Abstract
Nef is an HIV accessory protein that plays an important role in the progression of disease after viral infection. It interferes with numerous signaling pathways, one of which involves serine/threonine kinases. Here, we report the results of an NMR structural investigation on full-length Nef and its interaction with the entire regulatory domain of Hck (residues 72–256; Hck32L). A helical conformation was found at the N-terminus for residues 14–22, preceding the folded core domain. In contrast to the previously studied truncated Nef (Nef Δ1–39), the full-length Nef did not show any interactions of Trp57/Leu58 with the hydrophobic patch formed by helices α1 and α2. Upon Hck32L binding, the N-terminal anchor domain as well as the well-known SH3-binding site of Nef exhibited significant chemical shift changes. Upon Nef binding, resonance changes in the Hck spectrum were confined mostly to the SH3 domain, with additional effects seen for the connector between SH3 and SH2, the N-terminal region of SH2 and the linker region that contains the regulatory polyproline motif. The binding data suggest that in full-length Nef more than the core domain partakes in the interaction. The solution conformation of Hck32L was modeled using RDC data and compared with the crystal structure of the equivalent region in the inactivated full-length Hck, revealing a notable difference in the relative orientations of the SH3 and SH2 domains. The RDC-based model combined with 15N backbone dynamics data suggest that Hck32L adopts an open conformation without binding of the polyproline motif in the linker to the SH3 domain.
Keywords: NMR, HIV-1, Nef, Hck, Binding
Introduction
Human immunodeficiency virus type-1 (HIV-1) has evolved several mechanisms to evade the host’s immune defenses. This ability relies, to a large extent, on the activities of the virus' accessory proteins, Nef, Vif, Vpu, and Vpr, each of which have been found to interact with one or more host cell factors. Nef, in particular, has been reported to influence a plethora of different cellular proteins and processes.1,2 The three most prominent Nef activities are (i) engagement with components of the endocytic machinery, thereby down-regulating CD4 and major histocompatibility complex class I (MHC-I) antigens on the surface of infected cells3–8; (ii) enhancement of virus infectivity through unknown mechanisms after cell entry, as revealed by studies of long-term non-progressors that were found to be infected with virus carrying a defective nef gene9,10, and (iii) modulation of signaling pathways via direct regulation of kinase activities.11–14
HIV-1 Nef is a 23 kDa, 206 amino acid protein carrying an N-terminal myristoyl group.15 Nef proteins from different strains (>150) exhibit high sequence conservation (~85 % identity16) and mutational analyses have delineated sequence motifs important for its various functionalities. For example, an N-terminal region (residues 17–26), an acidic cluster (62-EEEE-65), and a polyproline motif (Pro69-Pro78) were shown to play a role in modulating MHC-I surface expression6,17,18 and these sequences are required for immunoprecipitation of Nef with the cytoplasmic tail of MHC-I.19 The dileucine motif (160-EXXXLL-165) in the C-terminal region is critical for downregulating CD4 surface expression7,20 and also mediates interactions with other adapter protein complexes.21 Kinase interactions22 involve the polyproline motif and the core domain11 and possibly the acidic cluster (62-EEEE-65) that plays a role in the PACS-src kinase-PI3K complex.23
Despite these remarkable and diverse activities, Nef has not been successfully exploited as a therapeutic target, primarily due to our incomplete understanding of the molecular mechanisms by which Nef exerts its action in the infected cell. In addition, although attempted in numerous laboratories, structural studies of Nef have remained problematic: the protein comprises large regions of unstructured polypeptide chain and has a pronounced propensity to aggregate at high concentrations, rendering NMR and X-ray crystallographic studies of full-length Nef a challenge. Therefore, only partial structures are available, with model building making up for the missing parts.16 The solution structure of the Nef core domain was solved first by NMR24 and soon thereafter by crystallography, both uncomplexed and complexed with the SH3 domain of the Fyn kinase R96I mutant25 or its wild-type counterpart.26 Structural features of the N-terminal domain (also called anchor domain) were deduced from NMR studies of a 25 amino acid peptide (residues 2–26)27 and of the full anchor domain (residues 2–57), with and without myristoylation.28 Both peptides exhibited little, or at best, nascent, helical structure in aqueous solution, and only in solutions of reduced water activity such as 100% methanol and 50% trifluoroethanol/50% water, stable helical structure for the Nef (2–26) peptide was found.27
Nef binds Hck, a Src-family kinase (SFK) expressed in macrophages, and this interaction promotes viral growth11 and stimulates Hck’s enzymatic activity.12 Like other SFKs, Hck has an N-terminal regulatory domain (referred to as Hck32L), comprising an SH3 domain, a short connector sequence, an SH2 domain, and a linker that joins the SH2 and the catalytic domains. A polyproline motif in the linker engages in an intra-molecular interaction with the SH3 domain in the crystal structures of the down-regulated, inactive Hck.29 Interestingly, Nef interacts with the Hck SH3 domain via its polyproline motif,11,25,30 suggesting that Nef modulates Hck kinase activity by displacing the intra-molecular SH3/PxxP interaction within Hck.12
In the present study, we used NMR to structurally characterize full-length HIV-1 Nef, alone and in complex with Hck32L. Most importantly, we present the first structural details of the N-terminal anchor domain in the context of full-length Nef in aqueous solution and its involvement in Hck binding. We also characterized NMR chemical shift perturbations of Hck32L induced by Nef binding. Substantial changes were observed for not only the SH3 domain but also the connector, the N-terminal end of the SH2 domain, and linker in Hck32L, indicating that the latter regions are also affected by Nef binding, either directly or indirectly. RDC-based structural modeling of Hck32L and 15N backbone dynamics studies of Hck32L revealed that Hck32L is in an open conformation, without contacts between the linker and the SH3 domain.
Materials and Methods
Expression and purification of proteins
The cDNA encoding full-length ‘consensus Nef’ (206 amino acids), derived by a multiple sequence alignment of 54 clinically isolated sequences from 12 patients showing low CD4 counts31, was obtained from the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. A vector containing the coding region for Hck32L (residues 72–256) was provided by T. Smithgall (University of Pittsburgh School of Medicine). The residue numbering of Hck in this work follows that of P08631 in the Swiss-Prot database. Both cDNAs were amplified and subcloned into pET21 (EMD chemicals, Inc. San Diego, CA) between NdeI/NotI and NdeI/XhoI restriction sites. For the Nef construct, Gly2 was substituted with Glu to prevent N-terminal methionine (+/−) heterogeneity. Proteins containing C-terminal hexa-His tags were expressed in E. coli, Rosetta 2 (DE3) (EMD chemicals, Inc. San Diego, CA), cultured in Luria-Bertani or modified minimal media, and induced with 0.5mM IPTG at 18°C for 14h. Uniform 2H/13C/15N labeling of Nef and Hck32L was carried out by growth in modified minimal medium using 15NH4Cl, [13C6,2H7]-glucose and 99.9% 2H2O as the sole nitrogen, carbon and deuterium sources, respectively. Uniform 15N labeling of Hck32L was carried out by growth in modified minimal medium using 15NH4Cl as the sole nitrogen.
Both Nef and Hck32L proteins were purified using a 5mL HisTrap Ni2+-chelating column (GE Healthcare, Piscataway, NJ) and aggregated material was removed by gel-filtration using a Hi-Load Superdex200 26/60 column (GE Healthcare, Piscataway, NJ), equilibrated with a buffer containing 25mM sodium phosphate, pH 7.5, 50mM NaCl, 1mM DTT, and 0.02% NaN3. The final purification step involved ion-exchange over a 5mL Hi-Trap Q HP column (GE Healthcare, Piscataway, NJ) with a 0 – 1M NaCl gradient in a buffer containing 25mM sodium phosphate, pH 7.5, 1mM DTT, and 0.02% NaN3. Proteins were concentrated to 0.1mM using Amicon Ultra-15 concentrators (Millipore, Billerica, MA) after buffer exchange to 10mM HEPES, 10mM DTT, 100mM NaCl, and 6%(v/v) D2O, pH 8.0 (NMR buffer). Molecular masses of the purified proteins were assessed by mass spectrometry, and labeling efficiency was found to be > 97% for 13C and 15N atoms and > 90% for 2H atom.
NMR spectroscopy
All NMR spectra were recorded at 300K using Bruker AVANCE 900, 700 or 600MHz NMR spectrometers, equipped with 5mm triple-resonance, three-axes gradient probes or z-axis gradient cryoprobes. The temperature was calibrated using 100% methanol, and the chemical shifts were referenced to external DSS. Spectra were processed with NMRPipe32 and analyzed using SPARKY33 (version 3.115) and CARA34. Backbone and Cβ chemical shift assignments of free Nef were obtained using TROSY-type 2D HSQC, 3D HNCACB and HN(CO)CACB35 experiments, employing deuterium decoupling, as well as conventional 3D HNCA, HN(CO)CA, HNCO, HN(CA)CO and 15N-edited NOESY experiments36,37 on a 0.1mM 2H/13C/15N labeled Nef sample in the NMR buffer. TROSY-type 3D HNCACB and HN(CO)CACB NMR experiments were recorded using 192 scans and a total data acquisition time of 131 hours per experiment. 3D HNCA, HN(CO)CA, HNCO and HN(CA)CO spectra were recorded with 96, 160, 48 and 64 scans, respectively, and total acquisition times of 88, 128, 85 and 114 hours.
