Abstract
Live-cell single mRNA imaging is a powerful tool, but has been restricted in higher eukaryotes to artificial cell lines and reporter genes. We describe an approach that enables live-cell imaging of single endogenous labeled mRNA molecules transcribed in primary mammalian cells and tissue. We generated a knock-in mouse line in which an MS2 binding site (MBS) cassette was targeted to the 3′UTR of the essential β-actin gene. As β-actin-MBS was ubiquitously expressed, we were able to uniquely address endogenous mRNA regulation in any tissue or cell type. We simultaneously followed transcription from the β-actin alleles in real-time and observed transcriptional bursting in response to serum stimulation with precise temporal resolution. We performed tracking of single endogenous labeled mRNA particles being transported in primary hippocampal neurons. The MBS also provided a means for high sensitivity Fluorescence In Situ Hybridization (FISH), allowing detection and localization of single β-actin mRNA molecules in various mouse tissues.
Introduction
The messenger RNA (mRNA) molecule is the first intermediate of gene expression1. At every stage of its lifetime, it is subject to regulation, both in space and time: transcription at the gene locus2, export through the nuclear pore3, diffusion and transport through the cytoplasm that in some cases result in localization of mRNA4, and eventually decay, perhaps in specialized bodies5. These levels of regulation are involved in many biological processes and diseases, 6. To achieve a complete understanding of gene expression regulation in physiological conditions as well as in the context of disease, one would ideally be able to visualize single mRNA molecules in real time and over their lifetime in a living organism.
Such an experiment has been out of reach, because it has not been possible to follow specific mRNAs transcribed from genes in their endogenous genomic context in primary cells or tissues. Therefore the vast majority our knowledge about mRNA regulation is derived from cultured cells. In culture, imaging of various stages of the mRNA life cycle has been possible using fluorescence microscopy techniques such as FRAP, FCS or wide field microscopy7. These are particularly powerful when coupled with the in vivo mRNA imaging approach using the MS2 system8. In this technique, a sequence derived from the bacteriophage MS2 genome is inserted into a gene of interest. When transcribed, the RNA immediately folds into a hairpin that forms the binding site (MS2 binding site, MBS) for the bacteriophage capsid protein (MS2 capsid protein, MCP). When cells co-express both a gene of interest carrying the MBS and a fusion of MCP with a fluorescent protein (FP), mRNAs are fluorescently labeled from the moment they are transcribed (Fig. 1a). Insertion of multiple MBS (24) copies increases the signal-to-noise ratio (SNR) so that single mRNAs can be amplified over the background of freely diffusing fluorescent MCP-FP8.
Fig. 1.
Schematic of the actin-MS2 system for live cell imaging. (a) As the gene gets transcribed by the RNA polymerase (RNAP), the RNA hairpins form and get bound by the co-expressed MCP-YFP. (b) Structure of the cassette. A unit containing two MBS sequences and the intervening linkers is repeated 12 times within the array, resulting in 24 MBS. [this should go to Methods]. The MBS array is inserted downstream of the zipcode regulatory region (green). (c) Top panel: Principle of the construct. The long homology arm (LA) encompasses the full β-actin gene, including a region 4 kbp upstream of the transcription start site (TSS); the positions of the exons (blue), introns (gray), start and stop codon, and polyadenylation site (polyA) are indicated. The short homology arm extends 1.3 kbp downstream of the Neo cassette (yellow, flanked by the two Lox sites indicated by purple arrows The 24x MBS cassette (red) is inserted within the 3′UTR in the sixth exon. Bottom panel: Resulting genomic locus in the β-actin-MBS mouse.
The MS2 system has been used in various contexts, such as bacteria9, yeast8, Dictyostelium10, Drosophila11 and mammalian cells12–16. These studies have yielded a wealth of information about transcription kinetics (such as on-off pulsing of gene expression, or the dynamics of elongation, including pausing), the dynamics of mRNPs and their intracellular localization. So far the MS2 system has only been applied to higher eukaryotes in the context of artificial reporter genes, usually inserted in many copies (10–1,000) within a random locus17. Imaging artificial reporters in live cultured cells constitutes a useful experimental model, but presents some limitations. In those systems consisting of many copies of the same gene, it is not possible to extract events happening at the single gene level due to the unsynchronized interactions taking place at the other genes within the locus. A recent improvement of the technique makes it possible to specifically insert single gene copies into the genome of a host cell line18. However, reporter genes might be prone to regulation artifacts. Importantly, the use of immortalized cell lines introduces an unknown factor in the analysis of gene expression, where regulatory events such as cell cycle controls are overshadowed by the transformed phenotype19. Finally, these systems do not permit the study of the expression of mRNA in primary cells or in tissue. As microscopy improves the ability to image in living tissue, gene reporters will be necessary for the monitoring of expression, for instance of disease-related defects.
Clearly, the next step in understanding mRNA regulation in its native state is to apply the MS2 approach to a single endogenous gene within a multicellular organism. We have generated a mouse that carries MBS repeats in the 3′UTR of both alleles of the β-actin gene. We show that in addition to its use for live-cell imaging, the repeated MBS stem-loops provide an efficient target for high sensitivity FISH in any tissue. We present a method to perform live-cell imaging in any cell type, by isolating the desired cells and by coexpressing a fluorescent MCP-FP either by transfecting the plasmid coding for the MCP-FP gene, or stably integrating the MCP-FP coding sequence in the genome using lentivirus infection. We demonstrate the utility of this approach using the examples of mouse embryo fibroblasts (MEF) and primary neuronal cell lines. This illustrates how single gene kinetics in native cells can reveal details of the molecular mechanisms characteristic of different cell types. The β-actin-MBS mouse offers a vast potential for imaging the lifecycle of an endogenous gene in its native environment, at the single molecule level.
Results
Generation of a homozygous β-actin-MBS mouse
We chose the β-actin gene to demonstrate the feasibility of an MBS mouse. β-actin mRNA is an essential, ubiquitous, highly expressed20, long-lived mRNA21. β-actin mRNA is known to localize to the leading edge of fibroblasts, a process involved in maintaining cell polarity22. The understanding of this localization mechanism is important, as enhanced localization is correlated with persistent motility and decreased metastasis6. β-actin mRNA is transcribed at a basal level in dividing cells, however its transcription can also be induced in response to serum addition20. Therefore it provides a good model to study both the regulation of a constitutively expressed gene and the transcriptional response to environmental cues.
