Abstract
We investigated the influence of the temporal resolution of dynamic contrast-enhanced MRI data on pharmacokinetic parameter estimation. Dynamic Gd-DTPA (Gadolinium-diethylene triamine pentaacetic acid) enhanced MRI data of implanted prostate tumors on rat hind limb were acquired at 4.7 T, with a temporal resolution of ~5 sec. The data were subsequently downsampled to temporal resolutions in the range of 15 sec to 85 sec, using a strategy that involves a recombination of k-space data. A basic two-compartment model was fit to the contrast agent uptake curves. The results demonstrated that as temporal resolution decreases, the volume transfer constant (Ktrans) is progressively underestimated (~4% to ~25%), and the fractional extravascular extracellular space (ve) is progressively overestimated (~1% to ~10%). The proposed downsampling strategy simulates the influence of temporal resolution more realistically than simply downsampling by removing samples.
Keywords: DCE-MRI, temporal resolution, k-space data, pharmacokinetic model, downsampling
Dynamic contrast-enhanced (DCE) MRI plays an essential role for cancer detection and characterization. Ideally, DCE-MRI data would be acquired with high spatial and high temporal resolution to fully exploit both the morphologic and kinetic information. However, there is always a tradeoff between spatial and temporal resolution with currently available equipment and techniques (1). Therefore, a wide range of temporal resolutions is encountered for clinical DCE-MRI. For instance, according to the European Society of Breast Imaging (2), a sampling time (Ts) of 60 sec – 120 sec is recommended for high spatial resolution bilateral acquisitions.
A widely used pharmacokinetic (PK) model to fit DCE-MRI data and extract physiologic parameters is the basic two-compartment model (3). To accurately fit this model to DCE-MRI data, Henderson et al. (4) recommended that contrast agent (CA) uptake in the breast be sampled every 16 sec to estimate the volume transfer constant (Ktrans) and the fraction of extravascular extracellular space (ve). For the extended model, taking the blood plasma fraction into account, their recommendation was to sample every 4 sec. The arterial input function (AIF), however, was suggested to be sampled every second. Other work usually did not make a distinction between CA uptake sampling and AIF sampling (5,6) or focused on AIF sampling (7). Most previous studies investigated the effect of low temporal resolution on PK-parameter estimation by downsampling high-temporal-resolution data while assuming an instantaneous collection of k-space data (4,5,8,9); except for Ramirez et al. (6), who used a sliding temporal window.
The purpose of this study is to investigate the effect of the temporal resolution of DCE-MRI data on PK-parameter estimation. Because the CA concentration levels are changing during a low-temporal-resolution acquisition, the CA uptake observed in the tissue is influenced by the phase-encoding order. To mimic this effect and simulate DCE-MRI data at different temporal resolutions, a realistic downsampling strategy is introduced that involves a recombination of k-space data. The method is applied to high-temporal-resolution rodent DCE-MRI data acquired with a 4.7-T animal scanner.
THEORY
Downsampling Strategy
In our downsampling strategy, the transient effect of CA uptake is incorporated by mimicking an MRI data acquisition. To simulate a linear phase-encoding order, equal fractions of k-space data are taken from a series of successive high-temporal-resolution k-space images. For instance, using 5-sec resolution k-space data to simulate the first image “acquired” at a Ts of 15 sec, we recombine the first one-third of k-space lines from the 5-sec image, the second one-third from the 10-sec image, and the third one-third from the 15-sec image. The second image at Ts of 15 sec is derived from the k-space images acquired at 20 sec, 25 sec, 30 sec, etc. The recombination for the above example is defined as:
| [1] |
where is the ith downsampled k-space image; NR is the number of dynamic repetitions; Si(k) is the kth phase-encoding line of the ith original k-space image; and Nφ is the total number of phase-encoding lines. We call this strategy “k-space-based sampling.”
The results obtained with k-space-based sampling are compared to the results of downsampling by taking the first out of every set of N images and discarding the rest, which implies the assumption of instantaneous k-space data collection (4,9). This strategy is referred to as “direct sampling” in the remainder of this work.
PK Model
In this study, the basic two-compartment model by Tofts et al. (3) is applied:
| [2] |
where ve is the fraction of extravascular extracellular space, Ktrans(min−1) is the volume transfer constant between blood plasma and extravascular extracellular space, Cp(mM) is the CA concentration in the blood plasma space, and Ct(mM) is the CA concentration in the tissue of interest.