Similar experiments were also carried out for a 0.1mM 2H/13C/15N labeled Nef sample in the presence of 0.12mM unlabeled Hck32L for deriving assignments of bound Nef. Unfortunately, the Hck32L-bound Nef sample provided much poorer 3D NMR data, even if more scans were used (256, 512 and 512 scans for the TROSY-type HNCACB, HN(CO)CACB and HNCA experiments, respectively) than for free Nef, preventing complete assignments in a straightforward manner. Therefore, TROSY-HSQC resonances of Hck32L-bound Nef were assigned by comparison with the free Nef spectrum only. Thus, we only obtained the "most likely" assignments, and not independently verified accurate ones, resulting in lower limit values for any chemical shift changes. Although imprecise, mapping of binding sites and assignment of bound resonances using this approach has been applied previously to other systems with unfavorable NMR properties.38
Backbone and Cβ chemical shift assignments of Hck32L, both without and with Nef, were obtained using a similar set of heteronuclear NMR experiments in the same buffer that was used for Nef alone. Hck32L samples (0.5mM 2H/13C/15N-labeled) were analyzed in the absence or presence of 0.6mM unlabeled Nef. Additional NMR experiments were carried out at pH 7.2 to detect fast exchanging amides.
Secondary chemical shifts were obtained using a term of ΔCα minus ΔCβ where the ΔCα and ΔCβ values were calculated by subtracting random coil Cα and Cβ shifts39 from measured values, with sequence-dependent correction.40
1H-15N residual dipolar couplings (RDC) were measured for 2H/13C/15N-labeled Hck32L at 700MHz using a sample in 5% C12E5/hexanol (r=0.96) as the alignment medium.41 IPAP 1H-15N HSQC spectra42 were recorded with 1024*128 complex points. RDC data were analyzed with PALES43 and MODULE.44
An RDC-based NMR structural model was obtained using MODULE44 where domains were subjected to rigid body rotation around the Asp137 C' - Ser138 N bond to match their alignment tensor. The connector (residues 138 to 143) between SH2 and SH3 and the linker (residues 242 to 256) were not treated as separate domains in the RDC data analysis since not enough unambiguous RDC data for these short segments were obtained. Instead, they were included in the SH2 domain. For the final NMR structural model, energy minimization in Cartesian coordinate space was carried out using XPLOR-NIH45 in which only residues 136–138 were allowed to move. An arbitrary x, y and z coordinate system was created using the 1AD5 structure to describe the orientation of the SH3 domain relative to the SH2 domain: the center of mass of the SH2 domain was placed at the origin of the coordinate system, the y axis was defined by the vector between centers of mass of the SH2 and the C-lobe of the catalytic domain, the yz plane was defined by three centers of mass (SH2 domain, N-lobe, C-lobe), and the x axis is the normal to the yz plane.
The structural model for the Hck32L/Nef complex was generated by superimposing the SH3 domain in the Hck32L NMR structure onto that in the Fyn SH3/Nef complex structure.25 The structural model for the full-length Hck in an open conformation was obtained by replacing the regulatory domain portion (residues 81–256) of the 1AD5 full-length crystal structure with the Hck32L model. The structural model for the full-length Hck/Nef complex was generated by superimposing the SH3 domain in the open full-length Hck model structure for onto that in the Fyn SH3/Nef complex structure.25
Backbone relaxation measurements of R1, R2 and heteronuclear {1H}-15N NOEs46 were carried out on 0.25mM 15N-labeled Hck32L at 700MHz. Relaxation delays in the R1 experiments were 50, 100, 200, 400, 700, 900 and 1500ms and spectra with 50ms and 400ms delays were collected twice for error estimation. The CPMG mixing times in the R2 experiments were 8, 16, 24, 32, 40 and 48ms using 1024*128 complex points and spectra with 16ms and 24ms were recorded twice for error estimation. Peak intensities were fitted using the CURVEFIT program (A.G. Palmer III, Columbia University). The heteronuclear {1H}-15N NOE experiment was recorded with an inter-scan delay of 6s (for the reference spectrum) and with presaturation using a 120 degree pulse train with 5ms spacing delay during the last 3s of the inter-scan delay (for the NOE spectrum). Relaxation data for the backbone amides for 49 SH3 residues (out of 65 non-proline total), all 6 connector amino acids, 74 SH2 residues (out of 94 non-proline total) and 12 linker residues (out of 13 non-proline) were obtained. For the remaining residues either significant resonance overlap or broadening due to chemical exchange precluded their use.
Results and Discussion
Sequential NMR assignment and structural characterization of full-length Nef
Structural characterization of non-myristoylated, full-length Nef (1–206) was carried out by NMR and compared to structural data available by NMR and X-ray crystallography (Figure 1A). Previously, NMR studies on full-length Nef had been difficult given the presence of large disordered/flexible regions that cause extensive resonance broadening and degeneracy24,47 as well as aggregation that hindered crystallization.25 Here we carried out extensive sample screening by dynamic light scattering and 1H-15N HSQC spectroscopy, varying protein concentration, buffer compositions, and temperature to derive conditions that were suitable for NMR analysis (data not shown). Nef was found to exhibit the best spectral properties in 10mM HEPES buffer, 100mM NaCl, 10mM DTT, pH 8.0, and it was possible to concentrate a 2H/13C/15N -labeled protein sample to 0.1mM without significant aggregation/NMR resonance broadening. At concentrations above 0.1mM, however, poor 1H-15N HSQC spectra and aggregation were noted (data not shown).
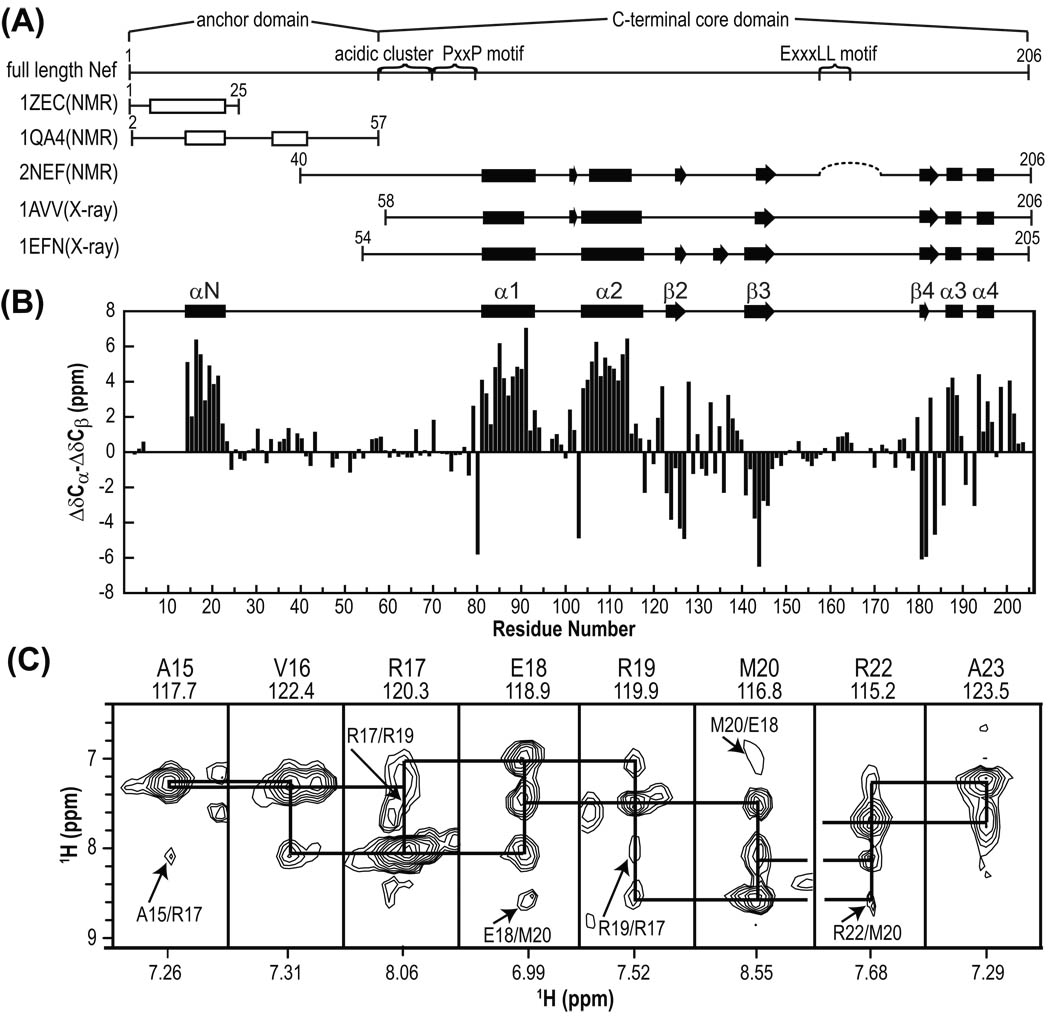
Figure 1.