The mouse β-actin gene consists of 6 exons spanning 3.5 kbp on chromosome 5, and codes for the 375 amino acid β-isoform of actin protein. The MBS motif can in principle either be introduced into the 5′UTR or the 3′UTR of the gene. A high number of MBS repeats increases the signal to improve detection over a background of free fluorescent protein. We inserted the 1.2 kbp cassette containing 24 repeats of the MBS motif 441 bp downstream of the stop codon and after the zipcode, a cis regulatory sequence important for mRNA localization23 (Fig. 1b–c). A construct carrying the modified gene and a Lox-NeoR cassette was linearized and electroporated into ES cells. After PCR screening, positive ES clones were transfected with MCP-GFP protein to verify the correct expression of the transgene and to ensure that single gene detection was feasible (data not shown). These ES cell lines were used to generate chimeric mice and these chimeras were mated and screened for germ-line transmission to create knock-in mice. Homozygous knock-in mice are viable and fertile, indicating that the presence of the MBS is not notably deleterious.
We first verified that the transgene expressed the expected β-actin-MBS mRNA. On a northern blot, the β-actin-MBS mRNA was a single band with the expected shift in migration due to the insertion of the MBS site (Supplementary Fig. 1). RNA isolated from various tissues demonstrates expression of the complete MBS-containing mRNA in all tissues examined (Supplementary Fig. 1).
FISH imaging of β-actin mRNA in tissue
In addition to its potential for live cell imaging, the MBS sites provide a target sequence that enhances its detection with Fluorescence in situ Hybridization (FISH). A mixture of three different probes complementary to the linker regions within the MBS repeats can potentially bind to 36 sites on the mRNA, providing strong signal amplification for detection within tissue sections (Fig. 1b). To demonstrate this potential, we sectioned paraffin embedded tissue and performed RNA FISH using the MBS probes (Fig. 2). In both the heterozygous (Actb+/M) and homozygous (ActbM/M) tissue we observed diffraction limited particles (Fig. 2b–c,e–f,h–i,k–l,n–o). No such particles could be observed in the wild type (Actb+/+) case (Fig. 2a,d,g,j,m), indicating that these correspond to β-actin-MBS mRNA particles. To demonstrate the potential of the technique to quantify expression across tissue, we counted single mRNA particles in the cerebellum (Fig. 2p). We found as expected that the heterozygous mice expressed about 50% less β-actin-MBS mRNA than the homozygous mice. Single molecule mRNA FISH enables accurate mapping of mRNA spatial localization in tissue. Although β-actin mRNA localization to the leading edge of cultured fibroblasts is well documented22 (see also Fig. 3), what happens in tissue is still unclear. Using the MBS FISH probes, we quantified the spatial variations in the concentration of mRNA within the liver and cerebellum (Fig. 2q). The nuclear β-actin-MBS mRNA concentration was on average lower than that outside of the nucleus, which probably reflects the difference between the time scales of transcription and export (few minutes15), and the lifetime of the β-actin mRNA (few hours). Interestingly, we observed that the concentration of mRNA decreased with increasing distance from the nucleus in the cerebellum. This was not the case in liver tissue (Fig. 2q, compare panels 2c and 2i). This demonstrates the potential of our technique to quantitatively assess the spatial variations of mRNA concentration, an element that is crucially needed to establish a complete picture of gene expression in vivo. The probes targeted at the MBS cassette provided a much higher RNA FISH signal to noise ratio than a set of probes complementary to the β-actin coding sequence. This demonstrates that in addition to its live cell applications, the MBS sequence constitutes a unique FISH signal enhancing tag for detecting β-actin transcription sites24 as well as mRNP particles in mouse tissue, opening avenues for studying β-actin mRNA regulation at the organ level.
Fig. 2.
RNA FISH in sections from various tissues. Merge of DAPI signal (blue) and Cy5 fluorescence from three FISH probes targeting the MBS cassette (red; bandpassed data, see Supplementary Fig. 7). Panels (a–l) bar: 10 μm; (m–o) bar: 5 μm. (p) Quantification of the expression levels of the β-actin-MBS allele in the cerebellum. Number of spots counted after thresholding the FISH signal. (q) Average mRNA concentration inside the nucleus (left panel), and outside the nucleus, displayed as a function of the distance from the nuclear boundary (right panel). Both concentrations were normalized to their value at the nucleus boundary. AU: arbitrary units.
Fig. 3.
β-actin mRNA localizes to the leading edge of primary fibroblasts isolated from MBS mice. (a) Differential interference contrast (DIC), (b) DAPI-stain, (c) FISH with Cy3-labeled probes to the β-actin coding region, and (d) FISH with Cy5-labeled probes to the MBS cassette. Scale bar, 10 μm. (e) Time-lapse images (min indicated) of a primary fibroblast migrating on a fibronectin substrate. Cells were infected with lentivirus that expresses NLS-MCP-GFP, and stained with membrane-permeable red cytoplasmic dye. The color map represents the fluorescence intensity of NLS-MCP-GFP (normalized by the red cytoplasmic dye intensity to account for the cell volume; AU: Arbitray Units). Cell migration was observed towards the direction of mRNA localization. Scale bar, 20 μm.
Live imaging in cell lines derived from the mouse
Any primary cell from the mouse is amenable to live cell imaging, which is a major advantage of the present technique over cell-line based methods. In the following we present applications to two cell types, fibroblasts and primary neurons. The fibroblast cell line described here has also been used to study the export of single mRNA particles through nuclear pores with nm resolution25.
We prepared E14 MEF cells from heterozygous and homozygous knock-in embryos and immortalized these using SV40 T antigen. The immortalized MEF cell lines were used to stably express NLS-MCP-YFP with a nuclear localization sequence (NLS). The NLS maintained a high NLS-MCP-YFP concentration in the nucleus and depleted the cytoplasmic background of unbound NLS-MCP-YFP protein. This ensured the mRNA was bound by fluorescent NLS-MCP-YFP as soon as it was transcribed and that single molecules would be visible in the cytoplasm since there would be no unbound NLS-MCP-YFP to decrease the contrast. Several cell lines were isolated with varying levels of stably expressed NLS-MCP-YFP, and without the NLS as well. We karyotyped one cell line that was determined to be tetraploid, a common consequence of T antigen immortalization26 (Supplementary Fig. 2).
