulated that functional loci may include different positions in the DNA sequence. As consequence, variants of the same functional locus may be located in different physical positions along the genome and, the observed effect of any particular genetic variant will be then reduced compared to its true effect. The differential use of regulatory regions such as gene promoters and enhancers would be a particular case of the proposed hypothesis. The hypothesis makes predictions that can be tested, offering potential paths of research to elucidate the genetic basis of complex human traits.
Keywords: Epigenetics, GWAS, missing heritability, gene expression, human disease
INTRODUCTION
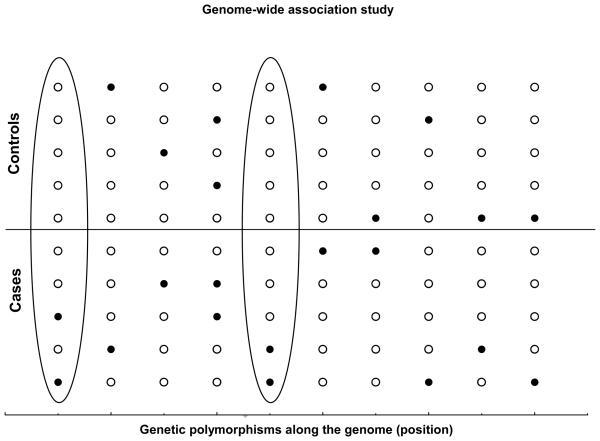
A basic assumption when studying how genetic variation may affect variability of human phenotypic traits is that functional variants correspond to physical variants, and their positions are fixed along the DNA. For example, a current approach to identify functional loci is the use of genome-wide association studies (GWAS), in which hundred of thousands of physical variants (single nucleotide polymorphisms, SNPs) across the genome are genotyped and compared among thousands of subjects to assess association with the phenotypic trait under study (for example a disease) (Figure 1). Because it is assumed that functional loci have fixed positions, differences of allele frequencies between cases and controls are tested within each genetic polymorphism. Thus, by genotyping hundred of thousands of physical loci we might be able to identify the subset of functional variants (or at least variants that are in linkage disequilibrium with the functional ones) that determine any phenotypic trait.
Figure 1. Genome-wide association study.
Hundred of thousands of single nucleotide polymorphisms (SNPs) are genotyped in cases of disease and controls to assess association with a particular disease. Differences on allele frequencies between cases and controls are tested for each genetic polymorphism. In this example, the first and fifth polymorphisms from left to right (in ovals) show differences between cases and controls: a genetic variant (filled circles) is more frequent in cases compared to controls.
I hypothesize in the present work that, contrary to current assumptions, positions of functional loci are not fixed in the DNA sequence; one particular functional locus may cover different physical loci.
THE PROBLEM
The last few years have seen the completion and publication of a great number of GWAS in different human diseases and phenotypic traits including cancers [1-3], psychiatric illnesses [4, 5], type-2 diabetes [6-8], weight [9-11], and height [12-14] among others. Although GWAS have the potential to identify genetic variants associated with new mechanisms of disease, results so far do not seem to explain much of total phenotypic variation due to genetic factors. For example, the identified genetic variants associated with weight explain only about 1% of the total expected genetic variation [15], 18 identified loci are associated with a sibling relative risk of just 1.07 (i.e., 7% increase in disease risk compared to the general population risk) for type-2 diabetes [16], and 40 newly identified loci explain just about 5% of the phenotypic variation in height [12-14]. As a consequence, genetic variants identified so far are also poor predictors of the occurrence of disease. For type-2 diabetes, the genotype score of the 18 identified single nucleotide polymorphisms (SNPs) does not offer additional predictive power when known risk factors (i.e., age, family history, weight, etc) are taken into account [16, 17]. Finally, a recent study found that the addition of 10 common genetic variants associated with breast cancer to a risk model containing reproductive, medical and family history did not provide additional information on risk of developing breast cancer [18].
Part of the problem may be due to technical limitations of current GWAS platforms that fail to capture rare variants contributing to variation of phenotypic traits [19, 20]. Current efforts such as the 1000 Genome Project will help to identify and catalogue rare variants to use in future GWAS. Other types of genetic factors, such as copy number variants (CNV) may also add to the genetic basis of human traits. However, I claim that a major part of the problem is due to our current limitations regarding the concept of functional variants. As Figure 1 shows, functional variants are currently defined in absolute positional terms or in other words, they are just considered as sequence variations in the same position along the DNA. Thus, comparisons of allele frequencies are made within each genetic polymorphism to assess whether particular genetic variants correlate with the phenotypic trait under study.
