Abstract
Myostatin is a transforming growth factor β family member that acts as a negative regulator of skeletal muscle growth. Myostatin circulates in the blood of adult mice in a noncovalently held complex with other proteins, including its propeptide, which maintain the C-terminal dimer in a latent, inactive state. This latent form of myostatin can be activated in vitro by treatment with acid; however, the mechanisms by which latent myostatin is activated in vivo are unknown. Here, we show that members of the bone morphogenetic protein-1/tolloid (BMP-1/TLD) family of metalloproteinases can cleave the myostatin propeptide in this complex and can thereby activate latent myostatin. Furthermore, we show that a mutant form of the propeptide resistant to cleavage by BMP-1/TLD proteinases can cause significant increases in muscle mass when injected into adult mice. These findings raise the possibility that members of the BMP-1/TLD family may be involved in activating latent myostatin in vivo and that molecules capable of inhibiting these proteinases may be effective agents for increasing muscle mass for both human therapeutic and agricultural applications.
Myostatin is a transforming growth factor β (TGF-β) family member that is essential for proper regulation of skeletal muscle growth (1). Mice lacking myostatin have a dramatic and widespread increase in skeletal muscle mass as a result of a combination of muscle fiber hypertrophy and hyperplasia. Naturally occurring mutations in the myostatin gene also cause the double-muscling phenotype in cattle (2-5). These findings have suggested that blocking myostatin activity may have applications for the treatment of human diseases in which increasing muscle growth may be desirable. Indeed, loss of myostatin signaling has been shown to have beneficial effects in mouse models of muscle degenerative (6, 7) and metabolic (8) diseases.
Like other TGF-β family members, myostatin is synthesized as a precursor protein that undergoes proteolytic processing at a dibasic site to generate an N-terminal propeptide and a disulfide-linked C-terminal dimer, which is the biologically active molecule. The circulating form of myostatin consists of a latent complex of the myostatin C-terminal dimer and other proteins, including the myostatin propeptide, which inhibit the biological activity of the C-terminal dimer (9-13). In this study, we have investigated the mechanisms by which this latent complex may be activated. We present evidence that members of the bone morphogenetic protein-1/tolloid (BMP-1/TLD) family of metalloproteinases may be involved in activating latent myostatin in vivo, raising the possibility that targeting these proteinases may be a novel strategy for the development of agents capable of promoting muscle growth.
Materials and Methods
Myostatin Purification and Analysis. The generation of Chinese hamster ovary (CHO) cell lines overexpressing myostatin has been described (9, 10). Similar strategies were used to generate CHO lines expressing mutant forms of full-length myostatin and propeptide/Fc fusion proteins. Myostatin propeptide/C-terminal dimer complexes were purified from the conditioned medium of CHO-expressing cells as described (9). Propeptide/Fc fusion proteins were purified by using a protein A Sepharose column. Antibodies directed against bacterially produced myostatin C-terminal domain and propeptide have been described (1, 9).
Proteinase and Reporter Gene Assays. Proteinases were purified and assayed as described (14). Myostatin activity was measured by using the pGL3-(CAGA)12-luciferase reporter assay in A204 rhabdomyosarcoma cells as described (10). A standard curve using purified myostatin C-terminal dimer was generated for each set of assays to quantify myostatin activity.
Injection of Mice. Female BALB/c mice (Charles River Breeding Laboratories) weighing 17-19 g were injected i.p. on days 1, 4, 8, 15, and 22 either with PBS alone or with various proteins diluted in PBS. Doses of proteins were: propeptide/Fc fusion proteins, 1 and 10 mg/kg; IgG2am (control antibody), 10 mg/kg; and JA16, 60 mg/kg. Mice were killed on day 29 for muscle analysis. Muscles from both sides of each animal were dissected and weighed, and the average weight was used for each muscle.
Results and Discussion
We previously showed that CHO cells engineered to overexpress myostatin secrete myostatin as a latent complex of the propeptide and C-terminal dimer (9, 10). In the course of characterizing the secretion of myostatin by these cells, we noted the presence of a discrete cleavage product of the propeptide, which could be detected by Western blot analysis using antibodies specific for the propeptide. This cleavage product was detected in the conditioned medium of CHO cells transfected with expression constructs containing either the full-length myostatin precursor protein (data not shown) or the myostatin propeptide alone in the absence of the C-terminal domain (Fig. 1A). Because the myostatin propeptide is capable of maintaining the C-terminal dimer in a latent state both in vitro (9, 10) and in vivo (11, 12) and because proteolytic cleavage of the TGF-β propeptide is believed to be one mechanism for activating latent TGF-β (15-19), we investigated the possibility that cleavage of the myostatin propeptide might be involved in regulating myostatin latency.
