Abstract
Relapsed acute lymphoblastic leukaemia (ALL) is a leading cause of death due to disease in young people, but the biologic determinants of treatment failure remain poorly understood. Recent genome-wide profiling of structural DNA alterations in ALL have identified multiple submicroscopic somatic mutations targeting key cellular pathways1,2, and have demonstrated substantial evolution in genetic alterations from diagnosis to relapse3. However, detailed analysis of sequence mutations in ALL has not been performed. To identify novel mutations in relapsed ALL, we resequenced 300 genes in matched diagnosis and relapse samples from 23 patients with ALL. This identified 52 somatic non-synonymous mutations in 32 genes, many of which were novel, including the transcriptional coactivators CREBBP and NCOR1, the transcription factors ERG, SPI1, TCF4 and TCF7L2, components of the Ras signalling pathway, histone genes, genes involved in histone modification (CREBBP and CTCF), and genes previously shown to be targets of recurring DNA copy number alteration in ALL. Analysis of an extended cohort of 71 diagnosis-relapse cases and 270 acute leukaemia cases that did not relapse found that 18.3% of relapse cases had sequence or deletion mutations of CREBBP, which encodes the transcriptional coactivator and histone acetyltransferase (HAT) CREB-binding protein (CBP)4. The mutations were either present at diagnosis or acquired at relapse, and resulted in truncated alleles or deleterious substitutions in conserved residues of the HAT domain. Functionally, the mutations impaired histone acetylation and transcriptional regulation of CREBBP targets, including glucocorticoid responsive genes. Several mutations acquired at relapse were detected in subclones at diagnosis, suggesting that the mutations may confer resistance to therapy. These results extend the landscape of genetic alterations in leukaemia, and identify mutations targeting transcriptional and epigenetic regulation as a mechanism of resistance in ALL.
Acute lymphoblastic leukaemia (ALL) is the commonest childhood malignancy5, and is a leading cause of cancer-related death in young people. Several structural chromosomal alterations in ALL, including rearrangement of MLL and the Philadelphia chromosome6 are associated with a high risk of treatment failure and relapse. However, many ALL cases that fail therapy lack these alterations, and the biologic basis of treatment failure in these cases is poorly understood. Genome-wide profiling of ALL has identified multiple recurring submicroscopic genetic alterations targeting lymphoid development, cell cycle regulation, tumour suppression and apoptosis1,2, and has identified genetic alterations that predict a high risk of relapse, including deletion of IKZF1 (IKAROS)7,8. Moreover, profiling of structural DNA alterations in matched diagnosis and relapse samples has demonstrated that in the majority of cases there are substantial differences in the complement of genetic lesions between diagnosis and relapse, although the predominant clones at both stages of disease share a common ancestral origin3,9. The predominant clones at relapse are commonly present at low levels at diagnosis, suggesting that specific genetic alterations may confer resistance to therapy. Frequently acquired lesions at relapse include deletions of CDKN2A/B, ETV6, and IKZF13,9. However, a detailed analysis of sequence variation in ALL has not been performed.
To identify novel sequence mutations in relapsed ALL, we resequenced 300 genes in matched diagnosis-relapse samples from 23 children with B-progenitor ALL. Cases studied included B-progenitor ALL with high hyperdiploidy (N=3), TCF3-PBX1 (N=1), ETV6-RUNX1 (N=3), rearrangement of MLL (N=3), BCR-ABL1 (N=3), and low hyperdiploid, pseudodiploid, or miscellaneous karyotypes (N=10) (Supplementary Table 1). We sequenced genes known to be mutated in leukaemia and cancer, and those that encode pathway components targeted by recurring copy number alteration in ALL, tumour suppressors, cell cycle regulators, tyrosine kinases, and genes encoding DNA repair proteins (Supplementary Table 2). We identified 52 somatic (non-inherited) protein coding variants in 32 genes in 20 cases (mean 2.5 variants per case, range 0–5) (Supplementary Table 3). Somatic mutations were identified in genes previously known to be mutated in haematopoietic malignancies and ALL, including the ETS-family transcription factor gene ETV6 (N=1); the Janus kinase gene JAK1 (N=1); the Ras pathway genes NRAS (N=5; Supplementary Table 4 and Supplementary Figure 1), KRAS (N=2), NF1 (N=3) and PTPN11 (N=2); the B-lymphoid transcription factor gene PAX5 (N=2); the U3 ubiquitin ligase gene FBXW7 (N=1); the histone methyltransferase gene EZH2 (N=1); and the tumour suppressor gene TP53 (N=2). In addition, we observed patterns of evolution of sequence variations between diagnosis and relapse that recapitulated those observed for DNA copy number alterations in ALL3 (Table 1, Supplementary Results and Supplementary Table 4).
Table 1.