Secondary structure elements of Nef. (A) Schematic representation of various Nef constructs whose structures have been determined. Locations of functionally important sequence motifs and domain boundaries in full-length Nef are indicated as well. Helices and sheets identified in the structures are depicted by rectangles and arrows, respectively. A helix, identified for the 1ZEC structure determined in a organic solvent mixture and the two nascent helices in 1QA4 in aqueous solution are depicted as open rectangles. The dotted line in 2NEF indicates the loop deletion in the protein used for NMR. (B) Secondary chemical shifts (ΔCα–ΔCβ) and secondary structure elements in full-length Nef determined here. (C) Strip plots of selected regions of the 3D 15N-edited NOESY spectrum at 800MHz of 2H/13C/15N-labeled Nef illustrating sequential HN-HN NOEs, with i, i+2 NOEs are marked by arrows and residue names. The residue name and number as well as amide 15N and 1H frequencies (in ppm) are shown at the top and bottom of each strip, respectively. The break between Met20 and Arg22 is caused by a missing assignment for the 15N resonance for Arg21.
For our optimized solution conditions, TROSY-type 3D HNCACB and HN(CO)CACB, conventional 3D HNCA, HN(CO)CA, HNCO, HN(CA)CO and 15N-edited NOESY experiments on 2H/13C/15N-labeled samples yielded ~90% backbone assignments for all 206 Nef residues (Figures 1B and 2). In addition, all 7 tryptophan indole side chain HNε1 resonances (located in the bottom left region of Figure 2A) were assigned using 3D 15N-edited NOESY (data not shown) and available assignments for truncated Nef.28,47
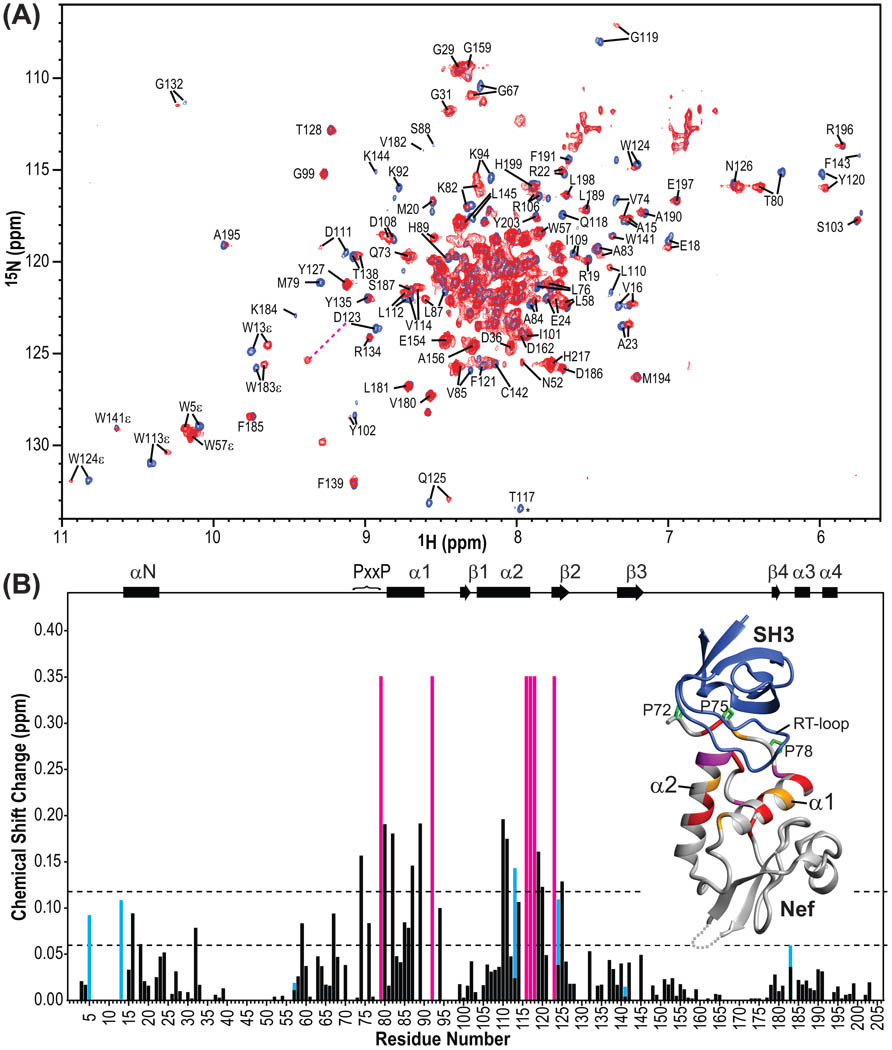
Figure 2.
Chemical shift changes of Nef upon Hck32L binding. (A) Superposition of the 900MHz TROSY spectra of 2H/13C/15N Nef in the absence (blue) and presence (red) of unlabeled Hck32L. Resonances are labeled by residue name and number. (B) 1H and 15N combined chemical shift differences between the free and Hck32L-complexed Nef resonances as a function of residue number. The combined chemical shift changes were calculated using Δδ=√(ΔH2+(0.15*ΔN)2). Residues that experience very large chemical shift changes and/or severe line broadening upon Hck32L binding, such as Met79, His116, Thr117, Gln118 and Asp123, are depicted in magenta bars (arbitrarily set to 0.35 ppm). The combined chemical shift changes for the Trp indole HNε groups are depicted in cyan bars. Locations of helices (cylinders) and β strands (arrows) as well as the PxxP motif in full-length Nef are displayed at the top. Inset: Mapping of the chemical shift changes onto the ribbon representation of the crystal structure of the Nef core/Fyn SH3 complex (PDB:1EFN25). The grey dotted line indicates the disordered loop (residues 149–178) in the crystal structure. Nef residues colored in red, orange and grey exhibit 1H,15N-combined chemical shift changes >0.12 ppm, between 0.06 and 0.12 ppm, and < 0.06 ppm, respectively. Residues associated with the magenta bars are colored magenta in the structure. Proline residues in the PxxP motif of Nef are shown in green stick representation. The Fyn SH3 structure is shown in blue ribbon representation.
The secondary structure of full-length Nef (Figure 1B) was derived using secondary chemical shifts.48 For the folded core region (residues 74–206) in full-length Nef, excellent agreement with the previous core NMR (2NEF; Δ2–39/Δ159–173 construct)24,47 and X-ray structures (1AVV; Δ2–57 construct and 1EFN; Δ2–53 construct) (Figure 1A and B)25,26 is noted. Some small differences, however, are observed. For example, the short β strand for residues Ile101-Tyr102 in 2NEF and 1AVV is not present, similar to what is seen in 1EFN. Both residues exhibit positive 13C secondary shifts (Figure 1B), incompatible with β-strand phi/psi angles. For residues 123–127 our data indicates a β strand in full-length Nef, identical to what is seen in 2NEF, and 1EFN, but not in 1AVV.
For the N-terminal anchor domain (residues 1–57), positive 13C secondary chemical shifts of significant size (1.5–6.3 ppm) are seen for residues Pro14-Arg22, indicating a well-defined helix.48 This alpha helical structure was corroborated by strong sequential (i, i+1) and medium-intensity, (i, i+2) HN-HN NOEs in the 3D 15N-edited NOESY spectrum (Figure 1C). These NMR data clearly indicate a well-structured helix in the anchor region of a full-length Nef in aqueous solution. Helical structure in this region for Pro14-Arg22 and Arg35- Gly41 of the anchor domain was previously observed in a Nef anchor peptide (residues 2–57) that exhibited very small (<0.2 ppm) Hα secondary shifts and a couple of sequential and medium-range NOEs.28 Our present data show that the only alpha-helical region in the N-terminal anchor domain resides between Pro14 and Arg22 while for the remaining residues including Arg35-Gly41, no helical propensity was seen. Therefore, the structure of the N-terminal anchor region in full-length Nef is not identical to that observed for the isolated peptide.
In the early NMR structure of the Nef core domain (2NEF) only a small portion of the N-terminal anchor domain was present (NefΔ2–39 or Δ2–39/Δ159–173).24,47 In this structure, Trp57 and Leu58 were found to interact with a hydrophobic patch on helices α1 and α2 of the core domain. In our current 3D 15N-edited NOESY data for full-length Nef, we did not find any NOEs to support this interaction. Indeed, the chemical shifts of the Trp57 and Leu58 NH resonances in full-length Nef are quite different from those in NefΔ2–39: the backbone 1H/15N chemical shifts of Trp57 and Leu58 of the full-length protein are 7.83/118.4 ppm and 7.66/122.4 ppm (Figure 2A), respectively, while those of NefΔ2–39 are resonating at 7.32/116.5 ppm and 6.82/120.9 ppm,24,47 respectively. Thus, Trp57 and Leu58 in the truncated Nef are structurally different compared to full-length Nef, possibly caused by the truncation of the sequence.
Resonances for residues in the C-terminal loop (Glu149-Lys178) exhibited very little chemical shift dispersion in their 1H and 15N chemical shifts (located in the crowded central region in Figure 2A) and exhibited only small (<1 ppm) 13C secondary shifts (Figure 1B), indicative of random coil structure. Again, this finding is in perfect agreement with previous results from NMR24,47 and X-ray25,26 studies, suggesting a flexible, unstructured chain in this region.