Homozygous cell lines expressed similar levels of β-actin-MBS mRNA and β-actin protein regardless of whether they coexpressed the NLS-MCP-YFP fusion protein (Supplementary Fig. 1b–c). β-actin protein levels were identical in all cell lines, although the homozygous mRNA levels were slightly lower than the wild type cells (Supplementary Fig. 3). These results confirmed that neither the MBS nor the NLS-MCP-YFP fusion protein prevented β-actin expression. The γ-actin protein levels were also identical in all cell lines indicating that there was no isoform compensation27 (Supplementary Fig. 1c). Finally, we verified that the actin cytoskeleton portrayed normal morphology in all cell lines by immunostaining MEFs with actin isoform specific antibodies (Supplementary Fig. 1d).
We then established that the repeats do not affect the transcription of the β-actin gene by comparing the numbers of β-actin mRNA nascent chains at the gene loci of the wild type and MBS-containing alleles in the heterozygous cell line. Quantitative RNA FISH showed that both alleles were transcribed with similar levels (Supplementary Fig. 4). In live homozygous primary fibroblasts, we were able to detect transcription sites as two bright nuclear spots and mRNA particles as they moved in the cell (Supplementary movie 1). Importantly, we confirmed that β-actin-MBS mRNA demonstrated the expected spatial localization to the leading edge of primary living fibroblasts28 (Fig. 3, Supplementary Fig. 5), demonstrating that the repeats did not interfere with recognition of the zipcode sequence and hence affect mRNA localization.
Transcriptional bursting in MEFs after serum stimulation
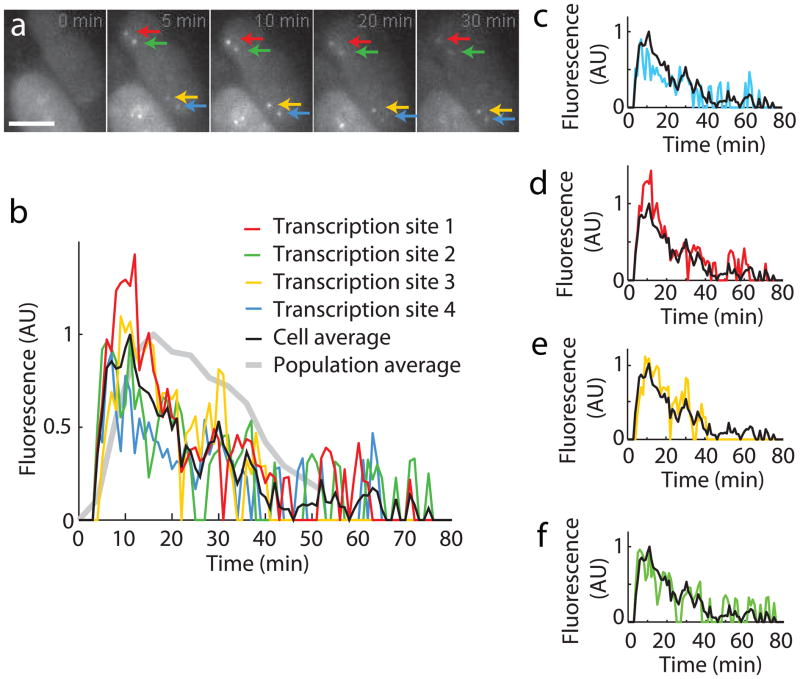
We used the labeled MEF cells to analyze serum-induced β-actin mRNA transcription 20. We cultured homozygous MEF cells overnight in low serum. After addition of serum-containing medium, we imaged cells over time to follow β-actin transcription. At the time of induction, we could only detect fluorescence background in the nucleus due to freely diffusing NLS-MCP-YFP (Fig. 4a). A few minutes after induction, four bright fluorescent spots corresponding to the β-actin loci appeared in the nuclei and increased in intensity until ~ 10 minutes, before gradually fading (Fig. 4a). RNA FISH probes targeted to both the β-actin coding region and the MBS cassette colocalized with the bright NLS-MCP-YFP spots observed in the nucleus (Supplementary Fig. 6). Quantification of the intensities at each gene locus showed that the cells responded to serum by a burst of transcription that started after a few minutes and lasted for ~ 1 h (Fig. 4b). The four alleles, although subject to the same regulatory features (identical loci and intracellular cues) displayed variations in their response (Fig. 4c–f). Interestingly, although the alleles displayed a high synchrony in the initial minutes of the response, they quickly started to diverge, and displayed a largely uncorrelated behavior after 40 minutes. Different cells displayed distinct response curves that produced a broader response curve when averaged together (Fig. 4b). The serum response kinetics observed here in the β-actin MBS system were consistent with a previous RNA FISH analysis20. However, live cell imaging offers many advantages such as finer time resolution than FISH and the possibility to follow the fate of each allele within an individual cell over long periods of time. Importantly, the expression of the endogenous alleles will allow a rigorous evaluation of the intrinsic and extrinsic factors regulating β-actin gene expression, a model essential constitutive gene, in its native state in a variety of tissues.
Fig. 4.
Live cell imaging of serum response in MBS immortalized MEFs. (a) Images of MEF during serum response taken from left to right 0, 5, 10, 20 and 30 min after serum was added (Maximum intensity projection of z-stacks). At the time of serum addition, no transcriptional activity is detected. After 5 minutes, all four transcription sites are detected as bright nuclear spots (arrows); immortalized MEFs are tetraploid, (Supplementary Fig. 2). The transcriptional activity reaches a peak after around 10 min, before gradually shutting down. Scale bar, 5 μm. (b) Quantification of the fluorescence intensity at the transcription sites. Red, green blue and yellow: fluorescent intensity at the transcription sites in panel (a). Black: average response of the four alleles in panel (a). Gray: average response over N = 11 cells. (c–f) individual plots of each of the alleles. AU: arbitrary units.