Recurrent mutations in 23 matched diagnosis-relapse ALL cases
| Gene | N | Present at diagnosis and relapse | Present at diagnosis only | Present at relapse only | Description/function |
|---|---|---|---|---|---|
| ASMTL | 3 | 2 | 1 | Acetylserotonin O-methyltransferase-like | |
| CREBBP | 4 | 3 | 1 | Transcriptional coactivator, histone and non-histone acetyl transferase, ubiquitin ligase | |
| ERG | 2 | 2 | ETS family transcription factor | ||
| FLT3 | 2 | 1 | 1 | Receptor tyrosine kinase, Ras pathway | |
| KRAS | 3 | 2 | 1 | Ras pathway | |
| NF1 | 2 | 2 | Ras pathway | ||
| NRAS | 5 | 2 | 2 | 1 | Ras pathway |
| PAX5 | 2 | 1 | 1 | B cell development | |
| PTPN11 | 2 | 1 | 1 | Ras pathway | |
| TP53 | 2 | 2 | Tumour suppressor | ||
| TUSC3 | 2 | 2 | Tumour suppressor candidate |
A novel finding was somatic coding mutations in CREBBP (or CBP, encoding CREB-binding protein) in four of 23 cases sequenced (Supplementary Table 5). CREBBP was selected for sequencing based on the identification of recurring focal deletions involving the gene. CREBBP and its paralog, EP300 (p300) are transcriptional coactivators with multiple functions in development and haematopoiesis4,10,11. Both are molecular scaffolds that interact with a diverse range of transcription factors, regulate transcription by acetylation of histone and non-histone targets, and may regulate protein turnover by E4 polyubiquitin ligase activity. CREBBP and EP300 are known targets of translocations in acute leukaemia (e.g. MLL-EP300, MLL-CREBBP, MOZ-CREBBP and MOZ-EP300)12. Inherited CREBBP mutations and deletions result in the Rubinstein-Taybi syndrome (RTS), a developmental disorder characterised by dysmorphology, intellectual impairment, and an increased susceptibility to solid tumours13,14. Homozygous deletion of Crebbp or Ep300 is lethal in mice due to developmental abnormalities, and Crebbp+/− mice show defects in B lymphoid development, and an increased incidence of haematopoietic tumours15. While CREBBP and EP300 sequence mutations have been reported in solid tumours16,17 and rare EP300 mutations have been detected in an ALL cell line and myelodysplasia18, there are no prior reports of CREBBP sequence mutations in haematologic malignancies.
To define the frequency of CREBBP and EP300 mutations in acute leukaemia, we sequenced these genes in an additional 318 pediatric leukemias. These additional cases included matched diagnosis and relapse samples from 48 children with ALL, and diagnosis samples from children with ALL (N=170) and acute myeloid leukaemia (AML, N=100)19 that did not relapse. Single nucleotide polymorphism (SNP) microarray DNA copy number alteration (CNA) and loss-of-heterozygosity (LOH) data were available for all cases sequenced1,20,21. We also examined DNA copy number alterations in a further 107 ALL cases that were not sequenced1,3,20. In addition, CREBBP was sequenced in 58 ALL and AML cell lines. Remarkably, 13 of 71 (18.3%) of relapsed ALL cases harboured either tumor-acquired (non-inherited) sequence alterations (N=13) or focal deletions (N=2) of CREBBP (Supplementary Table 5 and Supplementary Figure 2). In contrast, CREBBP alterations in cases of childhood acute leukaemia that did not relapse were rare, with only one additional CREBBP mutation identified in 200 AML and ALL cases sequenced (ALL Hyperdiploid-#22, C1408Y, Supplementary Table 5). Furthermore, three of 307 ALL cases with SNP array data had focal CREBBP deletions at diagnosis (Supplementary Table 5 and Supplementary Figure 2). CREBBP alterations were not observed in AML. Only one EP300 mutation was detected in the 71 cases that relapsed, a missense mutation (P925L) acquired at relapse. The identified CREBBP mutations resulted in amino acid substitutions, most commonly in the histone acetyltransferase (HAT) domain, or truncating frameshift or nonsense changes (Figure 1a). The HAT mutations involved highly conserved residues (Supplementary Figure 3). Modelling of the predicted effects of the CREBBP HAT mutations using the crystal structure of the highly homologous EP300 HAT domain22 demonstrated that the mutations are likely to disrupt the structure of the domain or its interaction with substrates (Figure 1b).
FIGURE 1.
CREBBP sequence mutations in relapsed ALL. a, Most variants are missense mutations in CREBBP domains involved in histone acetylation or transcription factor recruitment, or result in protein truncation. NRID, nuclear-receptor-interaction domain; TAZ1/2, transcriptional-adaptor zinc-finger 1/2; KIX, KID-binding domain; Bromo, bromodomain; HAT, histone acetyltransferase domain; ZZ, zinc-binding domain near the dystrophin WW domain; NCBD, nuclear-receptor coactivator-binding domain. b, The locations of CREBBP HAT mutations are shown using the crystal structure of the EP300 HAT domain complexed with its bisubstrate inhibitor, Lys-CoA (blue)22. CREBBP R1446 (equivalent to EP300 R1410) contacts phosphates of the CoA moiety of the inhibitor (salt bridges are shown as dashed lines), and the R1446H mutation is predicted to disrupt substrate binding. Q1500P (EP300 Q1464) is predicted to disrupt the alpha 4 helix, which stabilizes the substrate binding loop L1 between the beta 5 strand and the alpha 4 helix. C1408H (EP300 C1372) is predicted to disrupt the hydrophobic core that involves both the N and C termini of the HAT domain. R1563 cannot be shown as this residue lies in a proteolytically sensitive autoacetylation loop that was deleted in order to generate the crystal structure22. c, Duplication of the R1446H mutation at relapse (R). This mutation was heterozygous at diagnosis (D) and absent in the matched normal sample (N). There is copy neutral loss of heterozygosity of 16p at relapse but not at diagnosis. d, CREBBP mutations are present in subclones at diagnosis, and emerge in the predominant clone at relapse. The S1761* mutation is heterozygous in the relapse sample, absent in the matched normal sample, and appears as a minor peak in the diagnosis sample. Presence of this mutation in a subpopulation of cells at diagnosis was confirmed by PCR, cloning and bidirectional sequencing of multiple colonies of the diagnosis sample (data not shown).