Binding site mapping of Hck32L on full-length Nef
The interaction between Nef and Hck32L was mapped using chemical shift differences observed in the 1H-15N TROSY spectra of a 2H/13C/15N labeled Nef in the absence and presence of unlabeled Hck32L (Figure 2A). It was observed that exchange between bound and free Nef is slow on the chemical shift scale: adding aliquots of a stock solution of Hck32L to the sample of labeled Nef at sub-stoichiometric ratios, the intensities of the free Nef resonances gradually decreased whereas those of the bound resonances increased. This slow exchange is in agreement with the reported strong affinity (Kd = 0.25µM for Hck SH3 binding to Nef30). Unfortunately, very low resonance intensities were observed in the 3D TROSY-type HNCACB and HN(CO)CACB of the complex, preventing assignment in a straightforward manner. Therefore, assignments for the Hck32L-bound Nef (Figure 2A) were deduced by comparing the 2D 1H-15N TROSY spectrum of free Nef with that of bound Nef (see the details in Materials and Methods). Combined 1H and 15N chemical shift differences between the free and Hck32L-complexed Nef resonances along with residue number are depicted in Figure 2B. Chemical shift changes for all seven tryptophan indole HNε1 resonances of full-length Nef induced by Hck32L binding were also extracted (cyan bars in Figure 2B). Overall, the structural mapping of all chemical shift changes (Figure 2B inset) agrees well with the X-ray structure of the Nef core-Fyn SH3 complex.25 Specifically, the most substantial changes (>0.12 ppm) are associated with the PxxP motif, helixes α1 and α2 and the turn following α2.
Interestingly, several residues in the N-terminal anchor domain, such as Trp5, Trp13, Val16, Glu18 and Ala32, also exhibited some chemical shift changes (>0.06 ppm), upon Hck32L binding, suggesting the anchor domain may be involved or influenced by the interaction (Figure 2B). In particular, Val16 and Glu18 in the N-terminal helix (αN) are affected.
Structural characterization of the Hck regulatory domain
Resonance assignments (>95% of all backbone resonances) for the entire regulatory region of Hck32L (residues 72–256), comprising the SH3 domain, a short connector sequence, the SH2 domain, and a 15 residue linker that tethers the regulatory region to the kinase domain, were readily obtained (Figure 3A and B). Only the backbone 1H-15N resonances of residues 94 and 95 at the tip of the RT loop in SH3 and those of residues 175–178 in the phosphotyrosine-binding loop in SH2 could not be detected/assigned, due to severe line broadening, most likely caused by conformational averaging on the micro- to millisecond timescale. A similar lack of the RT loop resonances was noted in the NMR spectrum of the isolated SH3 domain of Hck.49
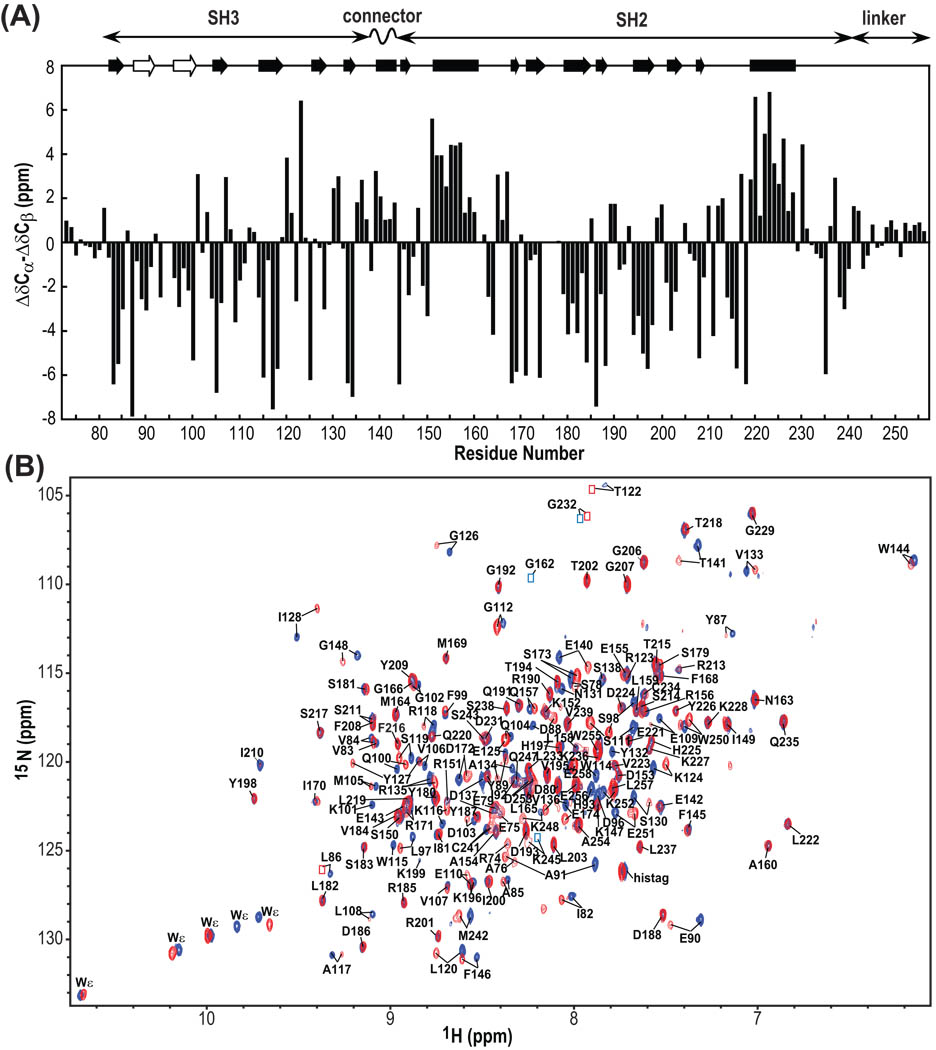
Figure 3.
13C secondary chemical shifts of Hck32L (A) and 900MHz TROSY spectra of free and Hck32L-complexed Nef (B). (A) Secondary chemical shifts (ΔCα–ΔCβ). At the top, helical and sheet segments that were identified from the crystal structure of a down-regulated full-length Hck (1AD5) as well as from our NMR work with Hck32L in solution (this study) are depicted by solid rectangles and arrows, respectively. The beta strands that are detected only in our NMR study of Hck32L are shown with open arrows. The domain locations of Hck32L are shown at the top. (B) Superposition of the 900MHz TROSY spectra of 2H/13C/15N Hck32L in the absence (blue) and presence (red) of unlabeled Nef. Assigned resonances are labeled with residue name and number. Resonances that are too weak for detection at this contour level, but clearly visible at lower levels, are indicated by blue (free Hck32L) and red (Nef-bound Hck32L) boxes.
Secondary structure characterization based on 13C secondary chemical shifts agrees well with the crystal structures of the down-regulated form of full-length Hck29 and Hck32L50, except for the RT loop region in the SH3 domain (Figure 3A). The negative secondary shifts observed for residues 87–91 and 96–100 clearly suggest that these residues are involved in forming β-strands in solution. Again, this is in agreement with NMR data for the SH3 domain complexed with a proline-rich peptide.51
The PxxP motif-containing linker (residues 242–256) and the connector (residues 138–143) regions in Hck32L were carefully analyzed due to their possible important role in regulation of the kinase activity.12,29,52,53 Only small 13C secondary chemical shifts were observed for linker residues (<1.4 ppm, Figure 3A), suggesting a mostly random conformation. The connector region, on the other hand, exhibited large, positive 13C secondary shifts (up to 3.2 ppm, Figure 3A), strongly implicating that the connector region is helical, similar to its conformation in full-length Hck.29
SH3-SH2 interdomain orientation in Hck32L
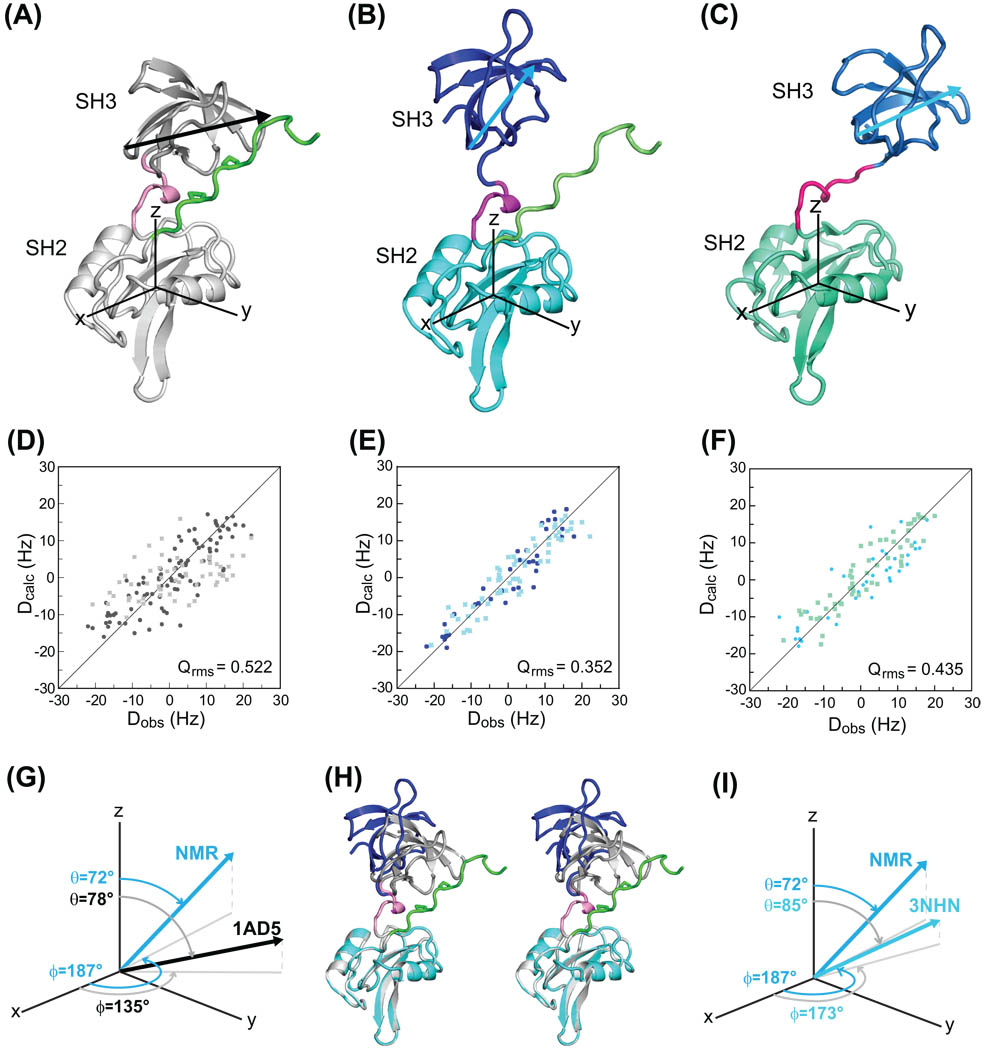