β-actin mRNA transport in neurons
Due to their extremely polarized structure, neurons provide an intrinsic challenge to gene expression: how, when and in what form do mRNAs reach the synapses where they undergo localized translation in order to strengthen specific synapses required for the stable interactions required for memory? We isolated primary neurons from MBS mouse embryos, and expressed the NLS-MCP-YFP. It was then possible to follow the motion of individual particles in neuronal processes with high (20 frames.s−1) spatiotemporal resolution (Fig. 5a–d, Supplementary movie 2 and 3). We observed several qualitatively different particle behaviors: unidirectional motion (anterograde and retrograde sometimes on the same process, Supplementary movie 4 and 5), bidirectional motion (one particle moving in one direction then back in the opposite direction, Supplementary movie 6), branching motion (one particle moving from one branch of a process to another, Supplementary movie 7), or immobile particle (Supplementary movie 8). Quantitative analysis of the tracks (Fig. 5e–f) yielded instantaneous transport rates in the 0.5–5μm.s−1 range, with averages around 3 μm.s−1. Interestingly, these rates are similar to the ones observed when tracking zipcode binding protein ZBP1 granules29, a protein that binds β-actin mRNA and is crucial for its localization. Previous work on granules has used transfected reporter genes to follow the movements of mRNAs in neurons, a process that inevitably leads to overexpression artifacts. This is the first detection of an endogenously expressed mRNA at its normal level and will allow for a more physiological analysis of the composition of the granules (are they single β-actin mRNAs, or clusters?) as well as details of their movements.
Fig. 5.
Live cell imaging of mRNP transport in primary hippocampal neurons. Neurons transfected with MCP-YFP were imaged at 20 frames.s−1. (a–d): images (1 s apart) showing an mRNP moving unidirectionally along a neuronal process. Bar: 10 μm. (e) trajectories of 2 particles observed successively along the same process (trajectories vertically shifted for clarity, heat map indicates earliest, blue, versus progressively later, latter, red image frames.). (f) Instantaneous rates of both mRNPs (resp. averages ± s.e.m.: 2.95 ± 0.14 μm.s−1; 2.90 ± 0.13 μm.s−1).
Discussion
Labeling of endogenous β-actin mRNA with the MBS cassette makes it possible to investigate how different tissues maintain a homeostasis of this structural protein. This approach is also a means to further develop high resolution, rapid imaging capabilities in tissue since single mRNA particles are visible in fixed tissue. Therefore it should be possible to record events of mRNA expression such as transport and localization in differentiating or regenerating tissues. This approach offers several advantages over existing techniques: the gene studied is endogenous, in its native chromosome locus (including promoter, terminator and any other regulatory sequences); it is present at a single copy (hence imaging the activity at the locus truly reflects the activity of a single gene versus the average activity of many copies of the same gene in the case of a gene array); both alleles of the gene are amenable to imaging, which constitutes a useful system to study transcriptional regulation and variability; it is expressed in the animal, and therefore amenable to studying gene expression within tissue or within any desired primary cell type. We have proven the feasibility of the MBS label on a crucial housekeeping gene, β-actin, without disrupting the animal physiology. We anticipate that the approach presented here is applicable to any gene of interest.
In addition to the increased sensitivity we demonstrated in tissue FISH and live cell experiments, the β-actin-MBS mouse can be used for many further developments. The obvious application is intravital mRNA imaging. The present mouse contains only the protein binding site, but the complete system (β-actin-MBS coexpressed with a fusion MCP-FP) can be obtained by various means. For example, one could cross the β-actin-MBS mouse with a mouse expressing a fluorescent MCP-FP fusion, or locally infect the β-actin-MBS mouse with viruses designed to express the fluorescent MCP-FP. Intravital studies of β-actin mRNA localization could lead to better understanding the role β-actin mRNA localization plays in cancer and metastasis6. Also of interest is the study of the modes of transcription regulation at the level of the organism, a crucial feature e.g. during development30.
Finally, the β-actin-MBS mouse also provides a powerful tool for biochemistry. Taking advantage of the 24 high affinity binding sites, one should be able to isolate the β-actin mRNA with a very high signal to noise ratio. This will make it possible to identify the binding partners of β-actin mRNA in various tissues.
We therefore anticipate that the β-actin-MBS mouse line will provide a valuable tool to study the mRNA lifecycle using multiple, complementary approaches (from fixed- and live-cell imaging to biochemistry). In addition, the present method can be adapted to study the expression of any gene of interest in its natural context.
Online Methods
Generation of the knock-in mouse
We targeted the MBS cassette to the 3′UTR of the mouse β-actin gene, downstream of the zipcode. We identified a suitable region containing appropriate restriction sites for integration of the 24 × MBS cassette. We used a ~ 9.6 kb genomic region to construct the targeting vector that was first sub-cloned from a positively identified BAC clone containing the mouse β-actin gene. We designed the region such that the long homology arm (LA) includes the full β-actin gene, extending from ~ 4 kbp upstream of the β-actin transcription start site to ~ 750 bp downstream of the end of the last exon. The short homology arm (SA) extends 1.3 kb 3′ downstream of the Neo cassette (Fig. 1c). The 1.3 kbp long MBS cassette is inserted 441 bps downstream to the stop codon.
We digested the MBS cassette on both ends using MluI and ligated the product into the MluI site of a vector containing the loxP flanked Neo cassette. We synthesized a PCR fragment from the genomic DNA using primers containing respectively MluI and AscI sites to generate the sequence between the site of the MBS cassette insertion and the site of the Neo cassette insertion. We ligated it into the vector between the MBS and the Neo cassettes. We excised the region including the MBS repeats, the intervening genomic sequence and the Neo cassette using BsiWI and ligated into the targeting construct. The exact genomic sequence is restored except for the insertion of the MS2 repeats and the floxed Neo cassette at the locations described above (Fig. 1c). We confirmed the targeting vector by restriction analysis and sequencing.
The generation of the targeting vector, the ES cells and the heterozygous mice was performed by Ingenious Targeting Laboratory Inc. The targeting vector was linearized and transfected by electroporation into 129SVev ES cells. After selection in G418, we expanded surviving clones for PCR analysis in order to identify recombinant ES clones. We obtained three stable ES clones and verified the expression of the β-actin-MBS by RNA FISH and by co-expression of MCP-GFP in both undifferentiated and differentiated state. We injected positive ES cells into foster mothers and obtained chimeric litter (based on coat color). Two males showing 90% chimerism were mated with female mice, and 7 positive F1 germline mice (4 males, 3 females) were obtained from clone 3C-C3. We verified F1 mice by genotyping and PCR of the MS2 sequences, as well as RNA FISH in primary mice cells derived from mouse tissues. Experiments on mice were approved by the Institutional Animal Care and Use Committee of Albert Einstein College of Medicine (Protocol Number 20100908).