Notably, CREBBP mutations present at diagnosis were retained or duplicated at relapse. Three cases had biallelic CREBBP mutations, either compound heterozygosity for different mutations, or homozygosity for a single mutation (Q1500P). Q1500P, but none of the other mutations identified in ALL, is also observed in RTS13. Homozygosity for this mutation was accompanied by DNA copy-neutral LOH (acquired uniparental disomy) of chromosome 16p in both diagnosis and relapse samples, but not the germline sample in this case. This was identified on analysis of SNP microarray genotype data for this patient, indicating that one copy of 16p containing the wild type CREBBP Q1500 allele has been deleted, whilst the remaining copy of 16p harbouring the Q1500P allele has been duplicated. One case (Hyperdiploid-#53) had a heterozygous HAT domain mutation (R1446H) at diagnosis that was homozygous at relapse with accompanying copy-neutral LOH of 16p, again indicating duplication of the mutated allele in the predominant relapse clone (Figure 1c). Furthermore, three mutations detected at relapse (T1070fs, S1761* and I2101) were detected at low levels in the diagnosis sample (Figure 1d).
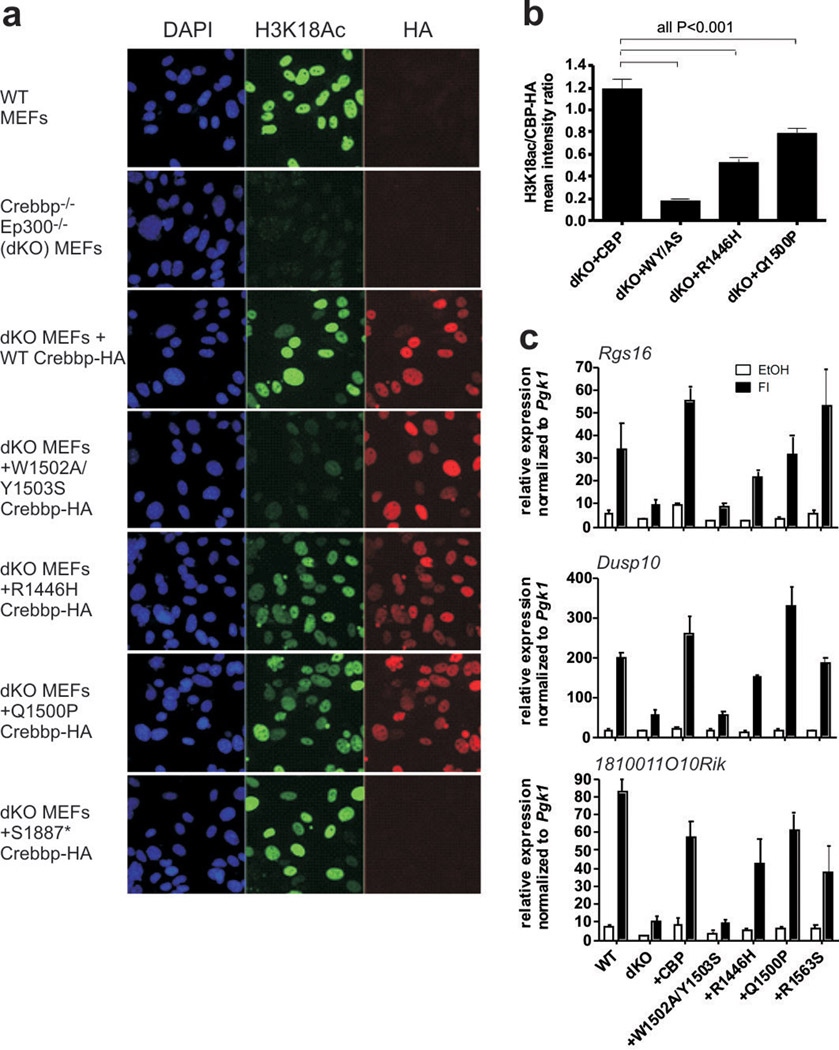
Together, the high frequency of CREBBP mutations in relapsed ALL, the persistence, reduplication or emergence of mutations from diagnosis to relapse, and the location of the mutations in key CREBBP functional domains suggest that these alterations impair CREBBP function and influence treatment responsiveness. CREBBP is expressed in leukaemic cells and normal B cell progenitors (Supplementary Figure 4), and the mutant CREBBP alleles are expressed in ALL cell lines harbouring mutations (Supplementary Table 6 and Supplementary Figure 5). To investigate the functional effect of the mutations we examined the histone acetylation and transcriptional activities of wild type and mutant Crebbp alleles expressed in CrebbpΔflox/Δflox;Ep300Δflox/Δflox (CBP/p300 Cre-deleted double knockout, or dKO) primary murine embryonic fibroblasts (MEFs)23–25 (Figure 2 and Supplementary Figures 6–10). The CREBBP HAT mutations resulted in diminished acetylation of histone H3 lysine 18 (H3K18), a known acetylation substrate of CREBBP25 (Figure 2a,b). Notably, the acetylation was attenuated but not blocked, and differed between HAT mutants (Figure 2a,b). The RTS Q1500P mutation resulted in only a modest reduction in acetylation despite being predicted to disrupt a key alpha helix in the HAT domain (Figure 1a). This suggests that HAT mutants have additional deleterious effects beyond impairing histone acetylation, that H3K18 is not the critical substrate, or that attenuated HAT activity is sufficient for the phenotype. To investigate these possibilities, we examined the effects of the CREBBP mutations on the expression of Crebbp target genes and cell proliferation in dKO MEFs transduced with retrovirus expressing wild type and mutant Crebbp cDNAs (Figure 2c and Supplementary Figures 6–10). We tested mutations in the HAT domain, as well as S1761*, S1887*, and I2101M that are predicted to affect the nuclear coactivator binding domain (NCBD, Figure 1a). Interestingly, the HAT mutants result in reduced expression of cAMP-responsive CREB target genes (Figure 2c and Supplementary Figures 6–8). Furthermore, many of the mutants tested also impaired cell proliferation (Supplementary Figure 11). Truncating mutations within the C-terminal region of the HAT domain affected expression of dsRNA and glucocorticoid-receptor-responsive genes (Supplementary Figure 8j–k and Supplementary Figures 9–10), consistent with the role of the NCBD in these pathways. The magnitude of reduced expression depended on the target gene, the specific