Residual dipolar couplings are ideally suited to establish relative domain orientations in multi-domain proteins. Since no a priory assumptions can be made about similarities or differences in the domain orientations in free Hck32L, we measured residual dipolar couplings and compared the experimental values to those predicted from the X-ray structure of the inactivated form of full-length Hck (PDB ID 1AD529). For alignment tensor determination, we used the coordinates of residues 81–137, 144–241, and 81–256 of 1AD5 as input models for the SH3, SH2 and SH3-connector-SH2-linker structural regions, respectively. For the SH3 and SH2 domains, the predicted RDCs agree well with the experimental ones: the quality factors, Qrms, for the SH3 and SH2 domains was 0.257 and 0.261, respectively, with an overall correlation coefficient of 0.950 for the SH3 and 0.944 for the SH2 (data not shown). The excellent agreement between predicted and measured RDCs confirms that within the domains no significant differences between the solution structure and the X-ray structure are present. However, using the overall structure of the entire regulatory domain of Hck of 1AD5 (residues 81–256, Figure 4A), a poor match between the predicted and measured values was noted, with a quality factor of 0.522 and a correlation coefficient of 0.851 (Figure 4D, bottom panel), indicating the relative SH3 to SH2 domain orientations of Hck32L in solution is different from that of the X-ray structure of inactivated full-length Hck (1AD5). In order to derive a RDC-based NMR structural model of Hck32L in solution, we used rigid body rotation of one domain with respect to the other to match the alignment tensors of both domains using MODULE.44 This NMR model (Figure 4B) exhibits a quality factor of 0.352 and a correlation coefficient of 0.936 (Figure 4E, bottom panel). Since the NMR model was generated using RDC data only, absolute positioning cannot be achieved since no translational information is available. In order to graphically depict the different relative orientations, we selected the vector connecting the Gly102 Cα and Ser111 Cα atoms in SH3 (depicted as arrows in Figure 4) for describing the orientational difference in the two structures: the rotation (ϕ) and tilt (θ) angles for this vector in the 1AD5 crystal structure are 135° and 78°, respectively, while the equivalent ones for the solution Hck32L structure are 187° and 72°, respectively (Figure 4G). As clearly seen in the superposition of these two structures (Figure 4H), the PxxP binding site of the SH3 domain in the Hck32L solution model faces away from the linker, in contrast to its conformation in the crystal structure of the down-regulated full-length Hck. The recent crystal structure of Hck32L, comprising the entire regulatory region (3NHN50, Figure 4C), was also used as the input model to predict RDCs, yielding fairly good agreement with a quality factor Qrms of 0.435 and a correlation coefficient of 0.9 (Figure 4F). Note that excellent agreement was obtained when the individual SH3 and SH2 domain structures in 3NHN were separately fit to the RDC data: Qrms values were 0.195 and 0.281 and the overall correlation coefficients were 0.98 and 0.96, respectively. Similar rotation and tilt angles for the vector connecting the G102 Cα and S111 Cα atoms in SH3 relative to the coordinate frame of the SH2 domain are observed (Figure 4I, NMR-Hck32L ϕ=187° and θ=72°; Xray-Hck32L ϕ=173° and θ=85°). Therefore, the difference in domain orientation in the solution Hck32L structure, compared to that in the X-ray structure of the down-regulated full-length protein, is predominantly determined by the nature of the shorter protein.
Figure 4.
Comparison of the relative orientations of the SH3 and SH2 domains in a down-regulated, full-length Hck and Hck32L. (A), (B) and (C) depict the structures for residues 82–260 in full-length Hck (PDB-ID: 1AD529), the NMR structure (this study) and the crystal structure of Hck32L (PDB-ID: 3NHN50), respectively, in ribbon representation. The two proline sidechains (246 and 249) are shown in stick representation in (A). The relative orientations of the SH3 and SH2 domains are graphically depicted with respect to a coordinate system whose origin resides at the center of mass of the SH2 domain and illustrated using a vector connecting the G102 Cα and S111 Cα atoms in SH3. (D), (E) and (F) are correlation plots between observed (Dobs) and calculated (Dcalc) backbone 1H-15N RDCs, using 1AD5 (D), the RDC-derived NMR model of Hck32L (E) and 3NHN Hck32L (F) as input models. (G) and (I), Tilt (θ) and projection angles onto the x,y plane (ϕ) are used to describe the G102 Cα - S111 Cα vector in 1AD5 and 3HNH compared to the NMR model, respectively. (H), Stereoview of the superposition of the X-ray structure of the regulatory domain from 1AD5 (grey) and the RDC-based NMR structure of Hck32L (blue/cyan), best fitting the SH2 domains (grey/cyan).
Dynamics properties of Hck32L
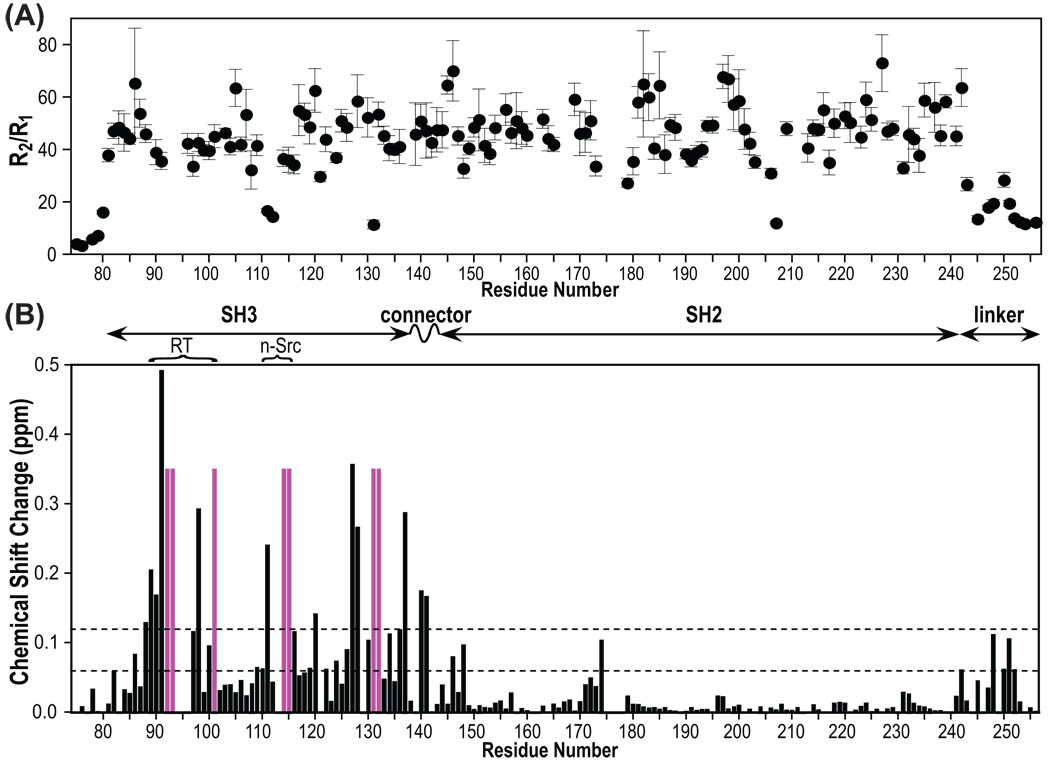
The backbone dynamics properties of Hck32L were assessed from {1H}-15N heteronuclear NOE, R1 and R2 relaxation data (Supplementary Material, Figure S1). As can be appreciated from the R2/R1 ratios for each residue (Figure 5A) flexible regions comprise the N-terminus (residues 72–80) and the C-terminal linker (residues 242–256). Flexibility for these parts is also indicated by smaller heteronuclear NOE values compared to the rest of the Hck32L protein (Supplementary Material, Figure S1). The SH3, connector and SH2 domains, on the other hand, exhibit similar low R1, high R2 and high NOE values, despite their differences in domain size (57, 6, and 98 residues, respectively), indicating that the protein behaves motionally as a single unit. The observed average R2/R1 value is ~46 (Figure 5A), yielding an estimated rotational correlation time of ~18ns, corresponding to what would be expected for a ~30kDa protein. 54,55 Somewhat lower than average R2/R1 values are observed for a couple of loop residues (Ser111, Gly112 and Asn131 in SH3 and Gly207 in SH2) within each domain, possibly indicating higher flexibility at theses sites.