Plasmids
We created a NLS-MCP-YFP gene using PCR. The final construct adds an NLS and an HA tag to the N-terminus of the previously reported construct MCP-YFP31. We cloned this expression cassette into the pHAGE-Ubc-RIG lentiviral vector32 from which the DsRed-IRES-GFP fragment had been excised using NotI and ClaI. We used this plasmid to create recombinant lentiviral particles generating expression of NLS-MCP-YFP driven from the human Ubiquitin C promoter in target cells.
Isolation of primary hippocampal neurons from mouse embryos
We isolated embryonic day E18 mice from the pregnant female. We excised the hippocampus from the brain and placed it into sterile cold Hank’s Balanced Salt Solution (HBSS, without Mg2+, without Ca2+ supplemented with 5 mM HEPES). We then dissociated tissue using a scalpel, resuspended it in 15 ml ice cold HBSS, and spun down (100 g for 1 minute). We resuspended the supernatant into 2 ml per brain HBSS supplemented with 1:10 of 2.5% Trypsin solution (Invitrogen), and incubated 20 minutes at 30°C. We added 1/9 volume of OMI (Ovomucid Trypsin inhibitor, 10 mg.ml−1 in PBS) and 1/19 volume of DNAseI (100 mg of DN25-100mg, Sigma resuspended in HBSS without Mg2+ or Ca2+ then sterile filtered) and the tube was left to incubate 5 min at room temperature. We resuspended the remaining tissue in DMEM + 10% FBS, further dissociated using a fire polished Pasteur pipette and allowed to settle 3 min (operation repeated up to 8 times). We counted and deposited cells on Ploy-Lysine coated coverslips, allowed them to attach (1 h at 37° C, 5% CO2), and replaced the culture medium by fresh Neurobasal medium (Invitrogen) supplemented with 1 × B27 (Gibco) 2 mM glutamax (Invitrogen), Primocin (Invivogen) and 25μM glutamate. We then half-changed the medium every 3–4 days (omitting glutamate), supplementing with 10μM Cytosine Arabinoside. To express NLS-HA-MCP-YFP in primary neurons, we applied lentiviral particles generated using the pHAGE vector to primary neurons by addition to the culture media 1 day after plating. We maintained cells under standard neuron culture conditions, and could see expression as early as 36 hours post infection.
Isolation of Mouse Embryonic Fibroblast Cell lines from mice
For the MEF cell lines, we isolated embryonic day E14 mice from the pregnant female. We separated the head from the body and used it to genotype each embryo. We removed the dark cardiac tissue from the body and digested the body with Trypsin EDTA for 20 min. We plated MEFs isolated from each embryo separately into a 10 cm dish and grew them for 1 day in DMEM + 10% FBS with antibiotics. We detached the cells with Trypsin and seeded them into a culture dish for immortalization or frozen in 10% DMSO in DMEM with 10% FBS. To express NLS-MCP-GFP in primary MEFs, we seeded cells on the day of their isolation on a fibronectin coated Labtek chamber (Thermo Scientific), and incubated them with lentiviral particles generated using the pHAGE vector.
To immortalize MEFs, we transfected the cells with a plasmid expressing SV40 Large T Antigen (pBSSVD2005, gift of David Ron) using Fugene 6 (Roche). We followed H.P. Harding protocol: after transfection cells were grown to confluence and then serially passaged at high and low densities at least 5 times in order to select transformed cells. To stably express MCP-YFP or MCP-GFP, we created recombinant lentiviral particles using the phage UbC plasmid (described above) and used them to infect these cells according to existing protocols. We used infected cultures after several passages to create a highly enriched population of stably expressing cells by FACS sorting of MCP-YFP fluorescent cells. We later selected individual clones from the MCP-YFP expressing cell lines.
Northern blotting
We scraped MEF cells grown in DMEM 10% FBS 1% pen-strep from their dish. For tissue, we snap froze tissue in liquid nitrogen and grinded it using a pestle and mortar. We then extracted total RNA using the RNeasy mini kit (Qiagen). We diluted 20 μg RNA in 50% formamide, denatured it 5 min at 65° C, chilled it on ice and then loaded it on a non-denaturing 1% agarose gel in 0.5 × TBE. The RNA was transferred overnight onto an immobilon N+ membrane (Millipore). After UV-crosslinking, we incubated the membrane 1 h at 37° C in pre-hyb solution (6 × SSC, 50 mM NaPO4 pH 7.0, 5 × Denhardt solution, 4% SDS). We generated random primed gamma-32P-ATP (Perkin Elmer) labeled DNA probes using Klenow enzyme (Roche). We prepared the DNA templates by PCR of MEF cDNA for the wild type Actb and GAPDH probes, and used a cleaved-out MBS insert for the MBS probe. We diluted the DNA probe in 10 ml pre-hyb solution and incubated it with the membrane for 3 h at 37° C. We washed the membrane twice 20 min at 37° C in 7 × SSC − 50 mM NaPO4 pH 7.0 - 1% SDS and exposed it overnight to a storage phosphor screen, before imaging on a Storm 860 phosphorimager (Molecular Dynamics).
RNA FISH
We performed RNA FISH on cultured cells to a protocol modified from 20, 33. The probes used were 50mers of ssDNA bearing each 4–5 fluorophores (probe list in Supplementary Methods). We grew MEF cells on coverslips in DMEM 10% FBS 1% pen-strep, then fixed them in 4% paraformaldehyde for 15 min at room temperature, before washing and storing in PBSM (PBS supplemented with 5 mM MgCl2) at 4° C. Prior to hybridization, we permeabilized the cells 10 min in 0.5% triton X100 in PBS, then washed them in PBS 10 min, and incubated 10 min in pre-hyb solution (50% formamide − 2 x SSC − 2 mg.ml−1 BSA - 0.2 mg.ml−1 E. coli tRNA - 0.2 mg.ml−1 sheared salmon sperm DNA). We then hybridized the probes to the cells for 3 h in pre-hyb solution supplemented with 10 ng DNA probe per locus per coverslip. We washed coverslips twice 20 min at 37° C with pre-hyb solution, then 10 min at room temperature in 2 × SSC, and 10 min at room temperature in PBSM. We counterstained DNA with DAPI (0.5 mg.l−1 in PBS). After a final wash in PBS, we mounted coverslips mounted on slides using ProLong gold reagent (Invitrogen). When 20mer probes were used, we replaced the pre-hyb solution by 10% formamide − 2 × SSC − 2 mg.ml−1 BSA - 0.2 mg.ml−1 E. coli tRNA - 0.2 mg.ml−1 sheared salmon sperm DNA − 10% dextran sulfate.