For the data used in this research, Cp is derived from the CA uptake in skeletal muscle, using the reference tissue approach, assuming literature values (10) for Ktrans (= 0.11 min−1) and ve (= 0.20). To minimize noise propagation, the muscle curve was fit with the empiric mathematical model (11) prior to this derivation.
To investigate the effect of a varying temporal resolution of the CA uptake in tissue as the only changing variable, we exclude the effect of a temporally downsampled AIF in this study. Therefore, the original high-temporal-resolution AIFs are used across all temporal resolutions.
MATERIALS AND METHODS
Animals and Imaging Protocol
Copenhagen rats (n = 6) with implanted AT6.1 prostate tumors on the hind limb were used in this study. Multislice T1-weighted gradient echo images (pulse repetition time/echo time = 40/3.5 ms, matrix size = 128 × 128, field of view × 40 mm × 40 mm, flip angle = 30°, phase encoding order = linear; slice thickness = 1 mm, number of slices = 3) were acquired through the center of the tumor along the long axis of the leg, with a temporal resolution of ~5 sec at 4.7 T (Bruker, Billerica, MA), before and after Gd-DTPA (Gadolinium-diethylene triamine pentaacetic acid; Omniscan, GE Healthcare, Piscataway, NJ) injection, for a total duration of ~10 min. The same amount of CA was manually injected (duration <5 sec) to all animals (based on a dose of 0.2 mmol/kg for a weight of 160 g). To account for variations in rat weight, the concentration of CA was normalized to the typical dose of 0.2 mmol/kg.
All the data were acquired at the University of Chicago, and procedures were carried out in accordance with the institution’s Animal Care and Use Committee approval. Animals were anesthetized prior to imaging experiments, and anesthesia was maintained during imaging at 1.5% isoflurane. The temperature was maintained at 37°C with a warm air blower. Heart rate, respiration rate, and temperature were monitored during the MRI experiments.
Contrast Concentration Calculation
All data processing was performed in Matlab (Mathworks, Natick, MA). To apply the PK model, DCE-MRI signal intensity curves were converted to CA concentration curves (Ct) using a previously published reference method (12). The precontrast reference was taken from a user-defined region of interest in muscle (9 × 9 voxels) that was not directly neighboring the tumor. A precontrast T1 value of 1285 ms for muscle (13) and a relaxivity value of 4.3 mM−1 sec−1 for Gd-DTPA (14) were used in the calculations.
Viable Tumor Segmentation
A single slice, showing most of the tumor area, was selected for further analysis and simulations. A background mask, based on a largest-connected-component and hole-filling algorithm, was first applied. Second, an image was constructed consisting of the maximum concentration level encountered in each voxel during the time course of the dynamic series. During the time course of 10 min, necrotic enhancement was not expected. Finally, a semi-automated connected-component algorithm was applied to this image to obtain a contiguous segmentation of the viable tumor tissue. The threshold was manually adjusted to ensure inclusion of most of the viable tumor tissue, resulting in a threshold range of 0.034 mM to 0.070 mM.
Fitting Routine
To fit the PK model to the data, a golden section search method was implemented (15). The driving force of the golden section search was the minimization of the goodness-of-fit measure R2. To facilitate the calculation of R2, the temporal resolution of Ct was increased to the temporal resolution of Cp by linear interpolation; i.e., in the event that Ct had a lower temporal resolution than Cp (Cp: Ts = 5 sec, Ct: Ts = 5 sec – 85 sec). The starting section for Ktrans was [0.001 min−1 – 1.0 min−1] and for ve it was [0.01 – 1.0]. The search section was narrowed down for Ktrans and ve until the difference between the lower and upper boundary was less than 10−3 min−1 or less than 10−3, respectively. The fitting routine was applied to voxels with significant CA uptake, i.e., to voxels with a maximum uptake higher than 0.02 mM.