THE HYPOTHESIS
I postulate that functional loci have not fixed positions along the DNA sequence. As consequence, variants of the same functional locus may reside in different physical positions in the genome. A single example will help to make clear the meaning and significance of the proposed hypothesis. The African trypanosome Trypanosoma brucei is a unicellular protozoa that infects the bloodstream of mammals and causes sleeping sickness in humans. T. brucei is able to elude the host immune system by periodically changing its protective coat, the variant surface glycoprotein (VSG) (see review in [21, 22]). T. brucei has more than 1000 VSG genes, but only one is transcribed at a given time with a switching rate of about 1% per trypanosome cell per generation [23]. As this example shows, we can consider the whole set of VSG genes as a single functional locus and therefore, its functional variants may be located in different positions along the T. brucei genome. As Figure 2 makes clear, the position of functional variants is not fixed rather it depends on which VSG gene is being expressed in a particular trypanosome parasite.
Figure 2. VSG genes in Tripanosome brucei.
The genome of the parasite T. brucei has more than 1000 VSG genes and only one gene (filled boxes) is expressed at a given time. In average, the genetic pattern of expression (i.e. which VSG gene is transcribed) will be transmitted through ~100 T. brucei generations before a switch of transcription. Because different T. brucei parasites may express different VSG genes, the assignment of functional loci to fixed genomic positions has no functional sense.
From the last example we can define a functional locus as the whole set of genomic regions that are alternatively used to carry out the same function. Regulatory regions may be also an example of different DNA regions belonging to the same functional locus. Different elements such as promoters and enhancers regulate gene expression, and a given gene may be under the action of multiple of such regulatory elements. For example, more than half of human genes have alternative promoters [24], with an average of 3.1 promoters per gene [25], and several enhancers may regulate the expression of the same gene [26-28]. The use of multiple regulatory elements is needed for the correct tissue- and developmental-specific gene expression, but it also opens the possibility that person-to-person differences in gene expression are mediated through the alternative use of regulatory elements. I propose the hypothesis that there is a heritable trend to preferentially use certain regulatory elements rather than others. There would be differences among persons in which regulatory element is used and such differences would be heritable. The use of alternative regulatory elements would be a particular case of the more general hypothesis of different genomic regions belonging to the same functional locus.
A SIMPLE MODEL
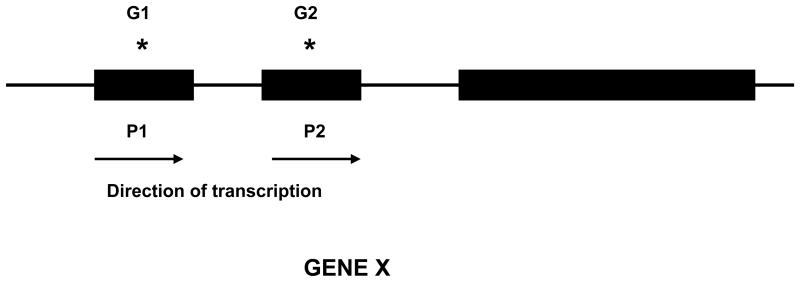
Let us know consider a simple model that may help us to understand why genetic variants detected so far through GWAS are poor predictors of disease. For simplicity, the model assumes a gene with two alternative promoters P1 and P2 (Figure 3), although the model can be generalized to more than two promoters and other regulatory elements may also be added to the model. The model assumes the existence of differences across subjects in which promoter is used; some persons will use the P1 promoter and others will use the P2 promoter. Which promoter is used may depend on epigenetic mechanisms such as methylation of cytosine residues in the DNA. A genetic polymorphism G1, with alleles A1 and A2, is located inside the P1 promoter; the A1 allele increases the risk of disease compared to the A2 allele. Another genetic polymorphism G2, with alleles B1 and B2, is located in P2 promoter; the B1 allele increases the risk of disease relative to the B2 allele.
Figure 3. Gene with two alternative promoters.