We performed RNA FISH in tissue section following a published method24. Immediately after extraction from euthanized mice, we fixed the tissues in formalin overnight and paraffin embedded them. We used 5 μm thick sections for FISH. We incubated the sections at 55° C for 30 min, then submitted them to a high pressure treatment (30 s at 125° C, then 10 s at 90° C under ~ 18 PSI) in decloaker reveal reagent (Biocare medical). After 5 min incubation in H2O, we briefly rinsed the slides in H2O and PBS. We incubated the sections 20 min at room temperature in 0.25% NH4OH + 70% EtOH, 50 min at 4° C in freshly prepared 0.5% NaBH4 in PBS, then rinsed them in water then PBS. We incubated the slides 10 min at room temperature in PBSM, then 3 times 5 minutes in PBS, and finally 30 minutes in pre-hyb solution. After a 2 hour hybridization at 37° C in a closed chamber, we washed the slides 20 min at room temperature in pre-warmed (37° C) pre-hyb solution, 20 min at room temperature in pre-warmed (37° C) 2 × SSC, 20 min at room temperature in pre-warmed (37° C) 1 × SSC, 15 min at room temperature in pre-warmed (37° C) 0.5 × SSC, and 5 min at room temperature in PBSM (all washes performed on a slow rotary shaker). We counterstained the slides with 0.5 mg.l−1 DAPI, washed them once in PBSM before mounting on coverslips using Prolong gold reagent (Invitrogen).
Immunofluorescence
We used as primary antibodies specific antibodies against β- and γ-actin isoforms (respectively 4C2F9H12/IgG1 and 2A3G8E2/IgG2b), a gift from Christine Chaponnier27. We used as secondary antibodies goat anti-mouse IgG1 – FITC (Southern Biotech 1070-02), and anti-mouse IgG2b – TRITC (Southern Biotech 1090-03). We used a custom protocol27: we incubated cells in prewarmed L-15 supplemented with 10% FBS, then fixed 20 min at room temperature in 1% paraformaldehyde in L-15, 10% FBS. We washed cells twice in PBS, permeabilized the cells with methanol (− 20° C, 5 min), and rinsed in PBS. We then incubated the cells 1 h in the primary antibody mix (PBS supplemented with 3% 0.2μm-filtered BSA fraction V, antibodies diluted 1:100 4C2F9H12/IgG1; 1:200 2A3G8E2/IgG2b) at 37° C in a closed chamber. We performed 5 rinses in PBS and incubated 1 h in the secondary antibody mix (PBS supplemented with 3% 0.2μm filtered BSA fraction V, 1:50 Southern Biotech 1070-02, 1:50 Southern Biotech 1090-03). After 5 rinses in PBS, we counterstained the DNA with 0.5 mg.l−1 DAPI and mounted on slides using Prolong gold reagent (Invitrogen).
Western Blotting
We used the following primary antibodies: β-actin (Sigma A1978), γ-actin (AB3265, Chemicon) and β-tubulin (Amersham N357), and secondary antibodies: Donkey anti-mouse 800 nm (Rockland 610-732-124) and Donkey anti-sheep 700 nm (Invitrogen A21102). We washed cells in ice-cold PBS and lysed them at room temperature 2 min in 1 ml lysis buffer per 10 cm dish (50 mM Tris-Hcl pH 8.0, 150 mM NaCl, 1% NP40, 5 mM DTT, 1 mM PMSF, half a mini tablet protease inhibitor). We spun the lysate 15 min at 14000 g at 4° C and loaded the supernatant on a Nupage 4–12% bis-tris gel using MOPS running buffer (Invitrogen). After transfer on a nitrocellulose membrane in Nupage transfer buffer (25 V for 1.5 h), we blocked non specific interactions by incubating the blot overnight at 4° C in PBS supplemented with 5% non-fat dry milk. After that, we rinsed the membrane and incubated it 1 h with the primary antibodies in PBS supplemented with 1% BSA (dilutions: 1:2,500 mouse anti-β-actin; 1:5,000 mouse anti-β-tubulin). We then washed the blot 5 times 10 min in PBS + 0.3% Tween-20, before incubation for 30 min with the secondary antibody (1:10,000 in PBS + 1% BSA). We then washed the membrane 5 times in PBS + 0.3% Tween-20, before exposure on an Odyssey infrared imaging system (2 color detection). We quantified the bands intensities using ImageJ software (NIH).
Imaging
For Immortalized cells, we plated homozygous MEFs expressing MCP-YFP on a 0.17 mm delta T dish (Bioptechs) and incubated 24 h in DMEM 10% FBS 1% pen-strep. We left cells to recover for 24 h and then starved them overnight. We replaced the media by L-15 supplemented with the Oxyfluor oxygen scavenging system (Oxyrase) prior to the experiment. We placed the cells on an Olympus IX-71 microscope equipped with a 150 × 1.45 NA objective (Olympus) and a Cascade II camera (Photometrics). We maintained a constant temperature of 37° C through the experiment using the Delta T chambers, a heated lid and objective heater (Bioptechs). We imaged cells in three dimensions over time using a 200 nm z-axis step size over a range of 4 μm every 4 min using a 488 nm argon laser for excitation of YFP fluorescence. We converted the three dimensional z-stacks into two dimensional movies using a maximum intensity projection. For primary fibroblasts, we plated the cells on fibronectin-coated MatTek dishes (MatTek), infected with lentivirus to express NLS-MCP-GFP, and incubated them for 48 h in DMEM 10% FBS, 1% pen-strep. We stained the cells with 5 μM CellTracker Orange CMRA (Invitrogen) and replaced the media with L-15 containing 10% FBS, 1% pen-strep, and 1% oxyrase prior to the experiment. We used an Olympus IX-71 inverted microscope equipped with a 40 × 1.35 NA oil-immersion objective (Olympus), and EMCCD camera (Andor). We maintained the temperature at 37° C using an environmental chamber (Precision Plastics). We excited GFP using a 488 nm line from an argon ion laser (Melles Griot) and CellTracker Orange (Invitrogen) using a 561 nm diode laser (Cobolt). For RNA FISH, we imaged the slides on an Olympus BX-61 microscope equipped with an X-cite 120 PC Mercury lamp (EXFO), a 100 × 1.4 NA objective (Olympus) and a Coolsnap HQ camera (Photometrics). For primary neurons, we changed neuron media into Hibernate Low Fluorescence (BrainBits LLC) with 1 × B27 supplement and 2 mM Glutamax. Post imaging, we restored cells to Neurobasal wtih 1 × B27, 2 mM Glutamax with 1 × primocin (in Vivogen). We used the same imaging setup as that used for immortalized cells.