RESULTS
Viable Tumor Segmentation
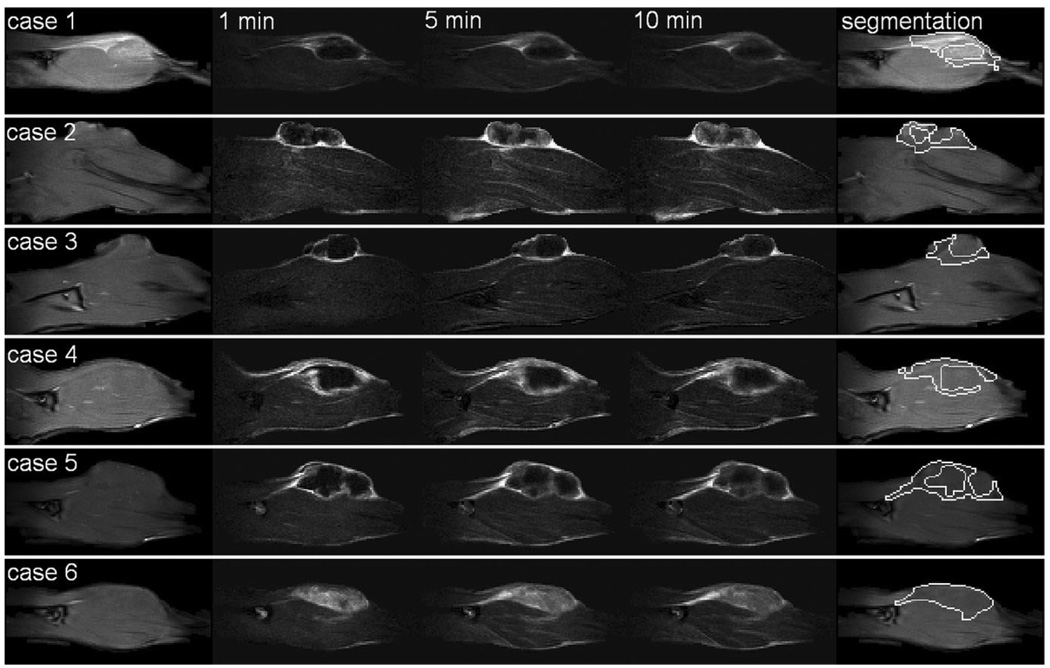
Figure 1 shows precontrast T1-weighted images and post-contrast subtraction images for all six cases. The viable tumor segmentations are shown in the last column. Except for case 6, all tumors developed a necrotic core. Therefore, only the periphery of the tumor was segmented for cases 1 to 5. The enhancement patterns vary from primarily early rim enhancement (case 2) to more global enhancement (case 4).
FIG. 1.
For six cases (rows), from left to right: the precontrast image, subtraction image at 1 min, subtraction image at 5 min, subtraction image at 10 min, and the viable tumor segmentation. The in-plane resolution is 0.31 mm × 0.31 mm. Images were cropped to a height of ~2.1 cm for display purpose. Except for case 6, all tumors developed a necrotic core.
k-Space-Based Sampling
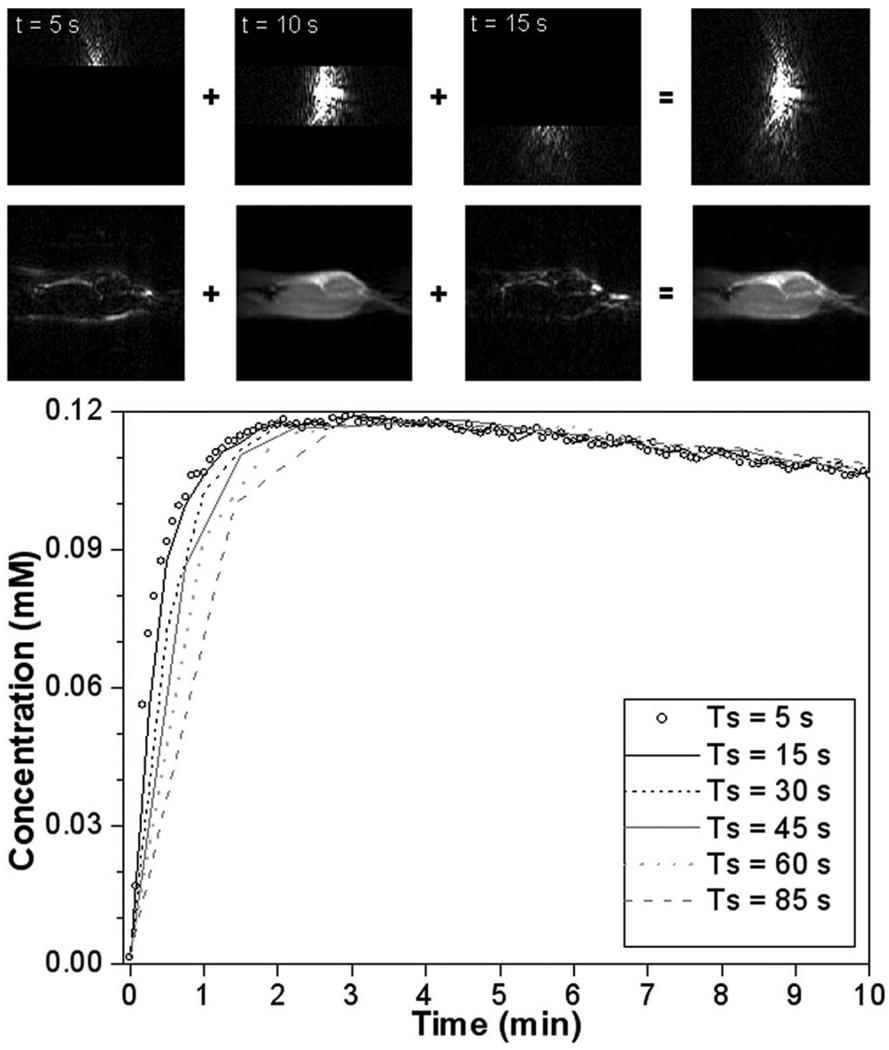
The k-space-based sampling strategy (Eq. 1) is illustrated in Fig. 2 for case 1. In this example we demonstrate down-sampling from Ts = 5 sec to Ts = 15 sec. The three consecutive sections of k-space data (top row images) add up to a new k-space image, which is equivalent to a k-space image acquired with Ts = 15 sec. Each section of k-space data, as well as the summation of the three, can be Fourier transformed to form an image (bottom row images). As expected, the central part of k-space data (low frequencies) is dominant in determining the overall signal intensity, whereas the outer parts of k-space data (high frequencies) define the details. In an equivalent way, smaller sections were combined to form new data sets at temporal resolutions of 30 sec, 45 sec, 60 sec, and 85 sec.
FIG. 2.
Illustration of the downsampling strategy (from Ts = 5 sec to Ts = 15 sec) mimicking a linear phase-encoding order, applied to case 1. First row of images: three consecutive parts of k-space data add up to a new set; second row: the corresponding images. Plot: the mean CA uptake in the viable tumor segmentation for case 1 at all temporal resolutions. Especially during the initial phase, the uptake curves do not overlap.
