Abstract
Large-scale perturbations unravel the complex networks of activated dendritic cells.
The body’s first line of defense against infection is the innate immune system, which recognizes conserved molecular patterns on microbes via receptors such as toll-like receptors (TLRs)1. TLRs transduce the information into pathogen-specific immune responses involving networks comprising ~2,000 genes2 (Fig. 1). A new study by Amit et al.3 in Science describes an important advance in elucidating these networks. By combining expression profiling and large-scale perturbations, the authors discover many novel regulators and interactions that might control the physiological processes induced by TLRs. These network components represent novel candidates for detailed analysis and potential targets for the development of vaccines and antimicrobial or anti-inflammatory drugs.
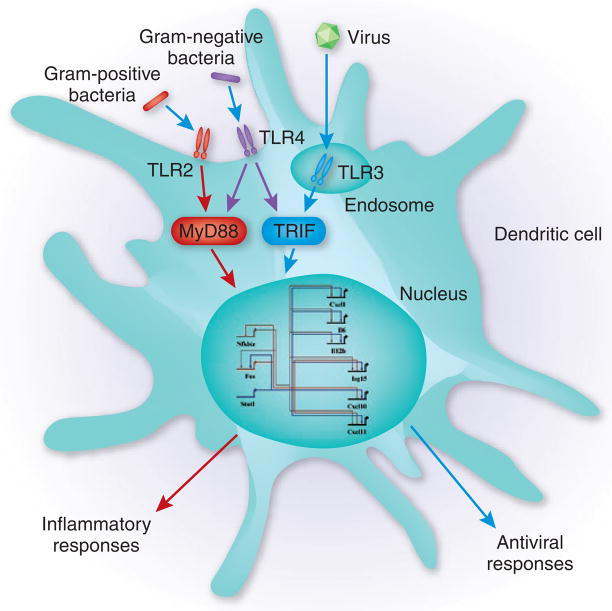
Figure 1.
Toll-like receptor (TLR)-activated gene regulatory networks. Innate immune cells, such as dendritic cells and macrophages, detect microbes through TLRs and other receptors. For example, TLR2 detects cell wall components common to Gram-positive bacteria, TLR4 recognizes lipopolysaccharides common to Gram-negative bacteria and TLR3 recognizes double-stranded RNA specifically associated with viral infection. Signals from TLRs are channeled through two major cytoplasmic adaptors, MyD88 and TRIF, which activate the transcriptional responses that control complex gene regulatory networks. The nature of the microbial component detected determines the complement of induced genes and the ultimate functional response (such as inflammatory or antiviral responses). The gene regulatory network depicted represents a subset of the interactions identified by Amit et al.3.
Addressing the complexity of innate immunity requires the large-scale approaches of systems biology. Previous work has focused primarily on the responses of the transcriptome to TLR activation. Our group has studied these responses in macrophages, integrating transcription factor binding site analysis and dynamic computational modeling to identify small novel regulatory networks that were validated in vitro and in vivo4,5. In this manner we showed that ATF3 is a negative regulator of TLR4 signaling4, whereas C/EBPδ, in conjunction with NF-κB and ATF3, forms a circuit that discriminates between transient and persistent TLR4 signals5.
Yet these sub-networks explain only a tiny fraction of TLR responses. Earlier studies have generated global network models by integrating microarray profiles and promoter scanning2,6, although they lacked experimental validation. More recently, Suzuki et al.7 investigated an experimentally tractable system— the differentiation of a macrophage-like cell line—and demonstrated the power of coupling computational network analysis with large-scale perturbations.
Amit et al.3 have now taken another step in global network analysis of the TLR pathways by applying large-scale perturbation analysis to dendritic cells—innate immune cells that prime adaptive immune responses. Their approach is straightforward: profile the transcriptomes of TLR-activated dendritic cells; mine the microarray data to identify candidate regulators that act on target genes; systematically perturb each regulator with shRNAs and measure the effects on target gene responses to TLR activation; and construct a regulatory network from the perturbation data.
