Abstract
We report that bufalin and other cardiac glycoside inhibitors of the sodium-potassium ATPase (sodium pump) potently inhibit the induction of the interferon-β (IFNβ) gene by virus, dsRNA or dsDNA. Cardiac glycosides increase the intracellular sodium concentration, which appears to inhibit the ATPase activity of the RNA sensor RIG-I, an essential and early component in the IFNβ activation pathway. This, in turn, prevents the activation of the critical transcription factors IRF3 and NFκB. Bufalin inhibition can be overcome by expressing a drug-resistant variant of the sodium pump, and knocking down the pump by shRNA inhibits IFNβ expression. Thus, bufalin acts exclusively through the sodium pump. We also show that bufalin inhibits tumor necrosis factor (TNF) signaling, at least in part by interfering with the nuclear translocation of NFκB. These findings suggest that bufalin could be used to treat inflammatory and autoimmune diseases where IFN or TNF are hyperactivated.
The production of Type I interferons (IFN), cytokines essential for the innate immune response, is induced in virtually every cell type by virus infection1, or exposure to double stranded RNA or DNA (dsRNA and DNA)2–4. Secreted type I interferons bind to cell surface receptors and induce the expression of hundreds of interferon stimulated genes (ISGs) that encode antiviral activities. These activities coordinate the establishment of a strong antiviral environment5. Type I interferons also play an essential role in the activation of immune cell activity in both the innate and adaptive immune responses1,5,6.
While required for antiviral immunity, high levels of IFN can be toxic. In fact, over-expression or aberrant expression of IFN has been implicated in several inflammatory and autoimmune diseases7,8. For example, overproduction of interferon is a critical factor in the autoimmune disease systemic lupus erythematosus (SLE)7. In addition, prolonged IFN production has been shown to contribute to AIDS virus infection9. Regulating the level and duration of IFN production is critical to the optimization of antiviral activities, while minimizing the detrimental effects associated with over-production or prolonged expression of these activities. Normally, IFN is only transiently expressed after infection10,11.
IFNβ gene expression is one of the most extensively studied eukaryotic gene regulatory systems2,12. Virus infection triggers the activation of a complex signal transduction pathway13 leading to the coordinate activation of multiple transcriptional activator proteins that bind to the IFNβ enhancer to form an enhanceosome, which recruits the transcription machinery to the gene12,14. The presence of viral RNA is detected by the RNA helicases RIG-I and MDA5, which are specific for different viruses15. Upon binding RNA, RIG-I or MDA5 dimerize, undergo a conformational change and expose a critical N-terminal caspase recruiting domain (CARD)16,17 that binds to a corresponding CARD domain in the downstream adaptor protein MAVS on the mitochondria membrane18. MAVS is also believed to form dimers on the surface of mitochondria19, leading to recruitment of downstream signaling molecules and kinases. The assembly of these signaling components ultimately leads to the activation of the key transcription factors Interferon Regulatory Factors IRF3/7 and NFκB. Phosphorylated IRF3/7 and NFκB translocate into the nucleus, and together with activated cJUN and ATF2 and the coactivators CBP/P300 form an enhanceosome complex upstream of the IFNβ gene promoter12. Histone modification and chromatin remodeling enzymes, and the RNA polymerase machinery are recruited to drive the transcription of the IFNβ gene14.
As mentioned above, the initial trigger of the IFN signaling pathway is the recognition of viral RNA. Recently, short double strand RNA (dsRNA) or panhandle RNA with a 5’-ppp group has been shown to be the RNA structure that activates RIG-I20. RIG-I dimerizes upon binding RNA16,17, and the dimer moves along the RNA, acting as a translocase21. This activity has been shown to be ATPase dependent21. Thus RNA binding and the ATPase dependent translocation along the RNA template are two critical activities of the RIG-I protein. Recent studies have revealed that RIG-I undergoes covalent modifications upon activation; its ubiquitination at lysine 172 by the E3 ligase Trim25 is important for signaling22, while phosphorylation of threonine 170 by an unidentified kinase antagonizes RIG-I activation23.
The activated RIG-I protein relays a signal to the mitochondria protein MAVS through CARD domains on both proteins. Since there is little mitochondria association of RIG-I after virus infection, the interaction between RIG-I and MAVS must occur transiently, and MAVS efficiently assembles the downstream signaling complex. The adaptor proteins, TRAF3, TRAF6 and TANK are thought to interact with MAVS, and activate the downstream kinases TBK1 and/or IKKε24,25, as well as the IKKα/β kinases18,26. Additional proteins have been reported to play roles in the activation of the IFN gene, including Sting/Mita, and DDX327–29. These proteins are thought to mediate interactions between RIG-I, MAVS or TBK1 proteins.
To further investigate the signaling pathways leading to the activation of the IFNβ gene, we have carried out a screen for small molecules that inhibit virus induction of IFNβ gene expression. Such molecules could provide mechanistic insights into the signaling pathways, and possibly lead to the development of drugs to treat diseases of IFN overproduction, such as SLE. Here we report the identification of cardiac glycosides as potent inhibitors of IFNβ gene expression. Analysis of the interferon activation pathway revealed that the ATPase activity of the RNA sensor RIG-I is inhibited by cardiac glycosides, as a result of their ability to increase the intracellular sodium concentration. Thus, cardiac glycosides could potentially serve as therapeutic agents for diseases involving IFN over-expression.