A gene X may be transcribed from two different alternative promoters P1 and P2. It is proposed the existence of person-to-person variation in which of the promoters is used. A polymorphism G1 is located inside the P1 promoter, and a different polymorphism G2 is located inside the P2 promoter.
Because current genetic association studies assume that functional variants have fixed positions, we will explore how this assumption may lead to underestimation of genetic effects. Let us suppose we carried out a genetic association study to explore the relationship between the G1 and G2 genetic polymorphisms with risk of disease. Assuming current thinking we will consider both genetic polymorphisms as different functional loci. Also we are unaware of the differential use of the P1 and P2 promoters across individuals. As Table 1 shows, the association between the G1 polymorphism and risk of disease (i.e. the A1 allele associated with higher risk of disease and the A2 allele associated with lower risk of disease) is only present when the P1 promoter is used (chromosome types 1 through 4 in Table 1). Because we do not know which promoter is being used we are unable to measure the true association between the G1 polymorphism and risk of disease; the observed association between the G1 polymorphism and risk of disease will be diluted by the presence of chromosomes in which the P2 promoter is being used. The same situation will happen for the G2 polymorphism given the fact that its association with risk of disease (i.e. the B1 allele associated with higher risk of disease and the B2 allele associated with lower risk of disease) is only present when the P2 promoter is used (chromosome types 5 through 8 in Table 1). The observed association between G2 and risk of disease will be diluted by the presence of chromosomes in which the P1 promoter is being used.
Table 1.
Risk of disease according to chromosome type
| Chromosome type |
Promoter being used |
G1 polymorphism |
G2 polymorphism |
Risk of disease |
|---|---|---|---|---|
| 1 | P1 | A1 | B1 | High |
| 2 | P1 | A1 | B2 | High |
| 3 | P1 | A2 | B1 | Low |
| 4 | P1 | A2 | B2 | Low |
| 5 | P2 | A1 | B1 | High |
| 6 | P2 | A1 | B2 | Low |
| 7 | P2 | A2 | B1 | High |
| 8 | P2 | A2 | B2 | Low |
To truly understand the genetic basis of the disease discussed in the last example we need to know which of the promoters is being used and, we need to consider both genetic polymorphisms, G1 and G2, as belonging to the same functional locus. Among subjects with both homologue chromosomes using the P1 promoter the relevant polymorphism is G1; among subjects with both homologue chromosomes using the P2 promoter the relevant polymorphism is G2; and for individuals using both promoters we need to define a new genotype consisting of one allele of the G1 polymorphism (from the chromosome using the P1 promoter) and one allele of the G2 polymorphism (from the chromosome using the P2 promoter).
BIOLOGICAL PLAUSIBILITY OF THE EPIGENETIC MODEL
The present work proposes that functional loci have no fixed positions along the genome. As discussed in the example regarding the alternative use of VSG genes in T. brucei, different physical (i.e. genetic) loci may be part of the same functional locus. Alternative promoters of the same gene in mammals offer the opportunity to search for empirical evidence of the proposed hypothesis. It is known that a considerable proportion of human genes have multiple promoters [24, 25], and the use of these alternative promoters tend to be tissue-specific [29-32]. However, there are no large-scale studies in human populations to assess inter-individual variation in the use of these alternative promoters. This study postulates that there is person-to-person variation in the use of alternative promoters and such variation can be inherited, at least in part, through epigenetic mechanisms such as DNA methylation. It is noteworthy that a recent study in twenty-six healthy subjects reported that five out of seven alternative promoters of the glucocorticoid receptor (GR) gene showed high inter-individual variability in methylation patterns [33] and there are evidence that methylation play a key role in the use of alternative promoters [34-37].
THEN, WHAT IS AN ALLELE?