For the same example, the mean CA uptake curves (all temporal resolutions) for the viable tumor segmentation are shown in the plot (Fig. 2). As anticipated, low temporal resolution had more effect on the early phase than on the late phase. The k-space-based sampling strategy demonstrates that low-temporal-resolution curves do not necessarily consist of points taken from the original curve at regular intervals, as is assumed by direct sampling.
PK Analysis
The k-space-based and direct sampling strategies were both applied to all six DCE-MRI experiments. The original 5-sec and the equivalent lower temporal resolution (15 sec up to 85 sec) CA uptake curves were all fit with the same PK model to estimate Ktrans and ve. Tables 1 and 2 show the results of fitting the mean CA uptake curves for k-space-based and direct sampling, respectively. It can be seen (Table 1) that the underestimation of Ktrans as a function of temporal resolution is quite consistent. It varies from ~3% (Ts = 15 sec) to ~25% (Ts = 85 sec). For ve, the overestimation varies from ~1% to ~10%. Although the central k-space portion did not necessarily come from one 5-sec k-space image (e.g., Ts = 30 sec), this did not affect the overall tendency of the estimation errors. While direct sampling (Table 2) roughly shows underestimation of Ktrans and overestimation of ve, the influence of temporal resolution appears smaller than with k-space-based sampling. This implies that the assumption of instantaneous k-space data collection (direct sampling) is inappropriate, as it prevents us from fully simulating the effect of low temporal resolution. For all fits, R2-values were within a range of 0.77 – 1.00 (mean R2 = 0.93 ± 0.05).
Table 1.