Results
Bufalin inhibits virus induction of IFNβ gene expression
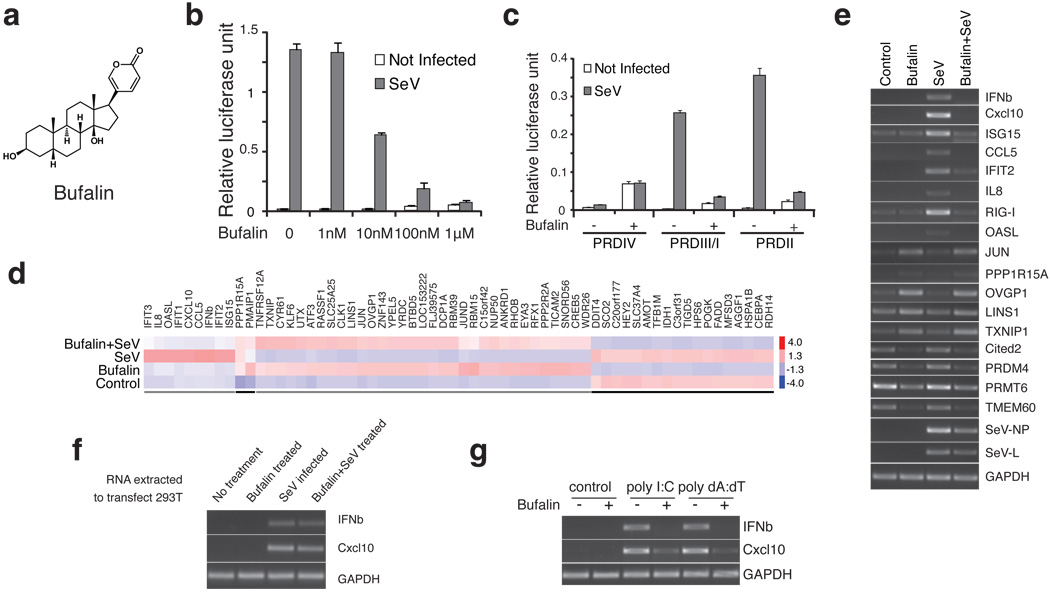
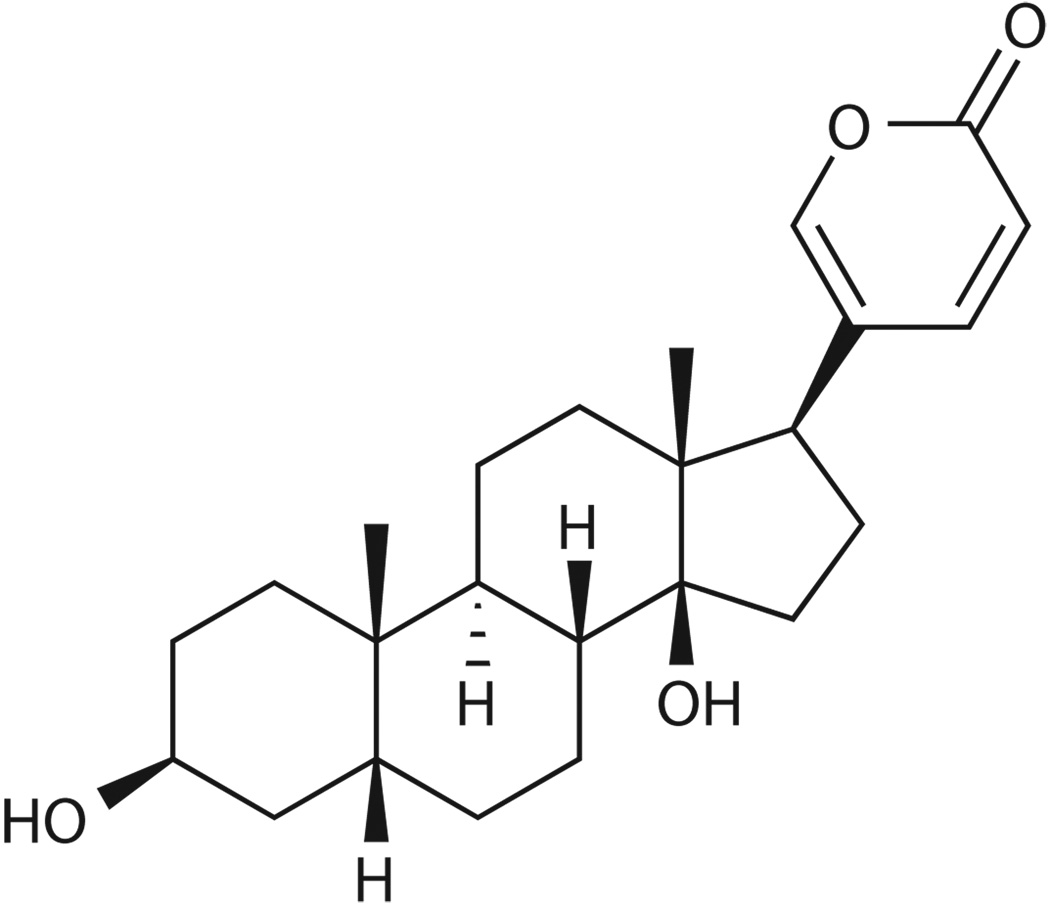
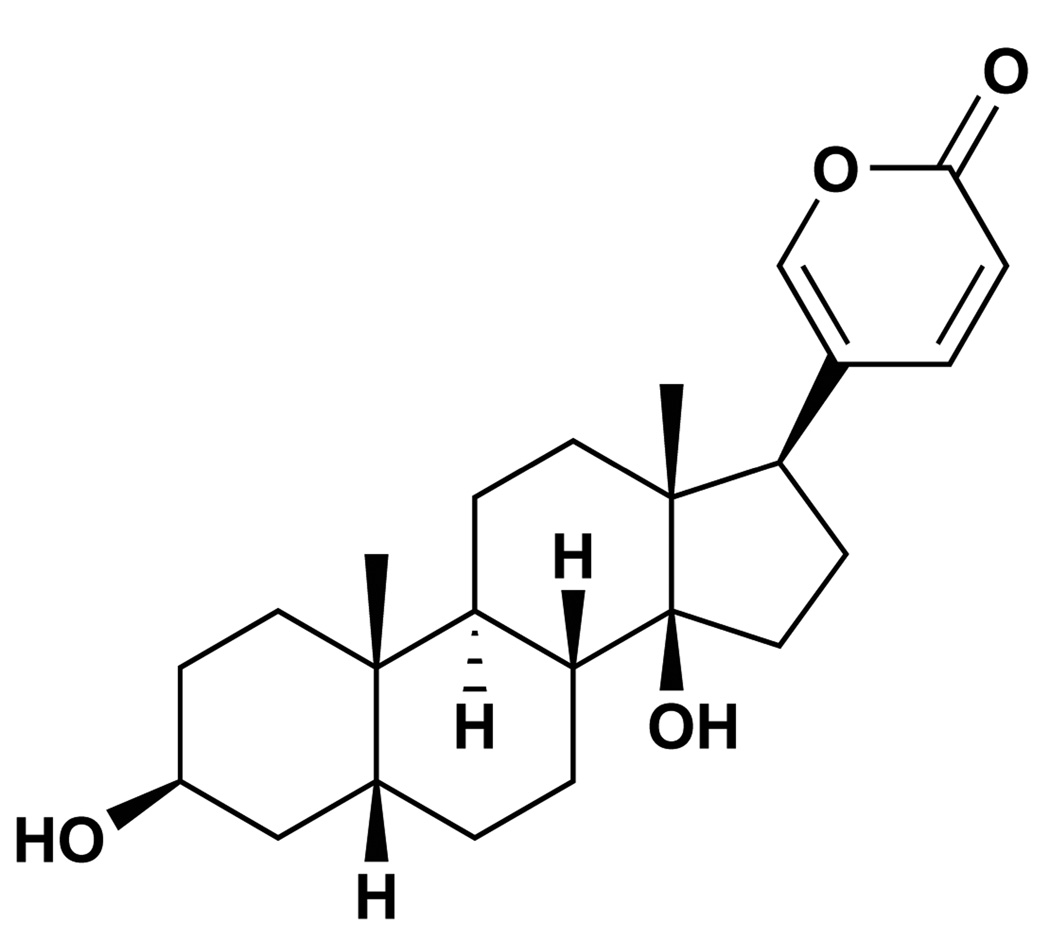
We utilized a virus inducible luciferase reporter assay to screen for small molecule inhibitors of IFNβ gene expression in response to various inducers. Human 293T cells were transfected with the IFNβ promoter driving the expression of a luciferase construct. The cells were treated with compounds from a chemical library, and infected with sendai virus (SeV) one hour later. Luciferase activity was measured after another 24 hrs of culture. Signals were normalized to the samples not treated with chemicals. Screening a chemical library containing 478 compounds identified six molecules with inhibitory effects on IFNβ gene expression (Supplementary Table 1). One compound, bufalin (Fig. 1a), a cardiac glycoside inhibitor of the sodium pump30, consistently and strongly inhibited virus induction of IFNβ gene expression. While the initial screen was conducted at a concentration of 10 µM of each molecule, subsequent tests with varying concentrations showed that bufalin exerts inhibitory effects in a dose-dependent manner (Fig. 1b). The 50% inhibition concentration (IC50) of bufalin was calculated to be 4.3 nM and a concentration of 1 µM is sufficient to inhibit IFNβ expression by more than 90%. It is important to point out that the inhibition of IFNβ induction by bufalin is not due to the loss of cell viability (Supplementary Fig. 1a, 1b, 1c) or to the induction of cell death by apoptosis or autophagy (Supplementary Fig. 1d, 1e).