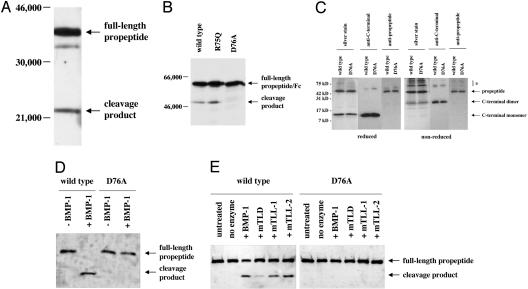
Fig. 1.
Cleavage of the myostatin propeptide by the BMP-1/TLD family of proteinases. (A and B) Detection of a propeptide degradation product in CHO cell-conditioned media. Conditioned media prepared from CHO cells expressing the propeptide (A) or WT and mutant forms of propeptide/Fc fusion proteins (B) were analyzed by SDS/PAGE followed by Western blot analysis using antibodies directed against either the myostatin propeptide (A) or IgG (B). Note that mutation of D76 to A resulted in the absence of the degradation product. (C) Purification of WT and mutant propeptide/C-terminal dimer complexes. Protein complexes were purified as described (9) and analyzed by SDS/PAGE in the presence or absence of 2-mercaptoethanol followed by Western blot analysis, as indicated. Note that, like the WT propeptide, the D76A mutant propeptide purified in a complex with the C-terminal dimer. The propeptide degradation product did not copurify with the C-terminal dimer and was thus not part of the complex. Bands denoted by * indicate misfolded myostatin species, which were evident under nonreducing conditions. (D and E) Cleavage of the propeptide by BMP-1/TLD proteinases. WT and mutant complexes were incubated with purified proteinases (14) and subjected to SDS/PAGE followed by Western blot analysis using antibodies directed against the propeptide. Incubations were carried out with 1 μg of latent complex and 250 ng of proteinase for 16 h at 37°C, except that in D the samples were incubated with an additional 250 ng of BMP-1 for 4 h more. In E, lanes labeled “no enzyme” indicate samples incubated for 16 h at 37°C in the absence of enzyme. Note that all enzymes were capable of generating the cleavage product and that the D76A mutant protein was completely resistant to cleavage.
N-terminal sequencing revealed that the propeptide degradation product detected in CHO cell-conditioned medium resulted from proteolytic cleavage between Arg-75 and Asp-76. To determine whether either of these amino acid residues is essential for proteolytic cleavage, we generated CHO cell lines expressing mutant versions of the propeptide in which either the arginine or aspartate residue was changed to glutamine or alanine, respectively. To enhance stability of these proteins for in vivo studies (see below), we fused the propeptides with an Fc domain. Although changing the arginine (R) to glutamine (Q) had no effect on proteolytic cleavage, no degradation product could be detected in conditioned medium prepared from CHO cells expressing the aspartate (D) to alanine (A) mutant propeptide/Fc fusion protein (Fig. 1B). The requirement for aspartate at the cleavage site suggested the possibility that members of the BMP-1/TLD family of metalloproteinases might be responsible for generating this degradation product. A number of substrates have been identified for mammalian members of the BMP-1/TLD family, and in nearly every case, proteolytic cleavage has been shown to occur immediately N terminal to an aspartate residue (14, 20). Furthermore, mutagenesis studies have documented the importance of the aspartate residue in rendering these sites susceptible to proteolytic cleavage (21). As we were unaware of other proteinases with a similar specificity or requirement for an aspartate residue just C terminal to the scissile bond in protein substrates, we investigated the ability of members of the BMP-1/TLD family to cleave the myostatin propeptide in vitro.