But if the concept is straightforward, its implementation is not. Knocking down individual genes in primary innate immune cells is notoriously difficult; knocking down >100 genes is a major accomplishment. Profiling the responses of target genes to the perturbations was facilitated by the nCounter system (from Nanostring Technologies), which enables highly parallel high-sensitivity transcript measurements from small amounts of sample.
In addition to finding well-established regulators of TLR responses (such as NF-κB, IRF, ATF and STAT family members), the authors identify many new factors that warrant further investigation. For example, the prediction that TLR4-induced Cbx4, a SUMO E3 ligase, negatively regulates the induction of IFNβ is intriguing. Similarly, the hypothesis that established regulators of the cell cycle (e.g., E2f5) or of circadian rhythms (e.g., Timeless) may moonlight as regulators of specific gene groups associated with antiviral responses is unexpected and compelling. It is highly unlikely that these novel interactions would have been discovered using conventional approaches.
This groundbreaking study does have a few limitations. First, after examining the effects of 125 regulators on 118 target genes, the authors conclude that nearly one-sixth of all possible interactions are significant, with 80% of the candidate regulators affecting four or more targets. By comparison, an earlier estimate based on forward genetic analysis is that ~50 genes in the entire genome play nonredundant roles in positively regulating TLR responses8. That so many of the candidate regulators suggested by Amit et al.3 are found to control TLR responsive genes is, in our opinion, unrealistic. Second, Amit et al.3 interpret their results in terms of the classical categories of ‘antiviral’ and ‘inflammatory’ responses, which does not take full advantage of a systems approach. The true power of systems biology lies in its ability to identify modules and networks directly from the data, extending beyond known signaling pathways. By casting their network in the canonical binary framework, predicted modules that do not fit neatly into either category are obscured. Finally, many of the network interactions, identified by gene knockdown instead of by direct measurements of transcription factor–promoter binding, are likely to be indirect.
That said, Amit et al.3 have produced a herculean study that pushes the limits of the approach. However, the systems biology field as a whole suffers from an inability to directly link inferred gene networks to function. Fortunately, several emerging technologies are likely to fill this gap. The scale and quality of networks will improve dramatically when large-scale perturbations in primary cells3 are coupled with next-generation sequencing9,10. Multiplexed RNA-Seq and ChIP-Seq approaches will allow thousands of transcriptomes to be directly linked to genome-wide binding profiles of thousands of transcription factors. It will be straightforward to discriminate direct from indirect gene regulation and to interpret networks in the context of additional mechanisms of RNA-dependent regulation, including alternative splicing and microRNA binding. Moreover, advances in high-throughput imaging and in microfluidics will allow much more precise definition of phenotype. Eventually, predictive networks linked to function will facilitate the translation of molecular interactions into therapies. In the particular case of dendritic cells, one might easily imagine how networks delineated by studying antigen presentation as a functional output could be reengineered to yield rationally designed vaccines.
References
- 1.Ishii KJ, Koyama S, Nakagawa A, Coban C, Akira S. Cell Host Microbe. 2008;3:352–63. doi: 10.1016/j.chom.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Ramsey SA, et al. PLoS Comput Biol. 2008;4:e1000021. doi: 10.1371/journal.pcbi.1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amit I, et al. Science. 2009;326:257–263. doi: 10.1126/science.1179050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilchrist M, et al. Nature. 2006;441:173–178. doi: 10.1038/nature04768. [DOI] [PubMed] [Google Scholar]
- 5.Litvak V, et al. Nat Immunol. 2009;10:437–443. doi: 10.1038/ni.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nilsson R, et al. Genomics. 2006;88:133–142. doi: 10.1016/j.ygeno.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki H, et al. Nat Genet. 2009;41:553–562. doi: 10.1038/ng.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beutler B, et al. Annu Rev Immunol. 2006;24:353–389. doi: 10.1146/annurev.immunol.24.021605.090552. [DOI] [PubMed] [Google Scholar]
- 9.Shendure J, Ji H. Nat Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 10.Meyer M, Stenzel U, Hofreiter M. Nat Protoc. 2008;3:267–278. doi: 10.1038/nprot.2007.520. [DOI] [PubMed] [Google Scholar]