Fig. 1. Bufalin potently blocks virus, double strand RNA and DNA induced gene expression.
a. Structure of the bufalin molecule. b. Bufalin blocks virus-induced IFNβ expression in reporter assays. 293T cells were transfected with the IFNβ promoter-driving firefly luciferase reporter together with a reference renilla luciferase reporter plasmid. 24 hrs later, cells were treated with increasing amounts of bufalin (1 nM to 1 µM) followed by infection with sendai virus. Firefly luciferase activities were measured after another 24hrs and normalized to that of renilla activities. Data represent mean values ± s.d. (n=3). c. Bufalin potently blocks the induction of PRDIII/I and PRDII elements of the IFNβ promoter. Experiments were conducted same as in b, but PRDIV, PRDIII/I and PRDII-driving luciferase reporter plasmids were used. Data represent mean values ± s.d. (n=3). d. Bufalin blocks the virus-induced gene expression program. 293T cells were treated with bufalin (1 µM) or SeV alone, or in combination for 8 hrs, and cellular RNA extracted and subjected to microarray analysis. Genes highly induced or repressed by bufalin or SeV are shown. e. RT-PCR confirms the microarray results of representative genes. f. Bufalin does not destroy sendai virus pathogen-associated molecular pattern (PAMP). RNA samples from Fig. 1d were transfected into new 293T cells, 6hrs later, cellular RNA was prepared and subjected to RT-PCR analysis. g. Bufalin also potently inhibits gene induction by dsRNA and dsDNA. 293T cells were treated with bufalin (1 µM), and then subjected to poly I:C (dsRNA) or poly dA:dT (dsDNA) transfection, total cellular RNA was prepared after 6 hrs and subjected to RT-PCR analysis.
The IFNβ virus-inducible enhancer contains four positive regulatory domains (PRD), corresponding to binding sites for the transcription factors cJUN/ATF2 (PRDIV), IRF3/7(PRDIII/I) and NFκB (PRDII) respectively. All of these sites are required for the activation of IFNβ gene expression12, and each transcription factor is activated through distinct signal transduction pathways. In order to identify which signaling pathway is the target for bufalin, we transfected 293T cells with luciferase reporters driven by multiple copies of the PRDIV, PRDIII/I or PRDII elements, and tested the effects of bufalin on viral induction. We noted that SeV did not induce the PRDIV reporter in 293T cells, however 1 µM bufalin treatment weakly induced the activity of PRDIV in the absence of virus infection (Fig. 1c), indicating that this element is not the target for inhibition. By contrast, viral induction of both the PRDIII/I and PRDII reporters was strongly inhibited by bufalin. With 1 µM of bufalin, viral induction of luciferase expression was >86% repressed compared with no drug treatment with both reporters (Fig. 1c). Thus, bufalin can either inhibit IRF3/7 and NFκB activation separately, or more likely it can target signaling events at or before the bifurcation of the IRF and NFκB signaling pathways.
Bufalin blocks the activation of virus-inducible genes
Virus infection leads to the activation of a large number of genes in addition to IFNβ31. We therefore investigated the effect of bufalin on this virus-inducible gene expression program. To accomplish this, we carried out a microarray analysis with 293T cells treated with bufalin and SeV either alone or in combination, and compared the genome wide expression profiles of all the samples. Interestingly, bufalin alone stimulated the expression of >80 genes to a level at least two-fold higher than the control (Fig. 1d, 1e), including the genes encoding transcription factors that bind to PRDIV (cJUN and ATF2). This may be a reason why the PRDIV reporter was weakly activated by bufalin (Fig. 1c). Treatment with bufalin alone also leads to a decrease in the expression of over 40 genes by at least 2-fold (Fig. 1d, 1e). By contrast, a typical cytokine and ISG expression profile was observed when cells were infected with virus (Fig. 1d). IFNβ, IL-8, IFIT1, IFIT2, IFIT3, ISG15, OASL, Cxcl10, and CCL5 genes are among the highest virus-induced genes. Strikingly, bufalin completely blocked the induction of these genes. The expression profile from the bufalin and virus treated condition is remarkably similar to that of bufalin treatment alone (Fig. 1d). Although slightly diminished, transcripts from the infecting virus (SeV nucleocapsid, NP and RNA polymerase gene, L) are readily detected in infected cells treated with bufalin (Fig. 1e). Moreover, when total RNAs extracted from virus infected cells or cells treated with bufalin and SeV were transfected into new 293T cells, both transfections strongly induced the expression of IFNβ and Cxcl10 genes (Fig. 1f). These data show that bufalin does not block virus infection, nor eliminates the viral pathogen-associated molecular pattern (PAMP) associated with virus infection, however, expression of the cellular innate antiviral genes was almost completely abolished by bufalin. We also found that bufalin similarly inhibits IFNβ gene expression when cells were infected with virus before treatment with bufalin (Supplementary Fig. 2).
We also tested the effects of bufalin on other inducers of IFNβ expression. For example, bufalin strongly inhibited IFNβ and Cxcl10 gene expression in response to treatment of cells with double strand RNA (poly I:C) and double strand DNA (poly dA:dT) (Fig. 1g). This inhibition is not due to a reduction in transfection efficiency, since Cy3 labeled dsRNA and dsDNA are similarly transfected into bufalin treated and non-treated cells (Supplementary Fig. 3).
To address the possibility that bufalin inhibition occurs only in 293T cells, we tested other human cell lines. Strong inhibition of virus-induced IFNβ gene expression was observed with 1 µM of bufalin in the human B cell line Namalwa (Supplementary Fig. 4a). We also found that 1 µM bufalin inhibits virus induced IFNβ expression in Hela and MG63 cells, but less than that observed with 293T and Namalwa cells (Supplementary Fig. 4c). Strong inhibition was observed, however, with higher concentrations of bufalin. By contrast, poly I:C and poly dA:dT induced IFNβ expression was strongly inhibited in Hela and Mg63 cells with 1 µM bufalin (Supplementary Fig. 4c). It is possible that these differences are due to variations in the cell-type specific expression levels of the sodium pump, or the signaling components affected by the drug. In fact, we found that the expression of the ATP1a1 gene, which encodes the alpha subunit of the sodium pump, is much higher in Hela and MG63 cells than in 293T and Namalwa cells. We also found that the expression of RIG-I and MDA5 followed the same trend (Supplementary Fig. 4d).