Myostatin was purified from the conditioned medium of overproducing CHO cells as described (9). After successive fractionation on hydroxyapatite, lentil lectin Sepharose, DEAE agarose, and heparin Sepharose, we obtained a purified preparation of the myostatin latent complex, which consisted of the N-terminal propeptide bound noncovalently to the C-terminal dimer (Fig. 1C). Incubation of the purified latent complex with purified BMP-1 resulted in complete cleavage of the propeptide (Fig. 1D) to generate a product with an electrophoretic mobility comparable to that detected in conditioned medium prepared from CHO cells engineered to overproduce myostatin. Indeed, N-terminal sequencing of BMP-1-treated propeptide confirmed that cleavage had occurred immediately N terminal to Asp-76. We also tested the ability of the other mammalian members of the BMP-1/TLD family, namely, mTLD, mTLL-1, and mTLL-2, to cleave the propeptide. For these experiments, we used enzyme concentrations that resulted in only partial cleavage, which allowed us to compare the relative activities of the four enzymes. As shown in Fig. 1E, incubation of the latent complex with each of the four proteinases resulted in cleavage of the propeptide. Three of the proteinases, BMP-1, mTLL-1, and mTLL-2, were approximately equally effective in cleaving the propeptide, whereas mTLD was consistently less active than the other three, even though the same mTLD preparation was found to be fully active against known substrates such as procollagen (data not shown). All four of these proteinases were also capable of cleaving propeptide that had been purified away from the C-terminal dimer (data not shown).
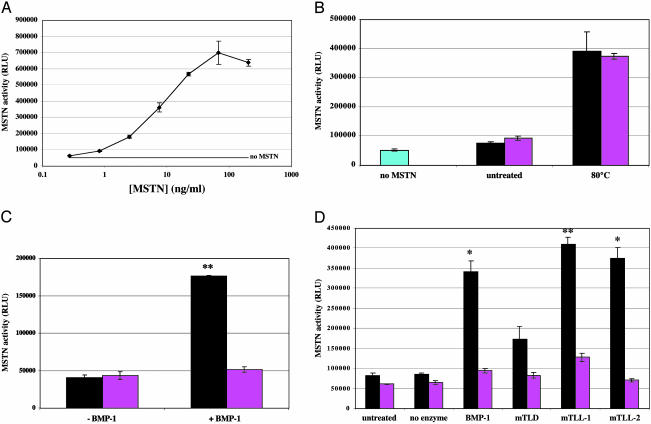
To determine the effect of proteolytic cleavage of the propeptide on myostatin latency, we measured myostatin biological activity in latent complexes treated with each of the four proteinases. For this purpose, we used a reporter gene assay in which A204 rhabdomyosarcoma cells were transfected with the pGL3-(CAGA)12-luciferase construct and incubated with myostatin (10). As described, the addition of purified myostatin C-terminal dimer to these cells caused an increase in luciferase activity above basal levels (Fig. 2A). In contrast, purified myostatin latent complex was inactive in this assay but could be activated by incubation at 80°C for 5 min (Fig. 2B). As shown in Fig. 2C, the latent complex was also activated by pretreatment with BMP-1. Based on quantification of myostatin activity relative to a standard curve, it appeared that cleavage of the propeptide by BMP-1 was approximately as effective as heat treatment in activating the latent complex. The latent complex was also activated by pretreatment with the other proteinases, and the extent of activation correlated roughly with the extent of proteolytic cleavage by these enzymes (Fig. 2D).
Fig. 2.
Activation of latent myostatin activity by BMP-1/TLD proteinases. (A) Activation of pGL3-(CAGA)12-luciferase reporter gene activity by purified myostatin C-terminal dimer. (B) Activation of the myostatin propeptide/C-terminal dimer latent complex by heat treatment. (C and D) Activation of the myostatin (MSTN) propeptide/C-terminal dimer latent complex by BMP-1/TLD proteinases. A standard curve using purified myostatin C-terminal dimer was generated for each set of assays to quantify myostatin activity. The samples used for the reporter assays in C and D are the same samples shown in Fig. 1 D and E, respectively. In B-D, black bars represent WT, and pink bars represent D76A mutant complexes. Note that, although heat treatment activated both the WT and mutant complexes, each proteinase was capable of activating only the WT complex. *, P < 0.05; **, P < 0.01. RLU, relative light units.
We also examined the requirement for aspartate at the cleavage site. We generated a CHO cell line expressing high levels of a mutant form of myostatin in which Asp-76 was changed to alanine, and we purified the latent complex from the conditioned medium of these cells. As shown in Fig. 1C, the mutation had no effect on the ability of the propeptide to bind to the C-terminal dimer, as the mutant propeptide and C-terminal dimer remained tightly associated throughout the purification. Moreover, the mutant propeptide was capable of maintaining the complex in a latent form that could be activated by heating, as assessed by the luciferase reporter assay (Fig. 2B). However, the mutant propeptide in the latent complex was completely resistant to proteolysis by each of the four proteinases, BMP-1, mTLD, mTLL-1, and mTLL-2 (Fig. 1 D and E) and resistant to activation by these proteinases (Fig. 2 C and D).