Change of Ktrans (Top) and ve (Bottom) Estimated From the Mean CA Uptake in the Viable Tumor Tissue Across All Temporal Resolutions (k-Space-Based Sampling)
| Case # | 5 sec | 15 sec | 30 sec | 45 sec | 60 sec | 85 sec | |
|---|---|---|---|---|---|---|---|
| Ktrans (min−1) | Percentage change: Ktrans (%) | ||||||
| 1 | 0.21 | −3.4 | −9.7 | −14.3 | −19.5 | −27.2 | |
| 2 | 0.16 | −3.3 | −9.7 | −13.2 | −17.3 | −22.8 | |
| 3 | 0.15 | −3.0 | −8.0 | −13.0 | −16.0 | −22.5 | |
| 4 | 0.32 | −4.0 | −9.0 | −13.0 | −18.0 | −26.0 | |
| 5 | 0.21 | −3.2 | −8.6 | −13.2 | −18.0 | −24.9 | |
| 6 | 0.23 | −4.2 | −12.0 | −14.8 | −20.8 | −28.8 | |
| ve | Percentage change: ve (%) | ||||||
| 1 | 0.54 | 0.7 | 2.1 | 3.4 | 5.0 | 8.8 | |
| 2 | 0.69 | 1.4 | 4.4 | 6.5 | 9.1 | 14.2 | |
| 3 | 0.58 | 0.8 | 3.0 | 5.6 | 8.1 | 12.8 | |
| 4 | 0.83 | 1.1 | 2.1 | 3.2 | 5.0 | 8.7 | |
| 5 | 0.62 | 0.8 | 2.1 | 3.8 | 5.4 | 8.8 | |
| 6 | 0.49 | 0.6 | 1.7 | 2.4 | 4.6 | 7.1 | |
Table 2.
Change of Ktrans (Top) and ve (Bottom) Estimated From the Mean CA Uptake in the Viable Tumor Tissue Across All Temporal Resolutions (Direct Sampling)
| Case # | 5 sec | 15 sec | 30 sec | 45 sec | 60 sec | 85 sec | |
|---|---|---|---|---|---|---|---|
| Ktrans (min−1) | Percentage change: Ktrans (%) | ||||||
| 1 | 0.21 | −0.6 | −3.0 | −6.6 | −11.8 | −19.7 | |
| 2 | 0.16 | −0.5 | −0.5 | −1.7 | −5.2 | −11.8 | |
| 3 | 0.15 | 0.5 | −0.6 | −4.3 | −6.8 | −15.8 | |
| 4 | 0.32 | −0.3 | −2.2 | −6.4 | −12.4 | −22.3 | |
| 5 | 0.21 | −0.6 | −3.2 | −6.8 | −12.1 | −19.3 | |
| 6 | 0.23 | −0.8 | −3.1 | −6.1 | −12.8 | −21.4 | |
| ve | Percentage change: ve (%) | ||||||
| 1 | 0.54 | 0.1 | 0.3 | 0.7 | 2.3 | 4.9 | |
| 2 | 0.69 | 0.1 | −0.1 | 0.4 | 1.9 | 5.9 | |
| 3 | 0.58 | −0.3 | 0.2 | 1.0 | 1.7 | 5.8 | |
| 4 | 0.83 | 0.2 | 0.3 | 1.0 | 2.4 | 5.0 | |
| 5 | 0.62 | −0.2 | 0.3 | 1.0 | 2.7 | 5.0 | |
| 6 | 0.49 | −0.1 | 0.5 | 0.9 | 2.0 | 3.4 | |
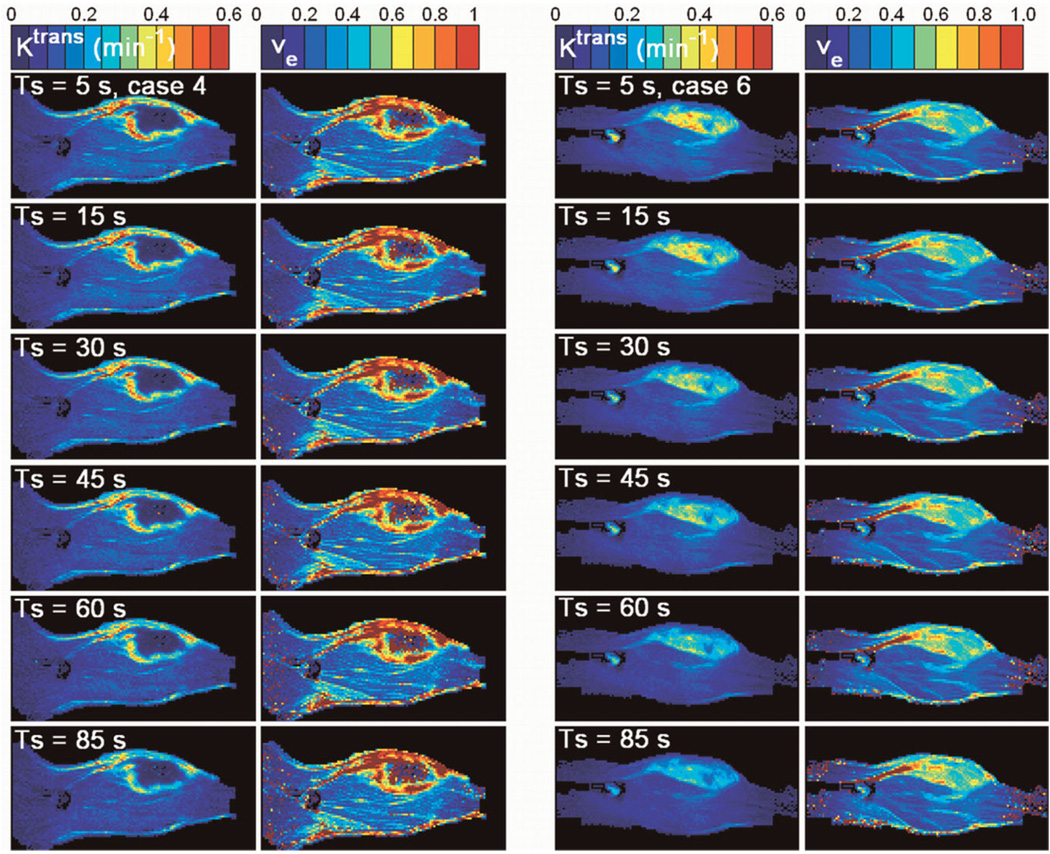
In addition to the above mean CA uptake analysis for the tumor region of interest, we also performed voxel-wise fitting using data derived with k-space-based sampling. The parametric color maps of Ktrans and ve for cases 4 and 6 were selected as examples (see Fig. 3) to show the difference between a tumor with (case 4) and without (case 6) a necrotic core. At first glance, the estimates of Ktrans and ve appear to be quite robust against temporal resolution, especially in muscle. A closer look shows that the hot spot areas present on the original Ktrans maps become less distinct with decreasing temporal resolution. The location of the tumor, however, remains visible up to a temporal resolution of 85 sec. For ve, there appears to be little influence of temporal resolution. The high ve values in the tumor rim (case 4) indicate the limitation of the model under study. For these curves, an unrealistically large ve is required to obtain an accurate fit.