The proposed hypothesis sheds new light to the concepts of gene and allele. As it has been widely noticed [38-40], our understanding of what is a gene has evolved through time because of our growing knowledge regarding the molecular biology of the DNA and RNA. Regardless of the definition of gene proposed for different authors [38-40], it is clear that the DNA sequence by itself is not enough to decide what a gene is and any meaningful definition of gene must include in some way reference to expression mechanisms. It is troublesome then, that even though the modern concept of gene involves more than the DNA sequence the allele is still defined mostly in positional terms; the single nucleotide polymorphism (SNP) representing the quintessential allele. The proposed hypothesis suggests that defining an allele just in terms of DNA physical variation without taking into account patterns of expression (for example, the alternative use of promoters) may be insufficient to grasp the whole spectrum of heritable variability. In particular, genetic polymorphisms in different physical positions may be functional variants of the same pattern of expression, and in some cases the same genetic polymorphism may be no equivalent when comparing different subjects in relationship to the phenotypic trait. The hypothesis has major implications in the fitting of risk models using genetic polymorphisms. Recent studies have reported that the addition of GWAS-identified polymorphisms to risk models of type-2 diabetes [16, 17] and breast cancer [18] does not significantly improve risk prediction when known non-genetic risk factors are taken into account. Although such results may due in part to the small effect of each individual polymorphism, the proposed hypothesis suggest that in presence of inter-individual variation of expression patterns we may be classifying subjects based on the wrong genotypes.
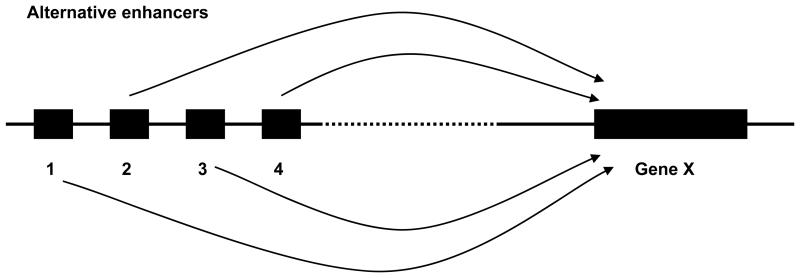
It must be noticed that the same framework of alternative promoters can be used for other types of alternative regulatory regions (for example enhancers) affecting expression of target genes (Figure 4). A ~500 kb region on chromosome 8q24 has been found to carry genetic variants associated with different types of cancer such as prostate, breast, colorectal, ovarian, and bladder cancers [1, 41-45]. It is noteworthy that the several polymorphisms identified in chromosome 8q24 are distributed in discrete regions that are associated with specific types of cancer [44]. These regions tend to overlap or be located close to putative enhancers identified through bioinformatics analyses and in vitro studies [46, 47], and at least one of those enhancers is able to physically interact with the promoter of the Myc gene located more than 300 kb far away [47]. It remains to be determined the in vivo activity of these different enhancers, and whether there are person-to-person variations in the use of the enhancers.
Figure 4. Gene under the regulation of alternative enhancers.
The gene X is regulated by alternative enhancers numbered 1 through 4. The hypothesis proposes that there is heritable person-to-person variation (mediated at least in part through epigenetic mechanisms) in the use of the alternative enhancers. Multiple potential enhancers have been found in the gene desert region in chromosome 8q24, that carry polymorphisms associated with different types of cancer.
In summary, a hypothesis is proposed that may explain in part why currently GWAS-identified genetic variants are poor predictors of complex human traits. The hypothesis makes a series of predictions that are testable, and shows new paths of research to assess the inter-relationship between genetic variants and individual patterns of expression.
ACKNOWLEDGMENTS
The author would like to thank to Julie R. Palmer, Lynn Rosenberg, and Jorge Azofeifa for their helpful comments on the manuscript. This work was supported by grants R01CA058420 and R01CA098663 from the National Cancer Institute, Division of Cancer Control and Population Science (http://www.cancercontrol.cancer.gov).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST
The author declares no conflict of interest.