Bufalin inhibits RIG-I activation
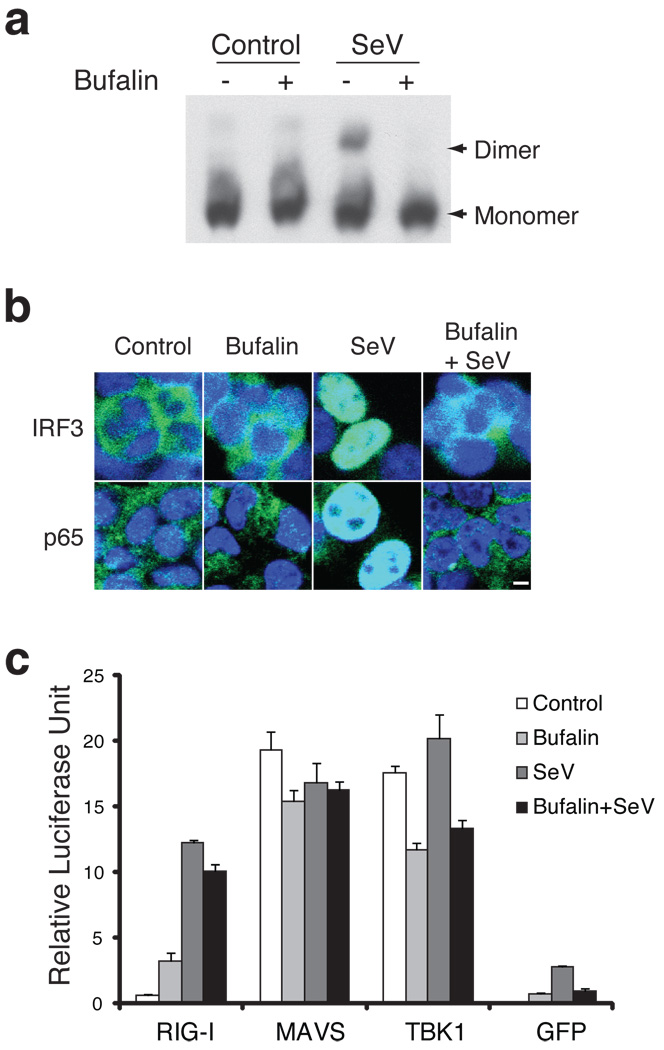
We next performed experiments to determine the step at which bufalin blocks the induction of virus-inducible genes. First, we examined the activation of the transcription factors IRF3 and NFκB. Native gel analysis, which detects virus-induced IRF3 dimerization revealed that IRF3 dimer formation is blocked by bufalin in 293T cells (Fig. 2a). Formation of the IRF3 dimer is also strongly inhibited by bufalin in Namalwa cells (Supplementary Fig. 4b). In addition, IRF3 nuclear translocation is completely blocked by bufalin treatment (Fig. 2b). A similar observation was made with the p65 subunit of NFκB, where bufalin blocked its nuclear localization in response to virus induction (Fig. 2b). These observations are consistent with reporter assays showing that virus induction of PRDIII/I (activated by IRF3) and PRDII (activated by NFκB) elements are strongly inhibited by bufalin (Fig. 1c).
Fig.2. Bufalin inhibits virus induced IRF3 and p65 activation.
a. Bufalin blocks IRF3 dimerization. 293T cells were treated with bufalin (1 µM) or SeV, either alone or in combination for 6hrs. Total protein was prepared and subjected to a native gel analysis for IRF3 dimerization. For the full image, see Supplementary Figure 12a. b. Bufalin blocks the virus induced nuclear translocation of both IRF3 and p65. 293T cells were treated same as in a, and cells were fixed with formaldehyde and immunofluorescent staining of IRF3 and p65 (green) were conducted. Blue: DAPI staining for nuclei. Scale bar, 5 µm. c. Over-expression of RIG-I, MAVS and TBK1 greatly relieved the blockage of bufalin of the IFNβ induction. 293T cells were transfected with RIG-I, MAVS and TBK1 expression plasmids together with IFNβ promoter-driving luciferase reporter in the presence or absence of bufalin (1 µM), and infected with SeV for 24hrs before measuring the luciferase activities. GFP plasmid was included as a control. Data represent mean values ± s.d. (n=3).
Thus, bufalin appears to act upstream of IRF3 and p65 activation. RIG-I, MAVS and TBK1 are known upstream factors, and over-expression of any of these proteins can activate the IFNβ reporter18,32,33. We therefore determined whether bufalin could block IFNβ expression induced by the over-expression of these proteins. Remarkably, bufalin only modestly inhibited the induction of IFNβ by any of these proteins. Over-expression of MAVS and TBK1 strongly induced the expression of the IFNβ reporter, and the addition of bufalin had no effect on MAVS induction and little effect on TBK1 induction (Fig. 2c). Over-expression of full-length RIG-I protein alone appeared to be a weak activator of the IFNβ reporter, but it greatly enhanced virus-induced IFNβ expression. Bufalin treatment only slightly reduced virus induction of the IFNβ reporter when RIG-I is over-expressed (Fig. 2c). These data suggest that the target of bufalin in the IFNβ signaling pathway lies upstream from RIG-I or at the RIG-I protein itself, but can somehow be compensated by over-expression of the protein. Since RIG-I is the most upstream sensor of SeV induction known, we considered the possibility that the activation of RIG-I is targeted by bufalin.