FIG. 3.
Columns 1 and 2 show Ktrans and ve for case 4. Columns 3 and 4 show Ktrans and ve for case 6. From top to bottom, the temporal resolution decreases (Ts = 5 sec, 15 sec, 30 sec, 45 sec, 60 sec, and 85 sec). These two cases are selected to show the difference between a tumor with and without a necrotic core.
Finally, to see if the observed trends for estimation errors in Ktrans and ve (Table 1) were statistically significant, one-sided sign tests (16) were applied to the parametric difference maps for subsequent Ts: (5 sec, 15 sec), (15 sec, 30 sec), (30 sec, 45 sec), (45 sec, 60 sec), (60 sec, 85 sec). For each rat and each combination of subsequent Ts, the null hypotheses that the median of the difference map Ktrans(Ts,2) − Ktrans(Ts,1) ≥ 0, and the median of the difference map ve(Ts,2) − ve(Ts,1) ≤ 0 were tested, with Ts,2 > Ts,1. Both hypotheses were rejected for all tests (P < 0.00001). This means that the progressive underestimation of Ktrans and the progressive overestimation of ve are statistically significant.
DISCUSSION
The influence of the temporal resolution of DCE-MRI data was investigated in the context of the basic two-compartment model, while using a high-temporal-resolution AIF. The original data were downsampled to lower temporal resolutions by making use of a k-space-based recombination technique. The downsampled data, as well as the original data, were fit with the PK model. According to both the mean CA uptake curve fitting and the voxel-wise fitting, Ktrans is progressively underestimated with decreasing temporal resolution, whereas ve is progressively overestimated. The estimation errors are larger for Ktrans than for ve, because the rapid uptake phase is especially affected by a drop in temporal resolution. Even though the “true” Ktrans values (at Ts = 5 sec) range from 0.15 min−1 to 0.32 min−1 (Table 1), the influence of temporal resolution appears to be consistent across all cases. For ve, this is less the case.
In comparing k-space-based sampling to direct sampling, the key difference is that the first strategy takes the effect of transient CA uptake into account, while the latter does not. Consequently, the effect of low temporal resolution appears smaller and less consistent with direct sampling than is actually the case. Using direct sampling and the basic two-compartment model, Henderson et al. (4) found that the required sampling time for the uptake in tissue should be shorter than 16 sec to obtain an error smaller than 10% in either Ktrans or ve. The sampling time at which we predict the same error level is about twice as large using k-space-based sampling (Ts ≈ 30 sec), and about three times as large using direct sampling (Ts ≈ 45 sec – 60 sec). The discrepancy between the two studies is probably due to the large differences in the experimental setups.