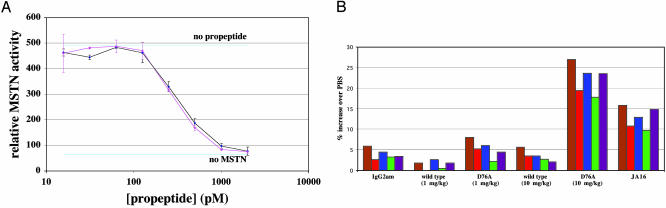
Finally, we investigated the role of proteolytic cleavage of the propeptide in vivo by examining the effect of injecting WT and mutant versions of the propeptide into mice. In previous experiments, we had shown that the half-life of WT propeptide after i.p. injections into mice could be increased from ≈2hto5-7 days by fusing the propeptide to an Fc domain (data not shown). For this reason, we generated CHO lines expressing WT or mutant (Asp-76 to alanine) propeptide fused to an Fc domain and purified the fusion proteins by using a protein A Sepharose column. The aspartate to alanine mutation did not affect the activity of the propeptide in vitro, as the purified WT and mutant propeptide/Fc fusion proteins were equally effective in inhibiting the activity of the purified myostatin C-terminal dimer in the reporter gene assay (Fig. 3A).
Fig. 3.
Muscle growth induced by injection of the D76A mutant propeptide/Fc fusion protein into mice. (A) Inhibition of reporter gene activity by WT and mutant propeptide/Fc fusion proteins in vitro. A204 cells transfected with the reporter construct were incubated with 10 ng/ml purified myostatin (MSTN) C-terminal dimer and various concentrations of WT (black) or D76A mutant (pink) propeptide/Fc fusion protein. Note that the WT and mutant proteins were equally effective in blocking myostatin activity. (B) Increased muscle mass in mice injected with mutant propeptide/Fc fusion protein. Numbers indicate percent increase in muscle mass compared with PBS-injected animals. Actual numbers used for these calculations are shown in Table 1. Only D76A at 10 mg/kg and JA16 at 60 mg/kg gave statistically significant increases in muscle mass (see Table 1). Brown, pectoralis; red, triceps; blue, quadriceps; green, gastrocnemius; purple, tibialis.
To assess the activities of these proteins in vivo, adult mice were given weekly injections of purified WT or mutant propeptide/Fc fusion proteins and killed after 4 weeks for muscle analysis. For comparison, we also injected a set of mice with the JA16 myostatin-neutralizing mAb, which had previously been shown to cause an ≈25-30% increase in muscle mass after 12 weeks of treatment (22). As shown in Table 1 and Fig. 3B, injection of WT propeptide/Fc fusion protein had no effect on muscle mass at doses of 1 and 10 mg/kg per week. Similarly, little or no effect was seen after injection of the aspartate to alanine mutant propeptide/Fc fusion protein at a dose of 1 mg/kg per week. However, injection of the mutant propeptide/Fc fusion protein at 10 mg/kg per week led to a statistically significant (P < 0.0001) increase of 18-27% in the weight of each skeletal muscle examined. This magnitude of increase in muscle weights seen at the higher dose of the mutant propeptide/Fc fusion protein was approximately twice that seen after injection of the JA16 myostatin-neutralizing mAb, which resulted in muscle weight increases of 10-16%.
Table 1. Muscle growth induced by injection of the D76A mutant propeptide/Fc protein into mice.