Image analysis
We performed spot detection and quantification from FISH and live cell imaging using custom designed IDL (ITT visual information solutions) and MATLAB (Mathworks) programs. We first detected high intensity pixels by 2D- or 3D- (when using 3D image stacks) bandpass filtering the image and then applied a threshold to the bandpassed image (we adjusted the threshold value depending on the quality of each individual image). We defined as spots the local maxima within a 2 pixel radius of the pixels above threshold. We then automatically quantified the individual spots intensities the original image. To do so, we first corrected the intensity profile in a square ROI surrounding each spot for the local background, calculated as a the linear fit of the intensity vs. position in the pixels adjacent to the ROI (we set the ROI size as 3 times the width of the point spread function, PSF). Then we calculated the position and intensity of each spot using a 2D or 3D Gaussian mask fit of the PSF34. In the 2D projections of the tissue FISH image stacks, we identified nuclei by either automatically thresholding or manually circling the DAPI signal. We then coarse-grained the image using 32 × 32 pixels (~ 2 × 2 μm) square bins. We counted the number of mRNA particles within each bin and computed the distance from that bin to the closest nucleus. We collected quantitative data over typically 100–150 cells.
Supplementary Material
Supplementary Figure 1. β-actin mRNA, β-actin protein and cell physiology are unaffected by the MS2 system
Supplementary Figure 2. Immortalized MEF cell line is tetraploid
Supplementary Figure 3. mRNA levels in the immortalized MEF cell lines
Supplementary Figure 4. Quantitative comparison of transcription by the WT and MBS β-actin alleles
Supplementary Figure 5. The knock-in MBS cassette does not affect the cytoplasmic localization of endogenous β-actin mRNA in primary fibroblasts
Supplementary Figure 6. Single transcription sites can be detected using MCP-GFP
Supplementary Figure 7. Spatial Filtering of RNA FISH images in cerebellum tissue effectively suppresses background
Supplementary Figure 8. PCR confirmation of correct targeting
Supplementary Video 1. Live imaging of a serum-induced primary fibroblast. MCP- GFP labeled mRNA particles can be detected moving around the cell; the two transcription sites of the primary cell appear as two bright spots in the nucleus. Note that the transcription sites intensity is intentionally saturated to allow visualizing the dimmer single particles.
Supplementary Video 2. Individual mRNP moving along a neuronal process
Supplementary Video 3. Individual mRNP moving along a neuronal process. The process is the same as the one imaged in Supplementary Video 2
Supplementary Video 4. Anterograde mRNP motion along a neuronal process
Supplementary Video 5. Retrograde mRNP motion along a neuronal process
Supplementary Video 6. Bidirectional mRNP motion along a neuronal process
Supplementary Video 7. Branching mRNP motion along a neuronal process
Supplementary Video 8. Immobile mRNP in a neuronal process
Supplementary Note 1. Description of clone screening, DNA FISH procedure and list of the FISH probes used.
Acknowledgments
We thank D. Ron (Skirball Institute of Biomolecular Medicine) for providing the SV40 large T antigen (pBSSVD2005) and the MEF immortalization protocol. The phage-UbC RIG vector for lentiviral expression was a generous gift from Gustavo Mostoslavsky and Gilad Vainer (Harvard University). We thank Christinne Chaponnier (Université de Genève) for the gift of antibodies. We thank Cristina Montagna (Albert Einstein College of Medicine Genomic Imaging Facility) for assistance in karyotyping and Rani S. Sellers (Albert Einstein College of Medicine Cancer Center Histopathology Facility) for assistance in tissue examination. Microscopy equipment for the live cell imaging experiments was provided by the Gruss Lipper Biophotonics Center. T.L. is supported by a Human Frontier Science Program long term fellowship. Supported by NIH GM84364, 86217 and EB2060 to RHS, and United States- Israel Binational Science Foundation to Y.S.T. and R.H.S. Y.S.T. is the Jane Stern Lebell Family Fellow in Life Sciences at BIU. H.Y.P. is supported by National Research Service Awards F32-GM087122. A.L.W is supported by a development grant from the Muscular Dystrophy Association MDA68802.
Footnotes
Authors contributions
T.L. performed the biochemistry experiments, the tissue FISH imaging, serum response live-cell imaging, quantitative mRNA FISH, analyzed the data and wrote the paper. K.C. generated cell lines and performed the neuron live-cell imaging. X.D. and Y.S.T. generated the mouse line. A.L.W. performed FISH mRNA localization experiments. J.A.C. performed the serum response live-cell imaging. H.Y.P. performed the live-cell localization experiments. V.d.T. generated cell lines. M.L.J. performed the biochemistry experiments. R.H.S. consulted on the research and helped write the paper.