mutation and the pathway, but the finding that CREBBP mutations result in impaired expression of glucocorticoid-receptor-responsive genes is particularly notable. The glucocorticoid dexamethasone is a mainstay of therapy for ALL, and poor responsiveness to steroid therapy is strongly associated with poor treatment outcome26. Accordingly, we examined responsiveness to dexamethasone and the class I/II HDAC inhibitor vorinostat in a panel of T-ALL cell lines with wild type or mutant CREBBP alleles. This demonstrated dexamethasone resistance in the majority of cell lines, but sensitivity to vorinostat at clinically useful concentrations (IC50 below 1µM) in the majority of cell lines tested (Supplementary Table 7 and Supplementary Figure 12). This suggests that multiple mechanisms are likely to influence glucocorticoid responsiveness, but that HDAC inhibitor therapy may be useful in steroid-resistant ALL.
FIGURE 2.
CREBBP mutations impair histone acetylation and multiple gene expression programs. a, Immunofluorescence to detect histone H3 lysine 18 acetylation (H3K18Ac) in nontransduced wild type MEFs, CrebbpΔflox/Δflox;Ep300Δflox/Δflox (dKO) MEFs and dKO MEFs transduced with retrovirus expressing wild type (+Crebbp) and mutant HA-tagged Crebbp. Four independent experiments with separate controls were performed, one of which is shown. S1887* truncates the protein before the C-terminal HA-tag, but is predicted to retain the HAT domain. b, Quantification of H3K18Ac mean signal intensity per nucleus relative to the HA-tagged Crebbp retrovirus mean signal intensity. Mean +/− SEM; 40–61 nuclei quantified per retrovirus. Only nuclei that have an HA-tag (Crebbp-HA) signal greater than 2.5 fold above background were included. Data is expressed as the ratio of the mean H3K18Ac signal intensity for each nucleus to the mean HA signal intensity for the same nuclei. P value shown is from Tukey post test of one way ANOVA. c, Quantitative RT-PCR gene expression data from dKO MEFs transduced with wild type or mutant CBP treated for 90 min with 10 µM forskolin + 100µM IBMX (FI). W1502A/Y1503S is a previously described dominant negative mutation30. Gene expression was normalized to expression of Pgk1. N=3–7.
These findings show that detailed analysis of sequence alterations in relapsed ALL can identify novel genetic alterations that are likely to be involved in the pathogenesis of treatment failure. Approximately 10% of the genes sequenced harboured somatic sequence mutations with multiple mutations involving transcriptional regulators and coactivators, many of which are targeted by other structural genetic alterations in ALL, including deletions and translocations (e.g. PAX5, ERG, ETV6, CREBBP and EP300). These results, together with those of Pasqualucci et al identifying a high frequency of CREBBP and EP300 mutations in diffuse large B-cell and follicular lymphoma27, also identify CREBBP and EP300 as new targets of recurring mutation in a range of lymphoid malignancies. The observation that the CREBBP mutations impair regulation of glucocorticoid-responsive genes, and that the mutations are selected for at relapse, suggests that these alterations may influence response to therapy and the likelihood of relapse. These results are also of clinical relevance, as they suggest that therapeutic approaches directed at modulating protein acetylation28 may be useful in high risk ALL, particularly as HDAC inhibitors may induce apoptosis in glucocorticoid resistant leukaemic cells29. Finally, these results indicate that comprehensive evaluation of sequence alterations and epigenetic modifications in relapsed ALL is likely to yield further biologic insights and potential therapeutic approaches into this disease.