Figure 5.
R2/R1 15N backbone relaxation rate ratios of Hck32L (A) and chemical shift perturbation of Hck32L upon full-length Nef binding (B). R1 and R2 data are provided in Supplementary Material. Chemical shift differences were extracted from the TROSY spectra provided in Figure 3B and calculated as described in the Figure 2B caption. Domain organization of Hck32L is depicted at the top.
Structural perturbation of Hck32L induced by Nef binding
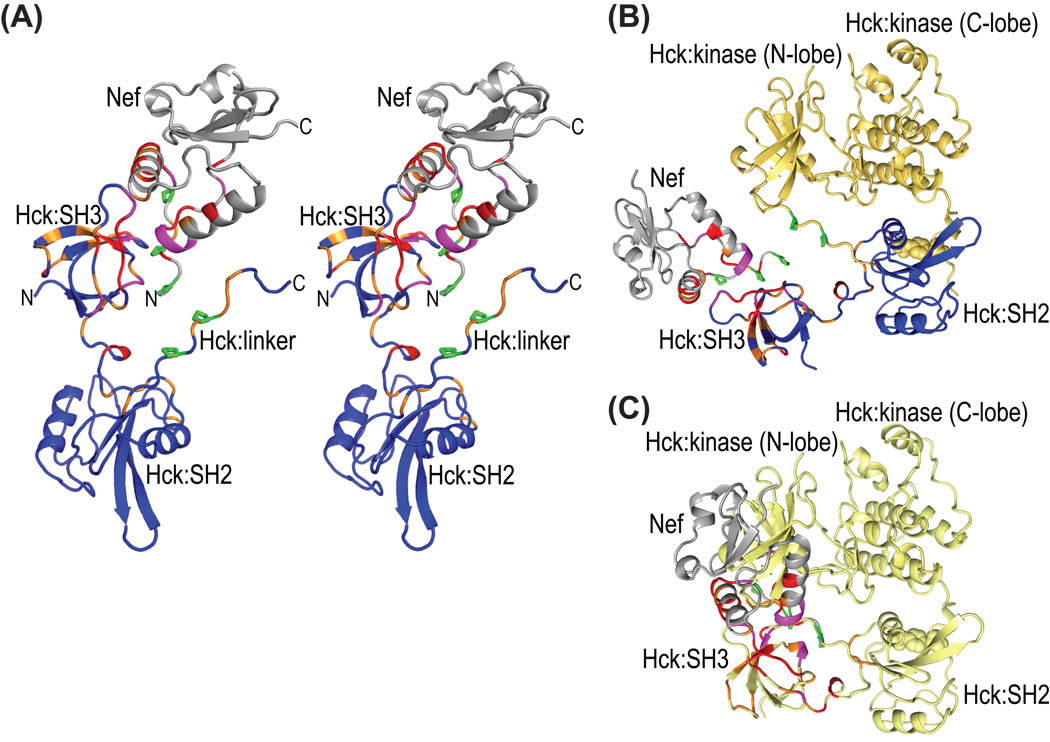
In contrast to the poor 3D NMR spectral data from triple-labeled Nef in complex with Hck32L, the 2H/13C/15N-Hck32L protein in the presence of Nef yielded high quality 3D TROSY-HNCACB and HN(CO)CACB data, allowing us to achieve almost complete backbone assignments (>90%; Figures 3). Therefore, chemical shift perturbations induced by Nef binding could be extracted in a straightforward manner (Figure 5B). Structural mapping of the chemical shift changes in Hck32L induced by Nef (Figure 5B) as well as those in Nef induced by Hck32L (Figure 2B) is provided in Figure 6A using a Hck32L-Nef complex model in which our NMR Hck32L structure (Figure 4B) was merged with a truncated Nef/Fyn SH3 complex crystal structure (1EFN).25 Residues exhibiting substantial chemical shift perturbations (>0.12 ppm) upon Nef binding are located mainly in the SH3 domain of Hck32L, particularly in the RT loop (residues 88–101), n-Src loop (Ser111-Lys116), and C-terminal region (residues 127–137) (Figure 5B). Gratifyingly, these spectral changes are all confined to areas of interaction in the Nef/Fyn SH3 complex (1EFN).25
Figure 6.
Structural mapping of all chemical shift changes for complex formation between full-length Nef and Hck32L onto a ribbon representation of the Nef-Hck model. (A) Stereoview of the structural model of the Hck32L/Nef complex in ribbon representation, colored using the same code as in Figures 2B and 5B based on the NMR binding data. Hck32L is shown in blue and Nef in grey with residues exhibiting changes >0.12 ppm and between 0.06 and 0.12 ppm colored red and orange, respectively. Those residues whose resonances experience severe line-broadening are colored magenta. Side chains of Pro72, Pro75 and Pro78 in Nef and Pro246 and Pro249 in the linker of Hck32L are depicted in green stick representation. (B) Structural model (ribbon representation) of the full-length Hck-Nef complex generated by assuming that the same relative SH3-SH2 orientation as in our current NMR model is present in full-length Hck. The same color scheme as in (A) is used for Nef and the SH3-connector-SH2 portion of Hck. The backbone structure of the linker and kinase region is shown in yellow. (C) Illustration of the steric interference created if Nef (grey) is added to the 1AD5 full-length Hck structure (yellow ribbon). The chemical shift changes upon complex formation (data and color-coding as in A) are also colored for Nef and Hck32L.
In addition to the well-established contact sites on the SH3 domain, the connector (Glu140 and Thr141, ~0.17 ppm), the N-terminus of the SH2 domain (Phe146 and Gly148, ~0.09 ppm), and Glu174 (~0.1 ppm) in the SH2 domain exhibit chemical shift changes upon Nef binding (Figure 5B). These regions are all close to the SH3 domain and near the N-terminal tail of Nef (Figure 6A). Indeed, in the complementary mapping experiment, the N-terminal Nef region experienced significant chemical shift perturbations upon binding Hck32L (Figure 2B). Therefore, the connector, the N-terminal portion of SH2, and the region around Glu174 may all be implicated in the interaction with the N-terminal region of Nef.
In the PxxP motif region (246-PQKPWE-251), notable spectral changes are seen for Lys248 and Glu251 (~0.11 ppm). In the crystal structure of the full-length Hck a large number of intra-molecular interactions are present between Pro246, Lys248, Pro249 and Glu251 and the SH3 domain and between Gln247, Lys248, Trp250, Glu251, Ala254 and Trp255 and the N-lobe of the kinase domain. These interactions, together with the intra-molecular contact between the phosphotyrosine 522 that gets buried in the SH2 domain, create the closed, inactivated conformation of Hck.29 Since Nef-Hck binding involves the interaction of the Nef PxxP motif with the SH3 domain of Hck (Figures 2 & 6A), one may expect that perturbations in the linker (Figures 5B & 6A) could be caused simply by the replacement of an intra-molecular Hck PxxP/SH3 interaction with an inter-molecular Nef PxxP/Hck SH3 interaction. However, there is no compelling experimental data that would suggest the presence of an intra-molecular interaction in the Hck32L protein in solution. Indeed, the linker appears to be highly mobile and in a random conformation (Figure 5A and Supplementary Material Figure S1). In addition, the NMR structural model of Hck32L does not support such an interaction (Figure 4B). Therefore, Hck32L exhibits an open conformation in contrast to the closed conformation present in the inactivated full-length Hck. Whether the spectral changes in the linker upon Nef binding (Figure 5B) are due to a direct or indirect effect cannot be ascertained unambiguously at present.
Structural modeling of the full-length Hck/Nef complex suggests that Nef could bind to full-length Hck without steric clash if the relative SH3-SH2 orientation of Hck would be similar to the one in the Hck32L NMR structure (Figure 6B). In contrast, severe steric clashes ensue when attempting to position Nef into the inactivated, closed full-length Hck (Figure 6C).
Conclusion
We used solution NMR methods to fill some of the gaps in our knowledge about full-length Nef. We identified a well-structured helical region (residues Pro14-Arg22) in the N-terminal anchor domain in aqueous solution, in contrast to a nascent helical conformation reported in NMR studies of anchor peptides. Given that the Nef anchor domain is essential for membrane binding and mediates the interaction with membrane bound partners56,57, the presence of helix in this region may aid in such binding events. In addition, the present data reveal a minor structural difference between full-length and truncated Nef, namely the lack of the contact between the Trp57 and Leu58 residues in the N-terminal anchor domain and the hydrophobic patch in the core domain that is seen in the Δ2–39 deletion mutant.47 We believe that the difference arises from the truncation and that in full-length Nef, this hydrophobic patch on the core domain can interact with other proteins partners, as has been reported for the TCR zeta ITAM fragment58 and a CD4 cytoplasmic peptide.59
In addition, our RDC-based analysis of the relative domain orientation of the SH3 and SH2 domains in the Hck32L protein revealed that the relative orientation of these domains is quite different from their arrangement in the X-ray structure of the full-length, down-regulated kinase (PDB-ID:1AD5). This difference (Figures 4G & H) may represent different functional states (open vs. closed conformation) and similar conformational variability involving the SH3 domain has been observed in the Fyn kinase.60 Based on the 15N heteronuclear relaxation data for Hck32L, we find that no intra-molecular interaction is present between the SH3 domain and the linker, consistent with an open conformation. This is similar to findings in the recent X-ray structure of Hck32L where no density was observed for the linker and an open conformation was proposed.50
In addition, the current NMR binding site mapping for complexation of full-length Nef with Hck32L not only confirmed previously established binding interfaces, but also identified the N-terminal domain of Nef and the connector, the N-terminus of SH2 and the linker in Hck32L as possible extended interaction sites.