REFERENCES
- 1.Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eeles RA, Kote-Jarai Z, Giles GG, Olama AA, Guy M, Jugurnauth SK, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 3.Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 4.Ferreira MA, O'Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L, et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008;40:1056–1058. doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Donovan MC, Craddock N, Norton N, Williams H, Peirce T, Moskvina V, et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008;40:1053–1055. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- 6.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 7.Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 8.Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scuteri A, Sanna S, Chen WM, Uda M, Albai G, Strait J, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3:e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, Helgadottir A, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 11.Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, Heid IM, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weedon MN, Lango H, Lindgren CM, Wallace C, Evans DM, Mangino M, et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat Genet. 2008;40:575–583. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lettre G, Jackson AU, Gieger C, Schumacher FR, Berndt SI, Sanna S, et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat Genet. 2008;40:584–591. doi: 10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gudbjartsson DF, Walters GB, Thorleifsson G, Stefansson H, Halldorsson BV, Zusmanovich P, et al. Many sequence variants affecting diversity of adult human height. Nat Genet. 2008;40:609–615. doi: 10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- 15.Hofker M, Wijmenga C. A supersized list of obesity genes. Nat Genet. 2009;41:139–140. doi: 10.1038/ng0209-139. [DOI] [PubMed] [Google Scholar]
- 16.Lango H, Palmer CN, Morris AD, Zeggini E, Hattersley AT, McCarthy MI, et al. Assessing the combined impact of 18 common genetic variants of modest effect sizes on type 2 diabetes risk. Diabetes. 2008;57:3129–3135. doi: 10.2337/db08-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meigs JB, Shrader P, Sullivan LM, McAteer JB, Fox CS, Dupuis J, et al. Genotype score in addition to common risk factors for prediction of type 2 diabetes. N Engl J Med. 2008;359:2208–2219. doi: 10.1056/NEJMoa0804742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wacholder S, Hartge P, Prentice R, Garcia-Closas M, Feigelson HS, Diver WR, et al. Performance of common genetic variants in breast-cancer risk models. N Engl J Med. 2010;362:986–993. doi: 10.1056/NEJMoa0907727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pritchard JK. Are rare variants responsible for susceptibility to complex diseases? Am J Hum Genet. 2001;69:124–137. doi: 10.1086/321272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCarthy MI, Hirschhorn JN. Genome-wide association studies: potential next steps on a genetic journey. Hum Mol Genet. 2008;17:R156–165. doi: 10.1093/hmg/ddn289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor JE, Rudenko G. Switching trypanosome coats: what's in the wardrobe? Trends Genet. 2006;22:614–620. doi: 10.1016/j.tig.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 22.Pays E. Regulation of antigen gene expression in Trypanosoma brucei. Trends Parasitol. 2005;21:517–520. doi: 10.1016/j.pt.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 23.Turner CM, Barry JD. High frequency of antigenic variation in Trypanosoma brucei rhodesiense infections. Parasitology. 1989;99 Pt 1:67–75. doi: 10.1017/s0031182000061035. [DOI] [PubMed] [Google Scholar]
- 24.Tsuritani K, Irie T, Yamashita R, Sakakibara Y, Wakaguri H, Kanai A, et al. Distinct class of putative “non-conserved” promoters in humans: comparative studies of alternative promoters of human and mouse genes. Genome Res. 2007;17:1005–1014. doi: 10.1101/gr.6030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimura K, Wakamatsu A, Suzuki Y, Ota T, Nishikawa T, Yamashita R, et al. Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 2006;16:55–65. doi: 10.1101/gr.4039406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim S, Yamazaki M, Zella LA, Meyer MB, Fretz JA, Shevde NK, et al. Multiple enhancer regions located at significant distances upstream of the transcriptional start site mediate RANKL gene expression in response to 1,25-dihydroxyvitamin D3. J Steroid Biochem Mol Biol. 2007;103:430–434. doi: 10.1016/j.jsbmb.2006.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ott CJ, Blackledge NP, Kerschner JL, Leir SH, Crawford GE, Cotton CU, et al. Intronic enhancers coordinate epithelial-specific looping of the active CFTR locus. Proc Natl Acad Sci U S A. 2009;106:19934–19939. doi: 10.1073/pnas.0900946106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zella LA, Meyer MB, Nerenz RD, Lee SM, Martowicz ML, Pike JW. Multifunctional enhancers regulate mouse and human vitamin D receptor gene transcription. Mol Endocrinol. 2010;24:128–147. doi: 10.1210/me.2009-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fajas L, Auboeuf D, Raspe E, Schoonjans K, Lefebvre AM, Saladin R, et al. The organization, promoter analysis, and expression of the human PPARgamma gene. J Biol Chem. 1997;272:18779–18789. doi: 10.1074/jbc.272.30.18779. [DOI] [PubMed] [Google Scholar]