The RIG-I ATPase is inhibited in bufalin treated cells
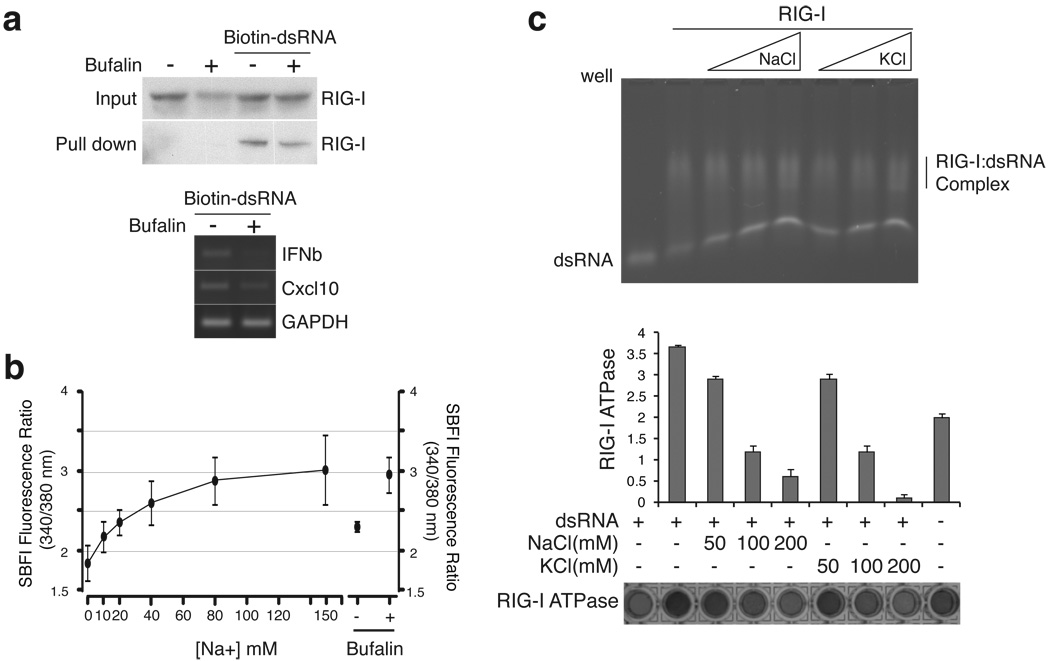
To determine whether the enzymatic activity of the most upstream sensor of virus infection is altered by bufalin treatment, we directly assayed the effects of bufalin on the activation of RIG-I. First, we examined the RNA binding activity of RIG-I. Since the basal expression level of RIG-I is very low in 293T cells (Supplementary Fig. 4d), we generated 293T cells stably expressing flag-tagged RIG-I protein. Bufalin treatment also significantly inhibited virus and dsRNA induced IFNβ expression in these cells (Fig. 3a). We then transfected these cells with biotin labeled dsRNA (also bearing a 5’-ppp group, as it was generated by in vitro T7 RNA polymerase transcription) in the presence or absence of bufalin. The associated proteins were captured with NeutrAvidin beads and separated on SDS-PAGE. RNA binding by RIG-I was detected by blotting with anti-RIG-I antibody. In contrast to the strong repression of IFNβ gene induction (Fig. 3a, bottom panel), bufalin treatment did not significantly decrease dsRNA binding by RIG-I (Fig. 3a, top panel).
Fig.3. RIG-I ATPase activity is inhibited by bufalin treatment.
a. Bufalin does not affect the RNA binding ability of RIG-I. 293T cells stably expressing RIG-I protein were transfected with biotin-labeled dsRNA (67bp, corresponding to 3’ end of GFP gene) in the presence or absence of bufalin (1 µM), 6 hrs later, total cellular protein was prepared and subjected to NeutrAvidin beads binding. Bound RIG-I protein was analyzed by western blot (top panel). The expression of IFNβ and Cxcl 10 genes in these cells was also analyzed by RT-PCR (bottom panel). For the full image, see Supplementary Figure 12b. b. Bufalin treatment increases the intracellular sodium concentration within 293T cells. The fluorescent dye SBFI was used to measure the intracellular sodium concentrations of 293T cells before and after bufalin treatment, concentrations were determined from the standard curve (See Supplementary Methods for details). Data represent mean values ± s.d. (n=3). c. High salt concentration inhibits RIG-I ATPase activities while the effect on RNA binding is minor. Recombinant RIG-I protein was incubated with dsRNA (67bp) in the presence of increasing NaCl and KCl concentrations. RNA binding was monitored by native agarose gel analysis (top panel). The ATPase activities were also measured (see Methods for details). The image of the ATPase assay wells was shown in the bottom panel, and signals from the reading were quantified and graphed in the middle panel. Data in the graph represent mean values ± s.d. (n=3). The lanes of the gel, bars of the graph and wells in the ATPase assay are aligned according to experimental conditions.
RIG-I has been shown to undergo dimerization after sensing its ligand16, and this dimerization can be studied by native gel electrophoresis34. We therefore compared the RIG-I mobility in native gel electrophoresis with and without bufalin treatment. We used both the stable line and Namalwa cells, where RIG-I expression is relatively high. No reproducible difference in gel mobility was detected with either cell type.
We also designed experiments to test the effects of bufalin on the ATPase activity of RIG-I, which is also critical for antiviral signaling21,34. Previous studies have shown that a major effect of bufalin treatment is the elevation of the intracellular sodium concentrations35. In 293T cells, the intracellular sodium concentration increased rapidly from about 20 mM to over 120 mM with 1 µM bufalin treatment (Fig. 3b). We therefore conducted in vitro experiments to directly test whether the RIG-I ATPase activity is affected by different salt concentrations. Flag-tagged recombinant RIG-I protein was purified from 293T cells by immunoprecipitation and eluted using a Flag peptide. The protein was incubated with dsRNA probe before addition of ATP and further incubated for 15 min. Free phosphates released from the ATPase hydrolysis were measured with the Biomol Green Reagent. Consistent with published results, addition of dsRNA boosted RIG-I ATPase activity17. Strikingly, increasing the concentration of both sodium and potassium (from 50 mM to 200 mM) strongly inhibited RIG-I ATPase activity (Fig. 3c, bottom panel), while the dsRNA binding assay of the same samples showed no change in binding efficiency (Fig. 3c, top panel). The binding of dsRNA increased proportionally to increasing amounts of RIG-I protein, suggesting it is specific for RIG-I binding (Supplementary Fig. 5a). These observations agree with our biotin labeled dsRNA pull down assay where similar binding of transfected dsRNA was detected with and without bufalin treatment (Fig. 3a). Control experiments with an ATPase dead mutant RIG-I protein (K270A)34 showed similar binding of the protein to dsRNA, while the ATPase activity was extremely low (Supplementary Fig. 5b). These data demonstrate that the observations with the wild type protein are specific. We conclude that the ATPase activity of RIG-I is strongly inhibited by bufalin treatment, and as a result, the antiviral signaling pathway is severely impaired. This inhibition is through a bufalin-induced major increase of intracellular sodium concentration, but not bufalin itself (Supplementary Fig. 5c).
Bufalin inhibits IFNβ expression through the sodium pump
Cardiac glycosides bind specifically to the sodium pump on the plasma membrane and inhibit its activity30. The normal function of the sodium pump is to maintain intracellular ion homeostasis (at the expense of ATP hydrolysis, it pumps out three sodium ions as it pumps in two potassium ions during each cycle). Therefore, the direct effect of inhibiting the sodium pump by cardiac glycosides is to change the intracellular ion concentrations and their gradients. For example, sodium and calcium concentrations are elevated and the potassium concentration decreases35. It is the increased concentration of sodium but not the decreased potassium directly leads to the inhibition of the ATPase activity of RIG-I (Fig. 3c).
The sodium pump is composed of the catalytic alpha subunit and the structural beta subunit, both are encoded by four genes in most species. Different isoforms of the alpha and beta subunits display tissue specific expression patterns36, with the ATP1a1 most widely expressed. The conserved aspartic acid (Asp) 376 in the active center of the alpha subunit is continuously phosphorylated and dephosphorylated during pumping activities37.