Methods summary
Sequencing of 300 genes was performed by PCR and capillary resequencing of whole genome amplified DNA extracted from leukaemic cells obtained at diagnosis and relapse from 23 B-progenitor ALL cases. CREBBP and EP300 mutation recurrence testing was performed in an additional 48 diagnosis-relapse B- and T-ALL samples, and CREBBP was also sequenced in 270 ALL and AML samples that did not relapse. All putative mutations were validated as somatic by sequencing of corresponding remission DNA samples, and by sequencing of unamplified tumour and matched normal DNA. All cases had DNA copy number alteration data from Affymetrix 500K or SNP 6.0 microarrays.
In vitro functional assays of CREBBP mutants
H13K18 acetylation, gene expression and cell proliferation were assayed in CrebbpΔflox/Δflox;Ep300Δflox/Δflox dKO MEFs23–25 transduced with retroviral supernatants expressing wild-type and mutant Crebbp alleles. Dexamethasone and vorinostat drug response assays and all methods are described in full in the full methods online.
Methods
Patients and samples
Seventy one children with matched diagnosis and remission ALL samples were studied, including 61 cases previously studied by SNP microarray analysis3. All samples had at least 80% blasts by immunophenotypic and/or morphologic analysis, or were flow sorted to at least 90% purity prior to DNA extraction. Samples obtained at diagnosis from two hundred and seventy children with AML and ALL that did not experience relapse were also sequenced for CREBBP variants. This cohort comprised AML cases with t(8;21) [RUNX1-RUNX1T1] (N=9), i/t(16;16) [CBFB-SMMHC] (N=16), t(15;17) or related rearrangements involving RARA (N=7), M7 morphology (N=9), rearrangement of MLL (N=15), normal or miscellaneous karyotype (N=44); B-progenitor ALL cases with high hyperdiploidy (N=13), ETV6-RUNX1 (N=13), TCF3-PBX1 (N=13), rearrangement of MLL (N=12), BCR-ABL1 (N=15), hypodiploidy with 44–45 chromosomes (N=10), normal, pseudodiploid or miscellaneous karyotype (N=12), and T-lineage ALL (N=82). The study was approved by the St Jude Children’s Research Hospital Institutional Review Board.
CREBBP was also sequenced in 58 acute myeloid and lymphoblastic leukaemia cell lines.
ALL cell lines
380 (MYC-IGH and BCL2-IGH B-precursor), 697 (TCF3-PBX1), ALL-SIL, AT1 (ETV6-RUNX1), B1 (MLL-AF4), BV173 (CML in lymphoid blast crisis), CCRF-CEM (T-ALL with TAL-SIL), CTV-1, DND41 (T-ALL with HOX11L2/TLX3-BCL11B alteration), HPB-ALL (T-ALL with HOX11L2/TLX3-BCL11B), HSB-2 (T-ALL), Jurkat (T-ALL), Kasumi-2 (TCF3-PBX1), KARPAS 45 (T-ALL with MLL-MLLT7), KE-37 (MYC-TRAD), KOPT-K1 (T-ALL with LMO2/TTG2-TRD), LOUCY (T-ALL), MHH-CALL-2 (hyperdiploid B-precursor ALL), MHH-CALL-3 (TCF3-PBX1), MHH-CALL-4 (IGH@-CRLF2), MOLT3 (T-ALL), MOLT 13 (T-ALL), MOLT 15 (T-ALL; may be identical to CTV-1), MKB-1 (T-ALL line; may be subline of CCRF-CEM), MOLT4 (T-ALL), MUTZ5 (IGH@-CRLF2), NALM-6 (B-precursor ALL), OP1 (BCR-ABL1), P12/Ichikawa (T-ALL), PEER (T-ALL with NKX2-5-BCL11B and NUP214-ABL1), PF-382 (T-ALL), Reh (ETV6-RUNX1), REX (T-ALL), RPMI 8402 (T-ALL with LMO1/TTG1-TRAD, SIL-TAL1/SCL), RS4;11 (MLL-AF4), SD1 (BCR-ABL1), SKW3 (T-ALL, may represent KE-37), SUP-B15 (BCR-ABL1), SUP-T1 (T-ALL with TRB-NOTCH1), SUP-T11 (T-ALL with TRA/D-TCL1), TALL-1 (T-ALL), TOM-1 (BCR-ABL1), U-937 (PICALM-AF10), UOCB1 (TCF3-HLF), YT (NK leukaemia).