Although we used a more realistic downsampling strategy than the one most commonly used, there are several limitations to this study.
The k-space-based sampling strategy presented in this study mimics a linear phase-encoding order, and the conclusions reached here are not valid for other acquisition schemes.
Temporal resolution was not reduced in exchange for higher image quality and/or larger image volume. In a scanner experiment, a reduction of temporal resolution (while keeping spatial resolution constant) would be expected to lead to an increase in signal-to-noise ratio.
Variations in the relative alignment of the sampling grid (4,17,18) were not taken into account.
The basic two-compartment model does not include a vascular contribution, which could be significant for certain tumors. Its exclusion could have introduced biases on the estimates of Ktrans and ve (19,20).
Emphasis was placed on the temporal resolution of the CA uptake in tissue, and therefore individual hightemporal-resolution AIFs were used. These high-temporal-resolution AIFs would not be available in case of an actual low-temporal-resolution acquisition. We are currently looking into the use of individual AIFs extracted from low-temporal-resolution data.
The study was designed to gain insight into estimation errors in Ktrans and ve for DCE-MRI data acquired at “suboptimal” temporal resolutions. It is important to study the modeling of CA uptake at low temporal resolution because, for instance, clinical DCE-MRI data of the breast are commonly acquired at a temporal resolution of about 60 sec to 120 sec. Under the applied model assumptions, we estimate that a temporal resolution of 60 sec would lead to an error margin of ~18% for Ktrans and ~6% for ve. Especially regions with very high Ktrans are significantly attenuated. However, the blood circulation of rats is about three times faster than that of humans. A similar study should therefore be performed using human DCE-MRI data. As we demonstrated by comparing our k-space-based downsampling strategy to the direct sampling strategy, the impact of the phase-encoding order on the appearance of CA uptake curves should be considered in both simulation and acquisition methods.
ACKNOWLEDGMENTS
We thank T. Twellmann and B. F. Coolen from the Eindhoven University of Technology for fruitful discussions on temporal downsampling; D. Mustafi and M. A. Zamora from the University of Chicago for acquiring the data; C. R. Haney from the University of Chicago for the segmentation algorithm; and T. Akinbiyi from the University of Chicago for consulting on the statistics.
REFERENCES
- 1.Kuhl CK, Schild HH, Morakkabati N. Dynamic bilateral contrast-enhanced MR imaging of the breast: trade-off between spatial and temporal resolution. Radiology. 2005;236:789–800. doi: 10.1148/radiol.2363040811. [DOI] [PubMed] [Google Scholar]
- 2.Mann RM, Kuhl CK, Kinkel K, Boetes C. Breast MRI: guidelines from the European Society of Breast Imaging. Eur Radiol. 2008;18:1307–1318. doi: 10.1007/s00330-008-0863-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tofts PS, Brix G, Buckley DL, Evelhoch JL, Henderson E, Knopp MV, Larsson HB, Lee TY, Mayr NA, Parker GJ, Port RE, Taylor J, Weisskoff RM. Estimating kinetic parameters from dynamic contrast-enhanced T(1)-weighted MRI of a diffusable tracer: standardized quantities and symbols. J Magn Reson Imaging. 1999;10:223–232. doi: 10.1002/(sici)1522-2586(199909)10:3<223::aid-jmri2>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 4.Henderson E, Rutt BK, Lee TY. Temporal sampling requirements for the tracer kinetics modeling of breast disease. Magn Reson Imaging. 1998;16:1057–1073. doi: 10.1016/s0730-725x(98)00130-1. [DOI] [PubMed] [Google Scholar]
- 5.Aref M, Handbury JD, Xiuquan JJ, Aref S, Wiener EC. Spatial and temporal resolution effects on dynamic contrast-enhanced magnetic resonance mammography. Magn Reson Imaging. 2007;25:14–34. doi: 10.1016/j.mri.2006.09.025. [DOI] [PubMed] [Google Scholar]