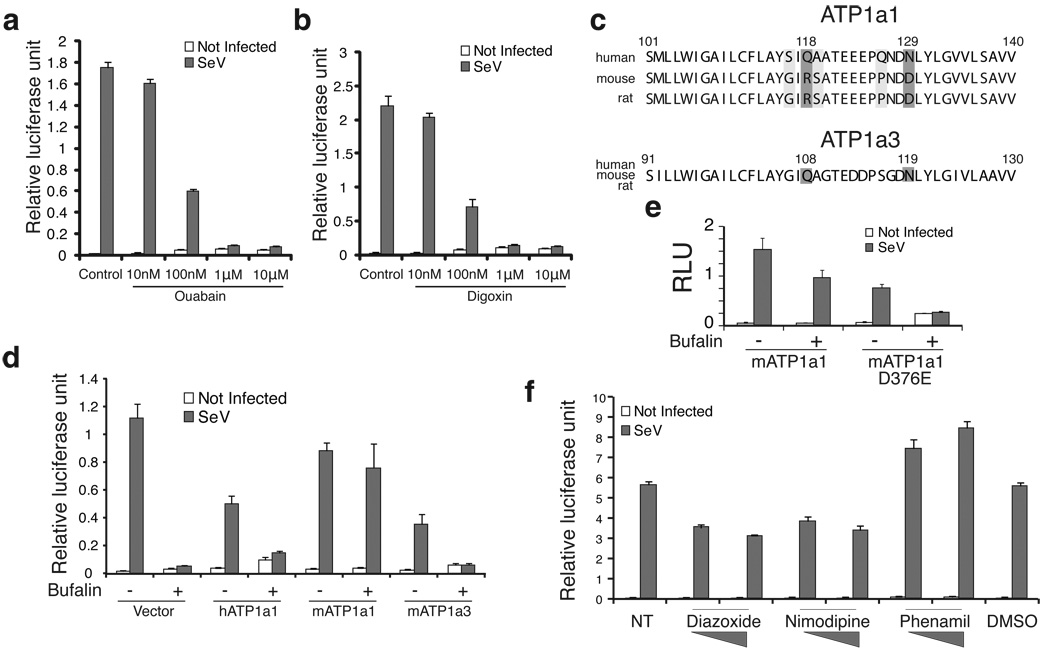
To show that inhibition of IFNβ expression by bufalin is exclusively through the sodium pump, we first tested whether other cardiac glycosides can also inhibit viral induced IFNβ expression. We found that ouabain and digoxin, two cardiac glycosides clinically approved to treat congestive heart failure and cardiac arrhythmias30, are also able to potently inhibit IFNβ induction, and this inhibition is also dose dependent (Fig. 4a, 4b). The calculated IC50 for ouabain and digoxin are 64.8 nM and 56.6 nM respectively, relatively higher than that of bufalin (4.3 nM).
Fig.4. Bufalin inhibits IFNβ induction exclusively through the sodium pump.
a–b. Ouabain and digoxin potently inhibit virus induction of IFNβ expression. 293T cells were transfected with an IFNβ promoter-driving luciferase reporter, 24 hrs later, increasing amounts (10 nM to 10 µM) of Ouabain (a) or digoxin (b) were added to cells before the Sendai virus infection. Luciferase activities were measured one day later. Data represent mean values ± s.d. (n=3). c. Sequence alignment of the cardiac glycosides binding sites in human, mouse and rat ATP1a1 and ATP1a3 proteins. Q118R and N129D mutations in mouse and rat ATP1a1 make the rodent protein insensitive to cardiac glycosides treatment. d. Mouse ATP1a1 gene fully rescued the inhibition of bufalin in human cells. 293T cells were transfected with various expression constructs together with IFNβ luciferase reporter. Cells were infected with SeV in the presence or absence of bufalin before measuring the luciferase activities. Data represent mean values ± s.d. (n=3). e. The catalytic activity of the ATP1a1 gene is required for the rescue. Experiments were conducted the same as in d, but a point mutation (D376E) of mouse ATP1a1 was tested together with the wild type expression construct. Data represent mean values ± s.d. (n=3). f. Modulating intracellular ion concentration affects IFNβ induction. 293T cells were transfected with IFNβ reporter construct, treated with various ion channel ligands and then infected with SeV. Firefly luciferase activities were measured and normalized to renilla luciferase activities. Data represent mean values ± s.d. (n=3).
We also performed rescue experiments with the mouse ATP1a1 gene38, which is not sensitive to bufalin due to the natural occurring Q118R and N129D single amino acid substitutions (Fig. 4c). In contrast, the ATP1a3 gene is highly conserved between human and mouse, and sensitive to the drug. We transfected plasmids encoding mouse ATP1a1 and ATP1a3 genes in parallel with human ATP1a1 gene, and infected the cells with virus in the presence or absence of bufalin. We found that only the mouse ATP1a1 transfection relieved the inhibition of IFNβ expression by bufalin, while little if any rescue was observed with the drug sensitive human ATP1a1 and mouse ATP1a3 proteins (Fig. 4d). These data show that bufalin inhibits IFNβ gene activation entirely through its binding and inhibition of the sodium pump.
To test whether the mouse ATP1a1 protein can rescue bufalin inhibition of IFNβ induction independent of its catalytic activity, we generated a catalytic inactive mutant (D376E) mouse ATP1a1 expression construct, and it failed to rescue the inhibition (Fig. 4e). These data demonstrate that the enzymatic activity of the sodium pump is required for the optimal induction of the IFN gene, presumably to maintain the appropriate concentrations of intracellular ions.
Intracellular ion concentrations modulate IFNβ expression
Since the major function of the pump is to maintain cellular ion homeostasis, we reasoned that it might be possible to inhibit or stimulate IFNβ induction by varying the intracellular ion concentration. We thus performed luciferase reporter assays with an ion channel ligand library, which contains 71 modulators for sodium, potassium, calcium and chloride channels. The experiments were conducted similarly to our initial screening assays: 293T cells were transfected with reporter plasmid, then chemical ligands were added before starting virus infection, and luciferase activity measured after another 24 hrs. A few ligands showed considerable negative effects on IFNβ induction, among these are nimodipine, a dihydropyridine-type voltage-sensitive (L-type) calcium channel blocker39, and diazoxide, a selective opener of ATP sensitive potassium channel40. As shown in figure 4f, increased inhibition of the IFNβ reporter was achieved with increasing concentrations of both nimodipine and diazoxide. The virus-induced expression of the endogenous IFNβ gene was similarly inhibited by nimodipine and diazoxide in 293T cells (Supplementary Fig. 6). These data show that not only sodium, but potassium and calcium can also modulate cellular antiviral responses. Interestingly, one type of amiloride-sensitive sodium channel inhibitor, phenamil41 consistently enhanced the activation of IFNβ gene expression in reporter assays (Fig. 4f). However, the stimulatory effect on endogenous IFNβ gene induction is less potent (Supplementary Fig. 6).