| Injectant | Pectoralis | Triceps | Quadriceps | Gastrocnemius | Tibialis |
|---|---|---|---|---|---|
| PBS (n = 10) | 82.8 ± 2.8 | 85.5 ± 1.6 | 142.0 ± 2.6 | 95.5 ± 1.5 | 32.6 ± 0.8 |
| IgG2am (10 mg/kg, n = 10) | 87.7 ± 1.9 | 87.8 ± 1.6 | 148.4 ± 2.3 | 98.7 ± 2.1 | 33.8 ± 0.9 |
| WT (1 mg/kg, n = 10) | 84.3 ± 1.6 | 85.3 ± 1.8 | 145.7 ± 2.2 | 96.0 ± 1.4 | 33.2 ± 0.4 |
| D76A (1 mg/kg, n = 9) | 89.4 ± 3.5 | 90.0 ± 2.0 | 150.7 ± 2.9* | 97.7 ± 2.2 | 34.1 ± 0.6 |
| WT (10 mg/kg, n = 10) | 87.5 ± 3.4 | 88.5 ± 2.7 | 147.1 ± 4.0 | 98.2 ± 2.2 | 33.3 ± 0.8 |
| D76A (10 mg/kg, n = 10) | 105.1 ± 1.2†‡ | 102.1 ± 1.2†§ | 175.6 ± 1.2†¶ | 112.6 ± 1.1†§ | 40.3 ± 1.2†§ |
| JA16 (60 mg/kg, n = 10) | 96.0 ± 1.2∥ | 94.8 ± 1.1∥ | 160.3 ± 1.1† | 104.9 ± 1.1∥ | 37.5 ± 1.1∥ |
, P < 0.05 (vs. PBS)
, P < 0.0001 (vs. PBS)
P < 0.05 (vs. JA16)
, P < 0.01 (vs. JA16)
, P < 0.001 (vs. JA16)
, P < 0.001 (vs. PBS)
Here, we have shown that members of the BMP-1/TLD family of metalloproteinases can cleave the myostatin propeptide bound to the C-terminal dimer and can thereby activate the latent complex. Furthermore, we have shown that a mutant form of the propeptide resistant to cleavage by BMP-1/TLD proteinases can cause increases in muscle mass when injected into adult mice, presumably by forming latent complexes incapable of being activated by this group of proteinases. This general mechanism for regulating the activity of the C-terminal dimer has been described previously for certain other TGF-β family members. In the case of TGF-β, proteolytic cleavage of its associated propeptide by plasmin (15, 16) or by matrix metalloproteinases (17-19) is believed to be one mechanism for activating latency in vivo. In the case of the BMPs, members of the BMP-1/TLD family appear to play an important role in regulating the activity of the C-terminal dimer by cleaving and inactivating the BMP antagonist chordin (14, 23-26).
Although all four mammalian proteinases in the BMP-1/TLD family are capable of cleaving the myostatin propeptide in vitro, we can only speculate as to which of these proteinases may be involved in regulating myostatin activity in vivo. In this regard, however, it is intriguing to note that mTLL-2, unlike the other three proteinases, is expressed specifically in skeletal muscle during embryonic development (14). The identification of the specific proteinase or proteinases involved in regulating myostatin latency will be an essential step in attempting to target these enzymes for the development of novel muscle enhancing agents for both human therapeutic and agricultural applications.
Acknowledgments
We thank Amanda L. Bemis, Ying Huang, and William S. Lane for N-terminal sequence analysis. This work was supported by National Institutes of Health Grants R01HD35887 and R01CA88866 (to S.-J.L.) and R01GM63471 and R01AR47746 (to D.S.G.) and funds from Wyeth Research. A.C.M. was supported by National Institutes of Health Training Grant 5 T32 CA09139. W.N.P. was supported by a predoctoral fellowship from the American Heart Association.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TGF-β, transforming growth factor β; BMP, bone morphogenetic protein; TLD, tolloid; CHO, Chinese hamster ovary.