Supplementary Material
Acknowledgements
We thank Drs. Teresa Brosenitsch, Rieko Ishima, and Joseph Walsh for useful discussions, Jason Concel and Hyun Lee for help with protein purification and Mike Delk for NMR instrumental support. Dr. Stephan Grzesiek kindly provided backbone resonance assignments of free and Hck SH3-bound NefΔ1–39 and Dr. Thomas Smithgall a vector containing the coding region for Hck32L (residues 72–256). Dr. Teresa Brosenitsch is also gratefully acknowledged for expert editorial help. This work was supported by the National Institute of General Medical Sciences (NIH Grant P50GM082251) and is a contribution from the Pittsburgh Center for HIV Protein Interactions.
Abbreviations
- HIV
human immunodeficiency virus
- Hck
Hemopoietic cell kinase
- Hck32L
N-terminal regulatory domain of Hck comprising the SH3 domain, the connector sequence, the SH2 domain and a linker between the SH2 and the catalytic domain
- MHC-I
major histocompatibility complex class I
- SFK
Src-family kinase
- TFE
2,2,2-Trifluoroethanol
- SDS
sodium dodecyl sulfate
- IPTG
isopropyl b-D-1-thiogalactopyranoside
- DTT
dithiothreitol
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- DSS
4,4-dimethyl-4-silapentane-1-sulfonic acid
- C12E5
pentaethylene glycol monododecyl ether
- TROSY
transverse relaxation optimization spectroscopy
- NOE
nuclear Overhauser effect
- NMR
nuclear magnetic resonance
- HSQC
heteronuclear single quantum coherence
- RDC
residual dipolar coupling
References
- 1.Roeth JF, Collins KL. Human immunodeficiency virus type 1 Nef: adapting to intracellular trafficking pathways. Microbiol Mol Biol Rev. 2006;70(2):548–563. doi: 10.1128/MMBR.00042-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Foster JL, Garcia JV. HIV-1 Nef: at the crossroads. Retrovirology. 2008;5:84. doi: 10.1186/1742-4690-5-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garcia JV, Miller AD. Serine phosphorylation-independent downregulation of cell-surface CD4 by nef. Nature. 1991;350(6318):508–511. doi: 10.1038/350508a0. [DOI] [PubMed] [Google Scholar]
- 4.Rhee SS, Marsh JW. Human immunodeficiency virus type 1 Nef-induced down-modulation of CD4 is due to rapid internalization and degradation of surface CD4. J Virol. 1994;68(8):5156–5163. doi: 10.1128/jvi.68.8.5156-5163.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwartz O, Marechal V, Le Gall S, Lemonnier F, Heard JM. Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 Nef protein. Nat Med. 1996;2(3):338–342. doi: 10.1038/nm0396-338. [DOI] [PubMed] [Google Scholar]
- 6.Greenberg ME, Iafrate AJ, Skowronski J. The SH3 domain-binding surface and an acidic motif in HIV-1 Nef regulate trafficking of class I MHC complexes. EMBO J. 1998;17(10):2777–2789. doi: 10.1093/emboj/17.10.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Craig HM, Pandori MW, Guatelli JC. Interaction of HIV-1 Nef with the cellular dileucine-based sorting pathway is required for CD4 down-regulation and optimal viral infectivity. Proc Natl Acad Sci U S A. 1998;95(19):11229–11234. doi: 10.1073/pnas.95.19.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kasper MR, Collins KL. Nef-mediated disruption of HLA-A2 transport to the cell surface in T cells. J Virol. 2003;77(5):3041–3049. doi: 10.1128/JVI.77.5.3041-3049.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirchhoff F, Greenough TC, Brettler DB, Sullivan JL, Desrosiers RC. Brief report: absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N Engl J Med. 1995;332(4):228–232. doi: 10.1056/NEJM199501263320405. [DOI] [PubMed] [Google Scholar]
- 10.Dyer WB, Geczy AF, Kent SJ, McIntyre LB, Blasdall SA, Learmont JC, Sullivan JS. Lymphoproliferative immune function in the Sydney Blood Bank Cohort, infected with natural nef/long terminal repeat mutants, and in other long-term survivors of transfusion-acquired HIV-1 infection. Aids. 1997;11(13):1565–1574. doi: 10.1097/00002030-199713000-00004. [DOI] [PubMed] [Google Scholar]
- 11.Saksela K, Cheng G, Baltimore D. Proline-rich (PxxP) motifs in HIV-1 Nef bind to SH3 domains of a subset of Src kinases and are required for the enhanced growth of Nef+ viruses but not for down-regulation of CD4. EMBO J. 1995;14(3):484–491. doi: 10.1002/j.1460-2075.1995.tb07024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moarefi I, LaFevre-Bernt M, Sicheri F, Huse M, Lee CH, Kuriyan J, Miller WT. Activation of the Src-family tyrosine kinase Hck by SH3 domain displacement. Nature. 1997;385(6617):650–653. doi: 10.1038/385650a0. [DOI] [PubMed] [Google Scholar]
- 13.Greenway AL, Dutartre H, Allen K, McPhee DA, Olive D, Collette Y. Simian immunodeficiency virus and human immunodeficiency virus type 1 nef proteins show distinct patterns and mechanisms of Src kinase activation. J Virol. 1999;73(7):6152–6158. doi: 10.1128/jvi.73.7.6152-6158.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malim MH, Emerman M. HIV-1 accessory proteins--ensuring viral survival in a hostile environment. Cell Host Microbe. 2008;3(6):388–398. doi: 10.1016/j.chom.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 15.Matsuura Y, Maekawa M, Hattori S, Ikegami N, Hayashi A, Yamazaki S, Morita C, Takebe Y. Purification and characterization of human immunodeficiency virus type 1 nef gene product expressed by a recombinant baculovirus. Virology. 1991;184(2):580–586. doi: 10.1016/0042-6822(91)90428-e. [DOI] [PubMed] [Google Scholar]
- 16.Geyer M, Peterlin BM. Domain assembly, surface accessibility and sequence conservation in full length HIV-1 Nef. FEBS Lett. 2001;496(2–3):91–95. doi: 10.1016/s0014-5793(01)02394-8. [DOI] [PubMed] [Google Scholar]
- 17.Akari H, Arold ST, Fukumori T, Okazaki T, Strebel K, Adachi A. Nef-induced major histocompatibility complex class I down-regulation is functionally dissociated from its virion incorporation, enhancement of viral infectivity, and CD4 down-regulation. J Virol. 2000;74(6):2907–2912. doi: 10.1128/jvi.74.6.2907-2912.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mangasarian A, Piguet V, Wang JK, Chen YL, Trono D. Nef-induced CD4 and major histocompatibility complex class I (MHC-I) down-regulation are governed by distinct determinants: N-terminal alpha helix and proline repeat of Nef selectively regulate MHC-I trafficking. J Virol. 1999;73(3):1964–1973. doi: 10.1128/jvi.73.3.1964-1973.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams M, Roeth JF, Kasper MR, Filzen TM, Collins KL. Human immunodeficiency virus type 1 Nef domains required for disruption of major histocompatibility complex class I trafficking are also necessary for coprecipitation of Nef with HLA-A2. J Virol. 2005;79(1):632–636. doi: 10.1128/JVI.79.1.632-636.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greenberg M, DeTulleo L, Rapoport I, Skowronski J, Kirchhausen T. A dileucine motif in HIV-1 Nef is essential for sorting into clathrin-coated pits and for downregulation of CD4. Curr Biol. 1998;8(22):1239–1242. doi: 10.1016/s0960-9822(07)00518-0. [DOI] [PubMed] [Google Scholar]
- 21.Craig HM, Reddy TR, Riggs NL, Dao PP, Guatelli JC. Interactions of HIV-1 nef with the mu subunits of adaptor protein complexes 1, 2, and 3: role of the dileucine-based sorting motif. Virology. 2000;271(1):9–17. doi: 10.1006/viro.2000.0277. [DOI] [PubMed] [Google Scholar]
- 22.Renkema GH, Saksela K. Interactions of HIV-1 NEF with cellular signal transducing proteins. Front Biosci. 2000;5:D268–D283. doi: 10.2741/renkema. [DOI] [PubMed] [Google Scholar]
- 23.Piguet V, Wan L, Borel C, Mangasarian A, Demaurex N, Thomas G, Trono D. HIV-1 Nef protein binds to the cellular protein PACS-1 to downregulate class I major histocompatibility complexes. Nat Cell Biol. 2000;2(3):163–167. doi: 10.1038/35004038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grzesiek S, Bax A, Clore GM, Gronenborn AM, Hu JS, Kaufman J, Palmer I, Stahl SJ, Wingfield PT. The solution structure of HIV-1 Nef reveals an unexpected fold and permits delineation of the binding surface for the SH3 domain of Hck tyrosine protein kinase. Nat Struct Biol. 1996;3(4):340–345. doi: 10.1038/nsb0496-340. [DOI] [PubMed] [Google Scholar]