Knocking-down pump expression suppresses IFNβ induction
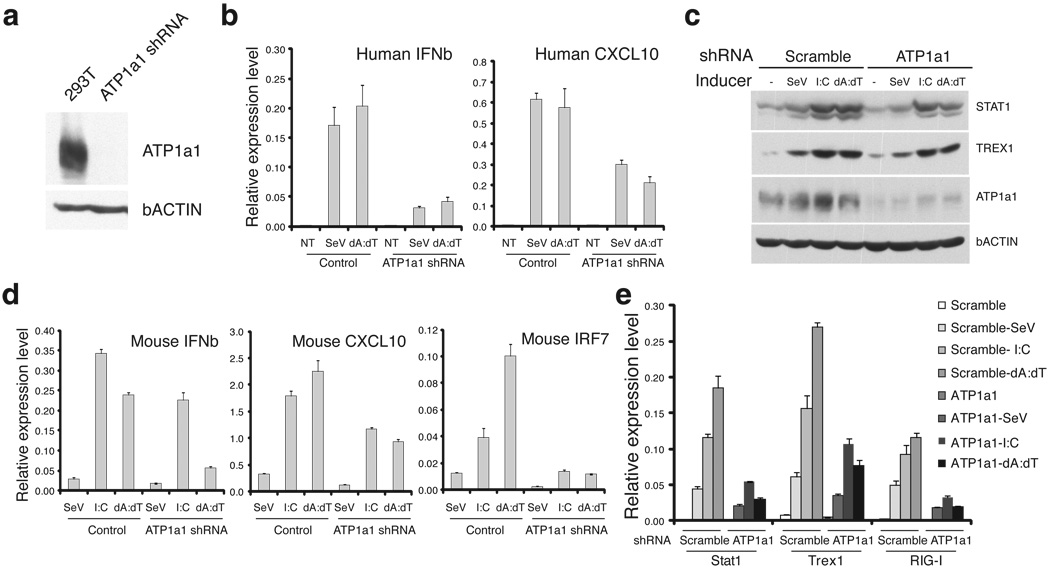
To further examine the role of the sodium pump in IFNβ gene expression, we conducted shRNA knockdown experiments. We first carried out the knockdown experiments in 293T cells, where the induction of IFNβ and Cxcl10 genes by virus and dsDNA was greatly inhibited when ATP1a1 gene expression was knocked-down by shRNA specifically targeting the gene (Fig. 5a,5b). Real time PCR quantification showed that a 4 fold reduction of IFNβ and at least 2 fold reduction of Cxcl10 (Fig. 5b) in knockdown cells compared to control cells for both inducers. The impaired IFNβ induction in ATP1a1 knock-down cells is not due to apoptosis (Supplementary Fig. 7a), and was similarly observed when compared to unrelated PARP1 knock-down cells (Supplementary Fig. 7b).
Fig. 5. Knocking-down sodium pump expression inhibits IFNβ induction.
a. Efficient knock-down of ATP1a1 expression in 293T cells. For the full image, see Supplementary Figure 12c. b. SeV and dsDNA induced gene expression was inhibited in ATP1a1 knock-down cells. Control or ATP1a1 knock-down cells were infected with SeV or transfected with dsDNA for 6 hrs. RNA was harvested and Q-PCR conducted to monitor the expression of IFNβ and Cxcl10 genes. The expression levels were normalized to that of GAPDH gene. NT: Not treated. Data represent mean values ± s.d. (n=3). c–e. Knocking-down ATP1a1 expression in MEFs also reduced virus, dsRNA and dsDNA induced gene expression. MEFs with shRNA targeting ATP1a1 or a scramble sequence as control were subjected to SeV, poly I:C and poly dA:dT treatment. 6 hrs later, cells were harvested for either protein analysis (c, blot for Stat1, Trex1, ATP1a1 and β-actin proteins) or Q-PCR analysis (d–e) for the expression of IFNβ, Cxcl10, IRF7 (d), and Stat1, Trex1 and RIG-I (e) genes. The expression of β-actin gene was used as the reference. Data represent mean values ± s.d. (n=3). For the full image, see Supplementary Figure 12d.
As mentioned before, the mouse ATP1a1 protein is not sensitive to cardiac glycoside binding. Not surprisingly, we found little effect of bufalin on the induction of IFNβ gene by various inducers in mouse embryonic fibroblasts (MEFs), which express only the ATP1a1 gene out of the four alpha subunits (Supplementary Fig. 8a). Interestingly, induction of IFNβ and Cxcl10 genes and other ISGs (IRF7, RIG-I, Trex1, STAT1 etc) were significantly reduced (>30% for IFNβ and Cxcl10 genes, and at least 50% for most other genes) at both mRNA and protein levels when the expression level of the ATP1a1 gene was reduced by shRNA (Fig. 5c–e). Genome wide analysis revealed that not only was the expression level of target genes affected, but also the number of genes induced by the stimulators tested (SeV, poly I:C,and poly dA:dT) was significantly affected (Supplementary Fig. 8b).
Taken together our data show that normal sodium pump activity is critically involved in the optimal induction of the IFNβ gene after pathogen challenge. The main mechanism is through maintenance of a balanced intracellular ion concentration. RIG-I, a key component of the virus activated signaling pathway, is sensitive to intracellular ion fluctuations. Our findings suggest that manipulating the intracellular ion concentration can effectively modulate the cellular antiviral signaling pathway. Since mis-regulation of cytokine production has been implicated in many human diseases, our results suggest that ion concentration modulators will serve as promising agents to treat these diseases.
Bufalin inhibits TNF signaling
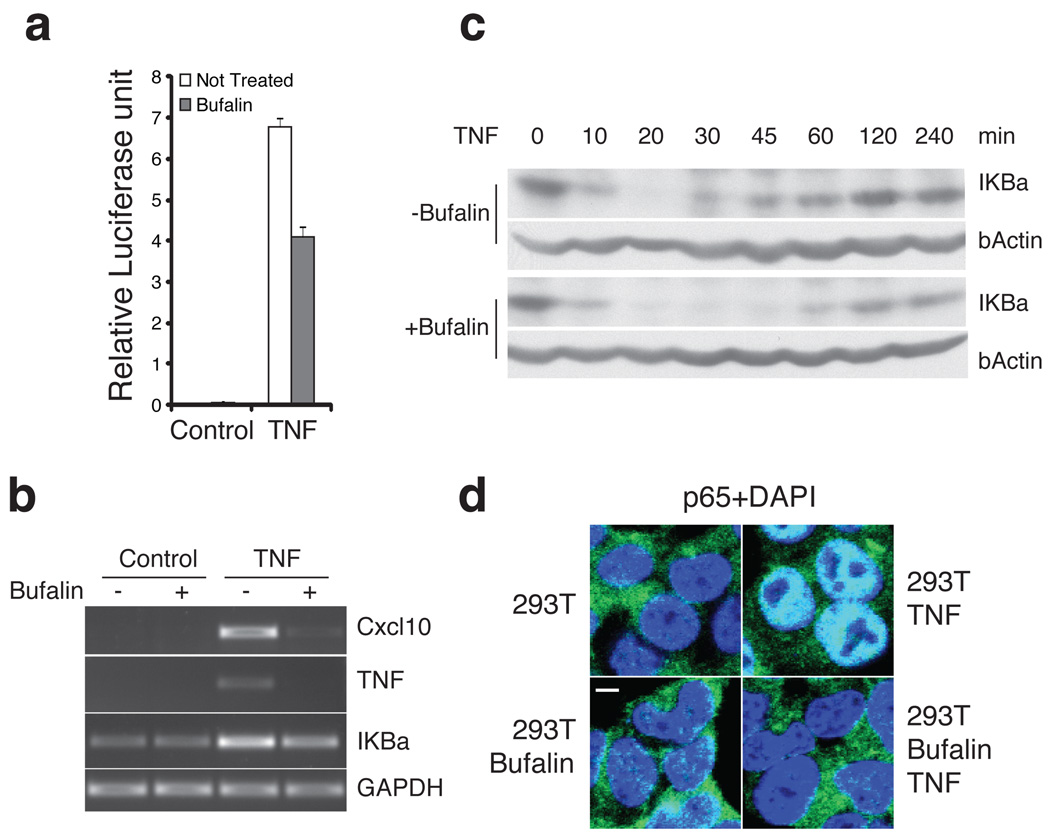
Since the sodium pump maintains physiological intracellular ion concentrations, any signaling event sensitive to changes in ion concentrations could be affected by cardiac glycosides. We therefore carried out experiments to test the effects of bufalin on other signaling pathways. Specifically, we tested interferon (IFN), tumor necrosis factor (TNF), epidermal growth factor (EGF) signaling, and treatment with Lipopolysaccharide (LPS). While effects of bufalin on IFN, EGF and LPS are selective and generally weaker (Supplementary Fig. 9), the effects on TNF signaling are strong. The latter inhibition was demonstrated using a luciferase reporter assay with the PRDII element (NFκB binding site) driving luciferase gene expression. Treatment with TNF induces the expression of the reporter gene. As shown in Fig. 6a the expression of the luciferase gene decreased by 40% after bufalin treatment. In addition, bufalin significantly inhibited the expression of endogenous TNF target genes (Fig. 6b).