References
- 1.McPherron, A. C., Lawler, A. M. & Lee, S.-J. (1997) Nature 387, 83-90. [DOI] [PubMed] [Google Scholar]
- 2.McPherron, A. C. & Lee, S.-J. (1997) Proc. Natl. Acad. Sci. USA 94, 12457-12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grobet, L., Martin, L. J. R., Poncelet, D., Pirottin, D., Brouwers, B., Riquet, J., Schoeberlein, A., Dunner, S., Ménissier, F., Massabanda, J., et al. (1997) Nat. Genet. 17, 71-74. [DOI] [PubMed] [Google Scholar]
- 4.Kambadur, R., Sharma, M., Smith, T. P. L. & Bass, J. J. (1997) Genome Res. 7, 910-915. [DOI] [PubMed] [Google Scholar]
- 5.Grobet, L., Poncelet, D., Royo, L. J., Brouwers, B., Pirottin, D., Michaux, C., Ménissier, F., Zanotti, M., Dunner, S. & Georges, M. (1998) Mamm. Genome 9, 210-213. [DOI] [PubMed] [Google Scholar]
- 6.Bogdanovich, S., Krag, T. O. B., Barton, E. R., Morris, L. D., Whittemore, L.-A., Ahima, R. S. & Khurana, T. S. (2002) Nature 420, 418-421. [DOI] [PubMed] [Google Scholar]
- 7.Wagner, K. R., McPherron, A. C., Winik, N. & Lee, S.-J. (2002) Ann. Neurol. 52, 832-836. [DOI] [PubMed] [Google Scholar]
- 8.McPherron, A. C. & Lee, S.-J. (2002) J. Clin. Invest. 109, 595-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee, S.-J. & McPherron, A. C. (2001) Proc. Natl. Acad. Sci. USA 98, 9306-9311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thies, R., Chen, T., Davies, M., Tomkinson, K., Pearson, A., Shakey, Q. & Wolfman, N. (2001) Growth Factors 18, 251-259. [DOI] [PubMed] [Google Scholar]
- 11.Zimmers, T., Davies, M., Koniaris, L., Haynes, P., Esquela, A., Tomkinson, K., McPherron, A., Wolfman, N. & Lee, S.-J. (2002) Science 296, 1486-1488. [DOI] [PubMed] [Google Scholar]
- 12.Hill, J. J., Davies, M. V., Pearson, A. A., Wang, J. H., Hewick, R. M., Wolfman, N. M. & Qiu, Y. (2002) J. Biol. Chem. 277, 40735-40741. [DOI] [PubMed] [Google Scholar]
- 13.Hill, J. J., Qiu, Y., Hewick, R. M. & Wolfman, N. M. (2003) Mol. Endocrinol. 17, 1144-1154. [DOI] [PubMed] [Google Scholar]
- 14.Scott, I., Blitz, I., Pappano, W., Imamura, Y., Clark, T., Steiglitz, B., Thomsa, C., Maas, S., Takahara, K., Cho, K. & Greenspan, D. (1999) Dev. Biol. 213, 283-300. [DOI] [PubMed] [Google Scholar]
- 15.Lyons, R. M., Keski-Oja, J. & Moses, H. L. (1988) J. Cell Biol. 106, 1659-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato, Y. & Rifkin, D. (1989) J. Cell Biol. 109, 309-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu, Q. & Stamenkovic, I. (2000) Genes Dev. 14, 163-176. [PMC free article] [PubMed] [Google Scholar]
- 18.D'Angelo, M., Billings, P., Pacifici, M., Leboy, P. & Thorsten, K. (2001) J. Biol. Chem. 276, 11347-11353. [DOI] [PubMed] [Google Scholar]
- 19.Maeda, S., Dean, D., Gay, I., Schwartz, Z. & Boyan, B. (2001) J. Bone Miner. Res. 16, 1281-1290. [DOI] [PubMed] [Google Scholar]
- 20.Scott, I. C., Imamura, Y., Pappano, W. N., Troedel, J. M., Recklies, A. D., Roughley, P. J. & Greenspan, D. S. (2000) J. Biol. Chem. 275, 30504-30511. [DOI] [PubMed] [Google Scholar]
- 21.Lee, S.-T., Kessler, E. & Greenspan, D. S. (1990) J. Biol. Chem. 265, 21992-21996. [PubMed] [Google Scholar]
- 22.Whittemore, L.-A., Song, K., Li, X., Aghajanian, J., Davies, M. V., Girgenrath, S., Hill, J. J., Jalenak, M., Kelley, P., Knight, A., et al. (2003) Biochem. Biophys. Res. Commun. 300, 965-971. [DOI] [PubMed] [Google Scholar]
- 23.Blader, P., Rastegar, S., Fischer, N. & Strahle, U. (1997) Science 278, 1937-1940. [DOI] [PubMed] [Google Scholar]
- 24.Piccolo, S., Agius, E., Lu, B., Goodman, S., Dale, L. & DeRobertis, E. M. (1997) Cell 91, 407-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marques, G., Musacchio, M., Shimell, M. J., Wunnenberg-Stapleton, K., Cho, K. W. & O'Conner, M. B. (1997) Cell 91, 417-426. [DOI] [PubMed] [Google Scholar]
- 26.Pappano, W., Steiglitz, B., Scott, I. C., Keene, D. R. & Greenspan, D. S. (2003) Mol. Cell. Biol. 23, 4428-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]