- 30.Bulun SE, Rosenthal IM, Brodie AM, Inkster SE, Zeller WP, DiGeorge AM, et al. Use of tissue-specific promoters in the regulation of aromatase cytochrome P450 gene expression in human testicular and ovarian sex cord tumors, as well as in normal fetal and adult gonads. J Clin Endocrinol Metab. 1993;77:1616–1621. doi: 10.1210/jcem.77.6.8263150. [DOI] [PubMed] [Google Scholar]
- 31.Esterbauer H, Oberkofler H, Krempler F, Strosberg AD, Patsch W. The uncoupling protein-3 gene is transcribed from tissue-specific promoters in humans but not in rodents. J Biol Chem. 2000;275:36394–36399. doi: 10.1074/jbc.M005713200. [DOI] [PubMed] [Google Scholar]
- 32.Neuhaus IM, Bone L, Wang S, Ionasescu V, Werner R. The human connexin32 gene is transcribed from two tissue-specific promoters. Biosci Rep. 1996;16:239–248. doi: 10.1007/BF01207338. [DOI] [PubMed] [Google Scholar]
- 33.Turner JD, Pelascini LP, Macedo JA, Muller CP. Highly individual methylation patterns of alternative glucocorticoid receptor promoters suggest individualized epigenetic regulatory mechanisms. Nucleic Acids Res. 2008;36:7207–7218. doi: 10.1093/nar/gkn897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheong J, Yamada Y, Yamashita R, Irie T, Kanai A, Wakaguri H, et al. Diverse DNA methylation statuses at alternative promoters of human genes in various tissues. DNA Res. 2006;13:155–167. doi: 10.1093/dnares/dsl008. [DOI] [PubMed] [Google Scholar]
- 35.Archey WB, Sweet MP, Alig GC, Arrick BA. Methylation of CpGs as a determinant of transcriptional activation at alternative promoters for transforming growth factor-beta3. Cancer Res. 1999;59:2292–2296. [PubMed] [Google Scholar]
- 36.Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet. 2000;25:315–319. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- 37.Ventura A, Luzi L, Pacini S, Baldari CT, Pelicci PG. The p66Shc longevity gene is silenced through epigenetic modifications of an alternative promoter. J Biol Chem. 2002;277:22370–22376. doi: 10.1074/jbc.M200280200. [DOI] [PubMed] [Google Scholar]
- 38.Gerstein MB, Bruce C, Rozowsky JS, Zheng D, Du J, Korbel JO, et al. What is a gene, post-ENCODE? History and updated definition. Genome Res. 2007;17:669–681. doi: 10.1101/gr.6339607. [DOI] [PubMed] [Google Scholar]
- 39.Gingeras TR. Origin of phenotypes: genes and transcripts. Genome Res. 2007;17:682–690. doi: 10.1101/gr.6525007. [DOI] [PubMed] [Google Scholar]
- 40.Pesole G. What is a gene? An updated operational definition. Gene. 2008;417:1–4. doi: 10.1016/j.gene.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 41.Amundadottir LT, Sulem P, Gudmundsson J, Helgason A, Baker A, Agnarsson BA, et al. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38:652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 42.Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjartsson D, Helgason A, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39:631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 43.Tomlinson I, Webb E, Carvajal-Carmona L, Broderick P, Kemp Z, Spain S, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39:984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 44.Ghoussaini M, Song H, Koessler T, Al Olama AA, Kote-Jarai Z, Driver KE, et al. Multiple loci with different cancer specificities within the 8q24 gene desert. J Natl Cancer Inst. 2008;100:962–966. doi: 10.1093/jnci/djn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wokolorczyk D, Gliniewicz B, Sikorski A, Zlowocka E, Masojc B, Debniak T, et al. A range of cancers is associated with the rs6983267 marker on chromosome 8. Cancer Res. 2008;68:9982–9986. doi: 10.1158/0008-5472.CAN-08-1838. [DOI] [PubMed] [Google Scholar]
- 46.Jia L, Landan G, Pomerantz M, Jaschek R, Herman P, Reich D, et al. Functional enhancers at the gene-poor 8q24 cancer-linked locus. PLoS Genet. 2009;5:e1000597. doi: 10.1371/journal.pgen.1000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sotelo J, Esposito D, Duhagon MA, Banfield K, Mehalko J, Liao H, et al. Long-range enhancers on 8q24 regulate c-Myc. Proc Natl Acad Sci U S A. 2010;107:3001–3005. doi: 10.1073/pnas.0906067107. [DOI] [PMC free article] [PubMed] [Google Scholar]