AML cell lines
CMK (FAB M7), HL-60 (FAB M2), K-562 (CML in myeloid blast crisis), Kasumi-1 (RUNX1-RUNX1T1), KG-1 (myelocytic leukaemia), ME-1 (CBFB-MYH11), ML-2 (MLL-AF6), M-07e (FAB M7), Mono Mac 6 (MLL-AF9), MV4-11 (MLL-AF4), NB4 (PML-RARA), NOMO-1 (MLL-AF9), PL-21 (FAB M3), SKNO-1 (RUNX1-RUNX1T1) and THP-1 (FAB M5). Cell lines were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany; the American Type Culture Collection, Manassas, VA, from local institutional repositories, or were gifts from Olaf Heidenreich (SKNO-1) and Dario Campana (OP1). Cells were cultured in accordance with previously published recommendations31. The paediatric BCR-ABL1 B-precursor ALL cell line OP132 was cultured in RPMI-1640 containing 100 units/ml penicillin, 100 µg/ml streptomycin, 2 mM glutamine and 10% fetal bovine serum. DNA was extracted from 5×106 cells obtained during log phase growth after washing in PBS using the QIAamp DNA blood mini kit (Qiagen, Valencia, CA).
Genomic sequencing
DNA from diagnosis, relapse and remission samples was amplified by Phi29 polymerase (RepliG, Qiagen, Germany). Resequencing of all coding exons was performed by PCR and capillary sequencing. Initial sequencing was performed by Beckman Coulter Genomics (formerly Agencourt Bioscience and Cogenics Danvers, MA). Mutations were validated by sequencing whole genome amplified remission DNA for variants not present in dbSNP33. CREBBP and EP300 variants not present in matched germline samples were validated by repeat PCR and sequencing of unamplified DNA. PCR primer sequences, PCR reaction conditions, thermal cycling parameters and sequence traces are available upon request. Expression of CREBBP variants was confirmed by reverse transcription using Superscript III (Life Technologies, Carlsbad, CA) and PCR using either Phusion HF DNA polymerase (New England Biolabs, Ipswich, MA) or BD Advantage DNA polymerase (Clontech, Mountain View, CA). To detect low levels of relapse-acquired mutations in diagnosis samples, (RT−) PCR products were cloned into pGEM-T-Easy (Promega, Madison, WI) and at least 24 colonies bidirectionally sequenced.
Sequence analysis
Base calls and quality scores were determined using the program PHRED34,35. Sequence variations including substitutions and insertion/deletions (indel) were analyzed using the SNPdetector36 and the IndelDetector37 software. A useable read was required to have at least one 30-bp window in which 90% of the bases have PHRED quality score of at least 30. Poor quality reads were filtered prior to variation detection. The minimum threshold of secondary to primary peak ratio for substitution and indel detection was set to be 20% and 10%, respectively. All sequence variations were annotated using a previously developed variation annotation pipeline38. Any variation that did not match a known polymorphism (defined as a dbSNP record that does not belong to OMIM SNP nor COSMIC somatic variation database33,39) and resulted in a non-silent amino acid change was considered a putative mutation. Sequence variants were visualized using Consed40.
Western Blotting
Western blotting of CREBBP/Crebbp of either whole cell lysates or nuclear extracts41 of leukemia cell lines and MEFs was performed using anti-HA and anti-CREBBP (A22, Santa Cruz Biotechnology, Santa Cruz, CA) antibodies.
Structural modeling of CREBBP and EP300 HAT domain mutations
The HAT domain amino acid sequences of CREBBP and EP300 are highly homologous. The mutations in the HAT domain in EP300, and the homologous EP300 mutations corresponding to the CREBBP HAT mutations were visualized using the solved crystal structure of a semi-synthetic heterodimeric EP300 HAT domain in complex with a bi-substrate inhibitor, Lys-CoA22 (http://dx.doi.org/10.2210/pdb3biy/pdb)42. Visualization was performed using Pymol43.
In vitro analyses of the effects of CREBBP mutations on target gene expression, histone acetylation, and cell proliferation
Murine embryonic fibroblast (MEF) isolation and culture were performed as previously described25. MEF treatments used were for 90 minutes with 10 µM forskolin + 100µM IBMX (FI) or ethanol vehicle (EtOH), 4 hours with 1µM dexamethasone or EtOH, or one hour treatment with 100µg/ml Poly I:C or PBS followed by a wash, medium change and incubation for three hours before harvest of cells into TRIzol (Life Technologies, Carlsbad, CA). Generation of mouse Crebbp (CBP)-HA retroviral constructs and retroviral transduction were performed as previously described25. Residue position numbering in the CBP-HA constructs is based on the conserved residue in human CREBBP. For all retroviral experiments except proliferation assays, retroviral transductions were 70% or higher. Quantitative RT-PCR (qRT-PCR), western blot and immunofluorescence (IF) protocols were described previously25. H3K18Ac (ab1191) (Abcam, Cambridge, MA), HA-11 monoclonal antibody against the HA epitope (Boehringer Mannheim), CREBBP (A-22) and EP300 (N-20) (Santa Cruz Biotechnology, Santa Cruz, CA) were used for IF. CBP/p300 antiserum (2574) used for IF was generated against GST-p300 1–328, but detects both CBP and p300 similarly by IF. For proliferation assays, Crebbpflox/flox;Ep300flox/flox;YFP MEFs were infected with CBP retrovirus, then endogenous CBP and p300 were deleted with adenovirus-Cre after 2 days (day 1 being the day following overnight ad-Cre treatment). YFP expression was activated in deleted cells. Proliferation assays were commenced with one million YFP+ MEFs on Day 1 and were passaged every 2–3 days. The total number of YFP+ cells on Day 11 was calculated from the total cell number and from YFP+ percentage as determined by flow cytometry.