References
- 1.Orphanides G, Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–451. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- 2.Sexton T, Schober H, Fraser P, Gasser SM. Gene regulation through nuclear organization. Nat Struct Mol Biol. 2007;14:1049–1055. doi: 10.1038/nsmb1324. [DOI] [PubMed] [Google Scholar]
- 3.Terry LJ, Shows EB, Wente SR. Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science. 2007;318:1412–1416. doi: 10.1126/science.1142204. [DOI] [PubMed] [Google Scholar]
- 4.Czaplinski K, Singer RH. Pathways for mRNA localization in the cytoplasm. Trends Biochem Sci. 2006;31:687–693. doi: 10.1016/j.tibs.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 5.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 6.Shestakova EA, Wyckoff J, Jones J, Singer RH, Condeelis J. Correlation of beta-actin messenger RNA localization with metastatic potential in rat adenocarcinoma cell lines. Cancer Res. 1999;59:1202–1205. [PubMed] [Google Scholar]
- 7.Darzacq X, et al. Imaging transcription in living cells. Annu Rev Biophys. 2009;38:173–196. doi: 10.1146/annurev.biophys.050708.133728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bertrand E, et al. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 9.Golding I, Paulsson J, Zawilski SM, Cox EC. Real-time kinetics of gene activity in individual bacteria. Cell. 2005;123:1025–1036. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 10.Chubb JR, Trcek T, Shenoy SM, Singer RH. Transcriptional pulsing of a developmental gene. Curr Biol. 2006;16:1018–1025. doi: 10.1016/j.cub.2006.03.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaramillo AM, Weil TT, Goodhouse J, Gavis ER, Schupbach T. The dynamics of fluorescently labeled endogenous gurken mRNA in Drosophila. J Cell Sci. 2008;121:887–894. doi: 10.1242/jcs.019091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shav-Tal Y, et al. Dynamics of single mRNPs in nuclei of living cells. Science. 2004;304:1797–1800. doi: 10.1126/science.1099754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janicki SM, et al. From silencing to gene expression: real-time analysis in single cells. Cell. 2004;116:683–698. doi: 10.1016/s0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darzacq X, et al. In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol. 2007;14:796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mor A, et al. Dynamics of single mRNP nucleocytoplasmic transport and export through the nuclear pore in living cells. Nat Cell Biol. 2010;12:543–552. doi: 10.1038/ncb2056. [DOI] [PubMed] [Google Scholar]
- 16.Ben-Ari Y, et al. The life of an mRNA in space and time. J Cell Sci. 2010;123:1761–1774. doi: 10.1242/jcs.062638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darzacq X, Singer RH, Shav-Tal Y. Dynamics of transcription and mRNA export. Curr Opin Cell Biol. 2005;17:332–339. doi: 10.1016/j.ceb.2005.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yunger S, Rosenfeld L, Garini Y, Shav-Tal Y. Single-allele analysis of transcription kinetics in living mammalian cells. Nat Methods. 7:631–633. doi: 10.1038/nmeth.1482. [DOI] [PubMed] [Google Scholar]
- 19.Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 21.Sharova LV, et al. Database for mRNA half-life of 19 977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA Res. 2009;16:45–58. doi: 10.1093/dnares/dsn030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shestakova EA, Singer RH, Condeelis J. The physiological significance of beta -actin mRNA localization in determining cell polarity and directional motility. Proc Natl Acad Sci U S A. 2001;98:7045–7050. doi: 10.1073/pnas.121146098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kislauskis EH, Zhu X, Singer RH. Sequences responsible for intracellular localization of beta-actin messenger RNA also affect cell phenotype. J Cell Biol. 1994;127:441–451. doi: 10.1083/jcb.127.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Capodieci P, et al. Gene expression profiling in single cells within tissue. Nat Methods. 2005;2:663–665. doi: 10.1038/NMETH786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grunwald D, Singer RH. In vivo imaging of labelled endogenous beta-actin mRNA during nucleocytoplasmic transport. Nature. 2010 doi: 10.1038/nature09438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ray FA, Peabody DS, Cooper JL, Cram LS, Kraemer PM. SV40 T antigen alone drives karyotype instability that precedes neoplastic transformation of human diploid fibroblasts. J Cell Biochem. 1990;42:13–31. doi: 10.1002/jcb.240420103. [DOI] [PubMed] [Google Scholar]
- 27.Dugina V, Zwaenepoel I, Gabbiani G, Clement S, Chaponnier C. {beta}- and {gamma}-cytoplasmic actins display distinct distribution and functional diversity. J Cell Sci. 2009;122:2980–2988. doi: 10.1242/jcs.041970. [DOI] [PubMed] [Google Scholar]
- 28.Lawrence JB, Singer RH. Intracellular localization of messenger RNAs for cytoskeletal proteins. Cell. 1986;45:407–415. doi: 10.1016/0092-8674(86)90326-0. [DOI] [PubMed] [Google Scholar]
- 29.Tiruchinapalli DM, et al. Activity-dependent trafficking and dynamic localization of zipcode binding protein 1 and beta-actin mRNA in dendrites and spines of hippocampal neurons. J Neurosci. 2003;23:3251–3261. doi: 10.1523/JNEUROSCI.23-08-03251.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boettiger AN, Levine M. Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science. 2009;325:471–473. doi: 10.1126/science.1173976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fusco D, et al. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr Biol. 2003;13:161–167. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mostoslavsky G, Fabian AJ, Rooney S, Alt FW, Mulligan RC. Complete correction of murine Artemis immunodeficiency by lentiviral vector-mediated gene transfer. Proc Natl Acad Sci U S A. 2006;103:16406–16411. doi: 10.1073/pnas.0608130103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophys J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. β-actin mRNA, β-actin protein and cell physiology are unaffected by the MS2 system
Supplementary Figure 2. Immortalized MEF cell line is tetraploid
Supplementary Figure 3. mRNA levels in the immortalized MEF cell lines
Supplementary Figure 4. Quantitative comparison of transcription by the WT and MBS β-actin alleles
Supplementary Figure 5. The knock-in MBS cassette does not affect the cytoplasmic localization of endogenous β-actin mRNA in primary fibroblasts
Supplementary Figure 6. Single transcription sites can be detected using MCP-GFP
Supplementary Figure 7. Spatial Filtering of RNA FISH images in cerebellum tissue effectively suppresses background
Supplementary Figure 8. PCR confirmation of correct targeting
Supplementary Video 1. Live imaging of a serum-induced primary fibroblast. MCP- GFP labeled mRNA particles can be detected moving around the cell; the two transcription sites of the primary cell appear as two bright spots in the nucleus. Note that the transcription sites intensity is intentionally saturated to allow visualizing the dimmer single particles.
Supplementary Video 2. Individual mRNP moving along a neuronal process
Supplementary Video 3. Individual mRNP moving along a neuronal process. The process is the same as the one imaged in Supplementary Video 2
Supplementary Video 4. Anterograde mRNP motion along a neuronal process
Supplementary Video 5. Retrograde mRNP motion along a neuronal process
Supplementary Video 6. Bidirectional mRNP motion along a neuronal process
Supplementary Video 7. Branching mRNP motion along a neuronal process
Supplementary Video 8. Immobile mRNP in a neuronal process
Supplementary Note 1. Description of clone screening, DNA FISH procedure and list of the FISH probes used.