- 25.Lee CH, Saksela K, Mirza UA, Chait BT, Kuriyan J. Crystal structure of the conserved core of HIV-1 Nef complexed with a Src family SH3 domain. Cell. 1996;85(6):931–942. doi: 10.1016/s0092-8674(00)81276-3. [DOI] [PubMed] [Google Scholar]
- 26.Arold ST, Franken P, Strub MP, Hoh F, Benichou S, Benarous R, Dumas C. The crystal structure of HIV-1 Nef protein bound to the Fyn kinase SH3 domain suggests a role for this complex in altered T cell receptor signaling. Structure. 1997;5(10):1361–1372. doi: 10.1016/s0969-2126(97)00286-4. [DOI] [PubMed] [Google Scholar]
- 27.Barnham KJ, Monks SA, Hinds MG, Azad AA, Norton RS. Solution structure of a polypeptide from the N terminus of the HIV protein Nef. Biochemistry. 1997;36(20):5970–5980. doi: 10.1021/bi9629945. [DOI] [PubMed] [Google Scholar]
- 28.Geyer M, Munte CE, Schorr J, Kellner R, Kalbitzer HR. Structure of the anchor-domain of myristoylated and non-myristoylated HIV-1 Nef protein. J Mol Biol. 1999;289(1):123–138. doi: 10.1006/jmbi.1999.2740. [DOI] [PubMed] [Google Scholar]
- 29.Sicheri F, Moarefi I, Kuriyan J. Crystal structure of the Src family tyrosine kinase Hck. Nature. 1997;385(6617):602–609. doi: 10.1038/385602a0. [DOI] [PubMed] [Google Scholar]
- 30.Lee CH, Leung B, Lemmon MA, Zheng J, Cowburn D, Kuriyan J, Saksela K. A single amino acid in the SH3 domain of Hck determines its high affinity and specificity in binding to HIV-1 Nef protein. EMBO J. 1995;14(20):5006–5015. doi: 10.1002/j.1460-2075.1995.tb00183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shugars DC, Smith MS, Glueck DH, Nantermet PV, Seillier-Moiseiwitsch F, Swanstrom R. Analysis of human immunodeficiency virus type 1 nef gene sequences present in vivo. J Virol. 1993;67(8):4639–4650. doi: 10.1128/jvi.67.8.4639-4650.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 33.Goddard TD, Kneller DG. SPARKY 3. 3.110 ed. San Francisco: University of California; 2004. [Google Scholar]
- 34.Keller RLJ. The Computer Aided Resonance Assignment Tutorial: CANTINA Verlag. 2004 [Google Scholar]
- 35.Salzmann M, Wider G, Pervushin K, Senn H, Wüthrich K. TROSY-type Triple-Resonance Experiments for Sequential NMR Assignments of Large Proteins. Journal of the American Chemical Society. 1999;121(4):844–848. [Google Scholar]
- 36.Bax A, Grzesiek S. Methodological advances in protein NMR. Accounts of Chemical Research. 1993;26(4):131–138. [Google Scholar]
- 37.Clore GM, Gronenborn AM. Determining the structures of large proteins and protein complexes by NMR. Trends Biotechnol. 1998;16(1):22–34. doi: 10.1016/S0167-7799(97)01135-9. [DOI] [PubMed] [Google Scholar]
- 38.Farmer BT, 2nd, Constantine KL, Goldfarb V, Friedrichs MS, Wittekind M, Yanchunas J, Jr, Robertson JG, Mueller L. Localizing the NADP+ binding site on the MurB enzyme by NMR. Nat Struct Biol. 1996;3(12):995–997. doi: 10.1038/nsb1296-995. [DOI] [PubMed] [Google Scholar]
- 39.Wishart DS, Bigam CG, Holm A, Hodges RS, Sykes BD. 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J Biomol NMR. 1995;5(1):67–81. doi: 10.1007/BF00227471. [DOI] [PubMed] [Google Scholar]
- 40.Schwarzinger S, Kroon GJ, Foss TR, Chung J, Wright PE, Dyson HJ. Sequence-dependent correction of random coil NMR chemical shifts. J Am Chem Soc. 2001;123(13):2970–2978. doi: 10.1021/ja003760i. [DOI] [PubMed] [Google Scholar]
- 41.Ruckert M, Otting G. Alignment of biological macromolecules in novel nonionic liquid crystalline media for NMR experiments. J Am Chem Soc. 2000;122(32):7793–7797. [Google Scholar]
- 42.Ottiger M, Delaglio F, Bax A. Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. J Magn Reson. 1998;131(2):373–378. doi: 10.1006/jmre.1998.1361. [DOI] [PubMed] [Google Scholar]
- 43.Zweckstetter M. NMR: prediction of molecular alignment from structure using the PALES software. Nature protocols. 2008;3(4):679–690. doi: 10.1038/nprot.2008.36. [DOI] [PubMed] [Google Scholar]
- 44.Dosset P, Hus JC, Marion D, Blackledge M. A novel interactive tool for rigid-body modeling of multi-domain macromolecules using residual dipolar couplings. J Biomol NMR. 2001;20(3):223–231. doi: 10.1023/a:1011206132740. [DOI] [PubMed] [Google Scholar]
- 45.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160(1):65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 46.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33(19):5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 47.Grzesiek S, Bax A, Hu JS, Kaufman J, Palmer I, Stahl SJ, Tjandra N, Wingfield PT. Refined solution structure and backbone dynamics of HIV-1 Nef. Protein Sci. 1997;6(6):1248–1263. doi: 10.1002/pro.5560060613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spera S, Bax A. Empirical correlation between protein backbone conformation and C.alpha. and C.beta. 13C nuclear magnetic resonance chemical shifts. Journal of the American Chemical Society. 1991;113(14):5490–5492. [Google Scholar]
- 49.Horita DA, Baldisseri DM, Zhang W, Altieri AS, Smithgall TE, Gmeiner WH, Byrd RA. Solution structure of the human Hck SH3 domain and identification of its ligand binding site. J Mol Biol. 1998;278(1):253–265. doi: 10.1006/jmbi.1998.1690. [DOI] [PubMed] [Google Scholar]
- 50.Alvarado JJ, Betts L, Moroco JA, Smithgall TE, Yeh JI. Crystal structure of the Src-family kinase Hck SH3-SH2-linker regulatory region supports an SH3-dominant activation mechanism. J Biol Chem. 2010;285:35455–35461. doi: 10.1074/jbc.M110.145102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmidt H, Hoffmann S, Tran T, Stoldt M, Stangler T, Wiesehan K, Willbold D. Solution structure of a Hck SH3 domain ligand complex reveals novel interaction modes. J Mol Biol. 2007;365(5):1517–1532. doi: 10.1016/j.jmb.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 52.Trible RP, Emert-Sedlak L, Smithgall TE. HIV-1 Nef selectively activates Src family kinases Hck, Lyn, and c-Src through direct SH3 domain interaction. J Biol Chem. 2006;281(37):27029–27038. doi: 10.1074/jbc.M601128200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Young MA, Gonfloni S, Superti-Furga G, Roux B, Kuriyan J. Dynamic coupling between the SH2 and SH3 domains of c-Src and Hck underlies their inactivation by C-terminal tyrosine phosphorylation. Cell. 2001;105(1):115–126. doi: 10.1016/s0092-8674(01)00301-4. [DOI] [PubMed] [Google Scholar]
- 54.Bruschweiler R, Liao X, Wright PE. Long-range motional restrictions in a multidomain zinc-finger protein from anisotropic tumbling. Science. 1995;268(5212):886–889. doi: 10.1126/science.7754375. [DOI] [PubMed] [Google Scholar]
- 55.Lee LK, Rance M, Chazin WJ, Palmer AG., 3rd Rotational diffusion anisotropy of proteins from simultaneous analysis of 15N and 13C alpha nuclear spin relaxation. J Biomol NMR. 1997;9(3):287–298. doi: 10.1023/a:1018631009583. [DOI] [PubMed] [Google Scholar]
- 56.Wonderlich ER, Williams M, Collins KL. The tyrosine binding pocket in the adaptor protein 1 (AP-1) mu1 subunit is necessary for Nef to recruit AP-1 to the major histocompatibility complex class I cytoplasmic tail. J Biol Chem. 2008;283(6):3011–3022. doi: 10.1074/jbc.M707760200. [DOI] [PubMed] [Google Scholar]
- 57.Gerlach H, Laumann V, Martens S, Becker C, Goody R, Geyer M. HIV-1 Nef membrane association depends on charge, curvature, composition and sequence. Nat Chem Biol. 2009 doi: 10.1038/nchembio.268. [DOI] [PubMed] [Google Scholar]
- 58.Kim WM, Sigalov AB, Stern LJ. Pseudo-merohedral twinning and noncrystallographic symmetry in orthorhombic crystals of SIVmac239 Nef core domain bound to different-length TCRzeta fragments. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):163–175. doi: 10.1107/S090744490904880X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grzesiek S, Stahl SJ, Wingfield PT, Bax A. The CD4 determinant for downregulation by HIV-1 Nef directly binds to Nef. Mapping of the Nef binding surface by NMR. Biochemistry. 1996;35(32):10256–10261. doi: 10.1021/bi9611164. [DOI] [PubMed] [Google Scholar]
- 60.Ulmer TS, Werner JM, Campbell ID. SH3-SH2 domain orientation in Src kinases: NMR studies of Fyn. Structure. 2002;10(7):901–911. doi: 10.1016/s0969-2126(02)00781-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.