Gene expression profiling of MEFs
Total RNA was extracted from untransduced dKO MEFs and dKO MEFs transduced with wild type and mutant Crebbp alleles treated with vehicle or 1µM dexamethasone for four hours. RNA was processed and hybridized to HT MG-430 PM gene expression arrays (Affymetrix, Santa Clara, CA) according to the manufacturer’s instructions. Probe sets were selected for visualization and validation that meet the criteria of 3-fold induction by dexamethasone in all four dKO + Crebbp-HA + dexamethasone samples and which had a minimal level of expression that is above twice background (≥3.1) for the same four samples.
Dexamethasone and vorinostat drug response assays
The activities of dexamethasone (APP Pharmaceuticals, LLC, Schaumburg, IL) and vorinostat (Selleck Chemicals LLC, Houston, TX) were evaluated against 9 T-ALL cell lines (Jurkat, HPB-ALL, SUPT1, MKB1, REX, P12/Ichikawa, TALL-1, LOUCY and PEER) using an MTT ((3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) cell viability assay. Cells were treated with the indicated drug concentration for 72 hours. Data were analyzed as the mean percentage of DMSO-treated control cells at each concentration. IC50 values were calculated from the dose-response curves using nonlinear regression analysis as implemented in the computer software program Prism version 5.0 (GraphPad Software, Inc., La Jolla, CA). For each drug and cell line, 3–4 independent experiments were performed with 6 replicates at each concentration.
Supplementary Material
Acknowledgements
The authors thank T Jeevan, S Orwick and A Gibson for technical assistance, B Schulman for assistance with structural modelling, and B Woolf and J Hartigan of Beckman Coulter Genomics for assistance with sequencing, the Tissue Resources Facility of St Jude Children’s Research Hospital for providing samples, and the following St Jude core facilities: Vector Development and Production, Flow Cytometry and Cell Sorting, Cell and Tissue Imaging, the Animal Resource Center, and the DNA sequencing and Macromolecular Synthesis laboratories of the Hartwell Center for Bioinformatics and Biotechnology. This study was supported by ALSAC of St Jude and Cancer Center support grant P30 CA021765, and DE018183 (PKB). C.G.M. is a Pew Scholar in the Biomedical Sciences.
Footnotes
Supplementary Information accompanies the paper on www.nature.com/nature
The authors declare no competing interests
Author contributions. C.G.M., P.K.B. and J.R.D. designed the study. S.L.H., L.H., C.G.M., L.A.P. and D.P.-T. performed PCR and sequencing. J.Z. and K.H.B. analyzed sequence data. L.H.K. and S.L. performed in vitro assays of the functional activity of Crebbp mutants. J.M. analyzed genomic data. S.L.H. and J.R.C.-U. performed cell line assays. S.D.B. designed and performed leukaemia cell line drug responsiveness assays. C.-H.P. provided samples and clinical data. C.G.M. wrote the manuscript. All authors reviewed the manuscript.
REFERENCES
- 1.Mullighan CG, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 2.Kuiper RP, et al. High-resolution genomic profiling of childhood ALL reveals novel recurrent genetic lesions affecting pathways involved in lymphocyte differentiation and cell cycle progression. Leukemia. 2007;21:1258–1266. doi: 10.1038/sj.leu.2404691. [DOI] [PubMed] [Google Scholar]
- 3.Mullighan CG, et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science. 2008;322:1377–1380. doi: 10.1126/science.1164266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 5.Pui CH, Robison LL, Look AT. Acute lymphoblastic leukaemia. Lancet. 2008;371:1030–1043. doi: 10.1016/S0140-6736(08)60457-2. [DOI] [PubMed] [Google Scholar]
- 6.Harrison CJ. Cytogenetics of paediatric and adolescent acute lymphoblastic leukaemia. Br J Haematol. 2009;144:147–156. doi: 10.1111/j.1365-2141.2008.07417.x. [DOI] [PubMed] [Google Scholar]
- 7.Mullighan CG, et al. Deletion of IKZF1 and Prognosis in Acute Lymphoblastic Leukemia. N Engl J Med. 2009;360:470–480. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuiper RP, et al. IKZF1 deletions predict relapse in uniformly treated pediatric precursor B-ALL. Leukemia. 2010;24:1258–1264. doi: 10.1038/leu.2010.87. [DOI] [PubMed] [Google Scholar]
- 9.Yang JJ, et al. Genome-wide copy number profiling reveals molecular evolution from diagnosis to relapse in childhood acute lymphoblastic leukemia. Blood. 2008;112:4178–4183. doi: 10.1182/blood-2008-06-165027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vo N, Goodman RH. CREB-binding protein and p300 in transcriptional regulation. J Biol Chem. 2001;276:13505–13508. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- 11.Blobel GA. CREB-binding protein and p300: molecular integrators of hematopoietic transcription. Blood. 2000;95:745–755. [PubMed] [Google Scholar]
- 12.Yang XJ. The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 2004;32:959–976. doi: 10.1093/nar/gkh252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schorry EK, et al. Genotype-phenotype correlations in Rubinstein-Taybi syndrome. Am J Med Genet A. 2008;146A:2512–2519. doi: 10.1002/ajmg.a.32424. [DOI] [PubMed] [Google Scholar]