- 6.Ramirez MS, Ragan DK, Kundra V, Bankson JA. Feasibility of multiple-mouse dynamic contrast-enhanced MRI. Magn Reson Med. 2007;58:610–615. doi: 10.1002/mrm.21348. [DOI] [PubMed] [Google Scholar]
- 7.Aerts HJ, van Riel NA, Backes WH. System identification theory in pharmacokinetic modeling of dynamic contrast-enhanced MRI: influence of contrast injection. Magn Reson Med. 2008;59:1111–1119. doi: 10.1002/mrm.21575. [DOI] [PubMed] [Google Scholar]
- 8.El-Khouli R, Jacobs M, Wei J, Barker P, Macura K, Bluemke D. Dynamic contrast enhanced MRI of the breast: kinetic curve analysis to determine the optimal temporal resolution. ISMRM 16th Scientific Meeting; 2008. p. 3751. [Google Scholar]
- 9.Gal Y, Mehnert A, Bradley A, McMahon K, Crozier S. An evaluation of four parametric models of contrast enhancement for dynamic magnetic resonance imaging of the breast. Conf Proc IEEE Eng Med Biol Soc. 2007;2007:71–74. doi: 10.1109/IEMBS.2007.4352225. [DOI] [PubMed] [Google Scholar]
- 10.Kovar DA, Lewis M, Karczmar GS. A new method for imaging perfusion and contrast extraction fraction: input functions derived from reference tissues. J Magn Reson Imaging. 1998;8:1126–1134. doi: 10.1002/jmri.1880080519. [DOI] [PubMed] [Google Scholar]
- 11.Fan X, Medved M, River JN, Zamora M, Corot C, Robert P, Bourrinet P, Lipton M, Culp RM, Karczmar GS. New model for analysis of dynamic contrast-enhanced MRI data distinguishes metastatic from nonmetastatic transplanted rodent prostate tumors. Magn Reson Med. 2004;51:487–494. doi: 10.1002/mrm.10737. [DOI] [PubMed] [Google Scholar]
- 12.Medved M, Karczmar G, Yang C, Dignam J, Gajewski TF, Kindler H, Vokes E, MacEneany P, Mitchell MT, Stadler WM. Seminiquantitative analysis of dynamic contrast enhanced MRI in cancer patients: Variability and changes in tumor tissue over time. J Magn Reson Imaging. 2004;20:122–128. doi: 10.1002/jmri.20061. [DOI] [PubMed] [Google Scholar]
- 13.Marzola P, Mocchegiani E, Nicolato E, Tibaldi A, Sbarbati A, Osculati F. Chemical shift imaging at 4.7 tesla of thymus in young and old mice. J Magn Reson Imaging. 1999;10:97–101. doi: 10.1002/(sici)1522-2586(199907)10:1<97::aid-jmri14>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 14.Furman-Haran E, Margalit R, Grobgeld D, Degani H. Dynamic contrast-enhanced magnetic resonance imaging reveals stress-induced angiogenesis in MCF7 human breast tumors. Proc Natl Acad Sci U|S|A. 1996;93:6247–6251. doi: 10.1073/pnas.93.13.6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Press WH, Teukolsky SA, Vetterling WT, Flannery BP. Numerical recipes in Fortran 77: the art of scientific computing. 2nd ed. New York: Cambridge University Press; 1992. p. 963. [Google Scholar]
- 16.Gibbons JD, Chakraborti S. Nonparametric statistical inference. 4th ed. New York: Marcel Dekker; 2003. p. 645. [Google Scholar]
- 17.Cheng HL. Investigation and optimization of parameter accuracy in dynamic contrast-enhanced MRI. J Magn Reson Imaging. 2008;28:736–743. doi: 10.1002/jmri.21489. [DOI] [PubMed] [Google Scholar]
- 18.Laue HO, Althaus M, Behrens S, Hahn HK, Peitgen H-O. Dependency of parameter estimates for the Tofts model on temporal sampling rate and on bolus arrival time. ISMRM 15th Scientific Meeting; Berlin, Germany. 2007. p. 1722. [Google Scholar]
- 19.Buckley DL. Uncertainty in the analysis of tracer kinetics using dynamic contrast-enhanced T1-weighted MRI. Magn Reson Med. 2002;47:601–606. doi: 10.1002/mrm.10080. [DOI] [PubMed] [Google Scholar]
- 20.Harrer JU, Parker GJ, Haroon HA, Buckley DL, Embelton K, Roberts C, Baleriaux D, Jackson A. Comparative study of methods for determining vascular permeability and blood volume in human gliomas. J Magn Reson Imaging. 2004;20:748–757. doi: 10.1002/jmri.20182. [DOI] [PubMed] [Google Scholar]