- 14.Miller RW, Rubinstein JH. Tumors in Rubinstein-Taybi syndrome. Am J Med Genet. 1995;56:112–115. doi: 10.1002/ajmg.1320560125. [DOI] [PubMed] [Google Scholar]
- 15.Kung AL, et al. Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. Genes Dev. 2000;14:272–277. [PMC free article] [PubMed] [Google Scholar]
- 16.Iyer NG, Ozdag H, Caldas C. p300/CBP and cancer. Oncogene. 2004;23:4225–4231. doi: 10.1038/sj.onc.1207118. [DOI] [PubMed] [Google Scholar]
- 17.Kishimoto M, et al. Mutations and deletions of the CBP gene in human lung cancer. Clin Cancer Res. 2005;11:512–519. [PubMed] [Google Scholar]
- 18.Shigeno K, et al. Disease-related potential of mutations in transcriptional cofactors CREB-binding protein and p300 in leukemias. Cancer Lett. 2004;213:11–20. doi: 10.1016/S0304-3835(03)00442-7. [DOI] [PubMed] [Google Scholar]
- 19.Radtke I, et al. Genomic analysis reveals few genetic alterations in pediatric acute myeloid leukemia. Proc Natl Acad Sci U S A. 2009;106:12944–12949. doi: 10.1073/pnas.0903142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mullighan CG, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453:110–114. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- 21.Mullighan CG, et al. Rearrangement of CRLF2 in B-progenitor-and Down syndrome-associated acute lymphoblastic leukemia. Nat Genet. 2009;41:1243–1246. doi: 10.1038/ng.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X, et al. The structural basis of protein acetylation by the p300/CBP transcriptional coactivator. Nature. 2008;451:846–850. doi: 10.1038/nature06546. [DOI] [PubMed] [Google Scholar]
- 23.Kang-Decker N, et al. Loss of CBP causes T cell lymphomagenesis in synergy with p27Kip1 insufficiency. Cancer Cell. 2004;5:177–189. doi: 10.1016/s1535-6108(04)00022-4. [DOI] [PubMed] [Google Scholar]
- 24.Kasper LH, et al. Conditional knockout mice reveal distinct functions for the global transcriptional coactivators CBP and p300 in T-cell development. Mol Cell Biol. 2006;26:789–809. doi: 10.1128/MCB.26.3.789-809.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kasper LH, et al. CBP/p300 double null cells reveal effect of coactivator level and diversity on CREB transactivation. EMBO J. 2010;29:3660–3672. doi: 10.1038/emboj.2010.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dordelmann M, et al. Prednisone response is the strongest predictor of treatment outcome in infant acute lymphoblastic leukemia. Blood. 1999;94:1209–1217. [PubMed] [Google Scholar]
- 27.Pasqualucci L, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2010 doi: 10.1038/nature09730. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 29.Tsapis M, et al. HDAC inhibitors induce apoptosis in glucocorticoid-resistant acute lymphatic leukemia cells despite a switch from the extrinsic to the intrinsic death pathway. Int J Biochem Cell Biol. 2007;39:1500–1509. doi: 10.1016/j.biocel.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 30.Bordoli L, et al. Functional analysis of the p300 acetyltransferase domain: the PHD finger of p300 but not of CBP is dispensable for enzymatic activity. Nucleic Acids Res. 2001;29:4462–4471. doi: 10.1093/nar/29.21.4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Drexler HG. The Leukemia-Lymphoma Cell Line Facts Book. 1st edn. Academic Press; 2001. [Google Scholar]
- 32.Manabe A, et al. Interleukin-4 induces programmed cell death (apoptosis) in cases of high-risk acute lymphoblastic leukemia. Blood. 1994;83:1731–1737. [PubMed] [Google Scholar]
- 33.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 35.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 36.Zhang J, et al. SNPdetector: a software tool for sensitive and accurate SNP detection. PLoS Comput Biol. 2005;1:e53. doi: 10.1371/journal.pcbi.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J, et al. Systematic analysis of genetic alterations in tumors using Cancer Genome WorkBench (CGWB) Genome Res. 2007;17:1111–1117. doi: 10.1101/gr.5963407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J, Rowe WL, Struewing JP, Buetow KH. HapScope: a software system for automated and visual analysis of functionally annotated haplotypes. Nucleic Acids Res. 2002;30:5213–5221. doi: 10.1093/nar/gkf654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bamford S, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer. 2004;91:355–358. doi: 10.1038/sj.bjc.6601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gordon D, Albajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 41.Andrews NC, Faller DV. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berman H, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat Struct Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 43.DeLano WL. The PyMOL Molecular Graphics System. 2002 http://www.pymol.org.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.