Fig.6. Bufalin inhibits TNF signaling.
a. Bufalin treatment reduced TNF induced NFκB activation in reporter assays. 293T cells were transfected with PRDII-driving luciferase reporter construct, treated with bufalin (1 µM) and TNF (10 ng/ml) 24hrs before measuring the luciferase activities. Data represent mean values ± s.d. (n=3). b. Bufalin inhibits TNF induced gene expression. 293T cells were treated with bufalin (1 µM) and TNF (10 ng/ml) for 6hrs, RNA extracted and subjected to RT-PCR analysis. c. Bufalin delays and decreases TNF induced NFκB activation. 293T cells were pretreated with bufalin (1 µM) for 30 min before addition of TNF (10 ng/ml) to the medium, cells were harvested at indicated times and the protein level of IKBα determined by western blot analysis. For the full image, see Supplementary Figure 12c. d. Bufalin treatment interferes with nuclear translocation of p65. 293T cells with and without bufalin (1 µM) pretreatment were stimulated with TNF (10 ng/ml) for 15 min, and then formaldehyde fixed and subjected to immunofluorescent staining with anti-p65 antibody (green). Blue: DAPI staining for nuclei. Scale bar, 5 µm.
To investigate the mechanism of the inhibitory effects of bufalin on TNF signaling, we monitored the degradation of IKBα protein in the presence or absence of bufalin. Bufalin did not affect the initial degradation of IKBα, as the degradation of IKBα is almost complete after 20 min of TNF treatment in the presence or absence of bufalin. However, the subsequent re-synthesis of IKBα is significantly delayed and the level reduced in cells treated with bufalin (Fig. 6c). The level of IKBα is at least twice as high in the control sample after 45–60 min compared to the bufalin treated sample. Monitoring the serine 32/36 phosphorylation of IKBα revealed similar levels of phospho-IKBα after 15–20 minutes of TNF stimulation in both bufalin treated and non-treated cells. By contrast, the level of IKBα phosphorylation (S32/36) was significantly higher 1 hr after TNF stimulation in control cells compared to bufalin treated cells (Supplementary Fig. 10a). We also observed that bufalin strongly inhibited the nuclear translocation of the NFκB p65 subunit after TNF stimulation. This effect was greater in the early time points: 15–20 minutes after TNF stimulation, p65 was in the nucleus in all control cells, while virtually no p65 was observed in the nucleus of the bufalin treated cells (Fig. 6d). Subsequently, a gradual increase in nuclear p65 was observed (Supplementary Fig. 10b). These data show that the nuclear translocation of p65 is sensitive to bufalin treatment, although the initial degradation of IKBα is not.
Discussion
We have shown that cardiac glycosides are potent inhibitors of IFNβ gene expression in response to virus infection, or treatment with dsRNA, or dsDNA. Although recent studies suggest that cardiac glycosides can induce cellular signaling events independent of their inhibition of the sodium pump30,42, we show here that inhibition of IFNβ expression by cardiac glycosides is exclusively through blocking the activities of the sodium pump rather than by an off-target effect (Fig. 4d, 4e). We also show that bufalin inhibits the activation of both IRF3 and NFκB, transcription factors required for IFNβ gene expression. Over-expression experiments with intermediates in the virus-induction signaling pathway reveal that bufalin acts at an early step of the pathway, with the inhibition of the ATPase activity of the RNA sensor RIG-I as a primary mechanism. This inhibition appears to be the consequence of the ability of bufalin to alter the intracellular ion concentrations by inhibiting the sodium pump. None of the downstream signaling components in the pathway appear to be directly inhibited by bufalin.
RIG-I may also be the target of inhibition by bufalin in the dsDNA activation pathway. Two recent studies show that AT-rich dsDNA can signal through RIG-I to activate the IFN gene. The mechanism involves a critical sensor: RNA polymerase III, which transcribes 5’-ppp bearing “panhandle” RNA from the dsDNA template3,4. These nascent RNAs then activate IFNβ expression through the RIG-I-MAVS pathway. We provide evidence that the activity of cytoplasmic RNA polymerase III is also inhibited by bufalin (Supplementary Figure 11). When total RNA from dsDNA transfected cells treated with bufalin was transfected into new cells, it failed to induce the expression of IFNβ gene (Supplementary Fig. 11).
We found that the RNA binding and ATPase activities of RIG-I can be uncoupled, as demonstrated by in vitro RNA binding and ATPase assays (Fig. 3c). Cardiac glycosides inhibit the function of the sodium pump, and subsequently lead to elevated intracellular sodium and calcium concentrations. Although the RNA binding activity of RIG-I is not affected by bufalin treatment (Fig. 3a), the high salt concentrations induced by bufalin inhibits the ATPase activity. This observation is in agreement with a previous report showing that the ATPase activity of the recombinant helicase domain of RIG-I is inhibited by high concentrations of sodium chloride43. Although the function of the ATPase activity of RIG-I in signaling is not understood, an ATPase dependent translocase activity is required to ratchet RIG-I repeatedly along the dsRNA ligand21. It is possible that this activity is required for the unwinding of the RNA and the binding of RIG-I to the MAVS protein on the mitochondrial membrane.
We have also shown that bufalin inhibits the activation of NFκB by TNF. Remarkably, bufalin inhibits the nuclear translocation of NFκB in response to TNF, but not the degradation of IKBα. The nuclear translocation of NFκB is regulated by post-translational modifications like phosphorylation or sumoylation44. We examined the effects of bufalin on the phosphorylation of p65, and found little difference on the initial phosphorylation of S276, S468 and S536 residues between bufalin treated and non-treated samples. However, S536 phosphorylation appeared weaker in later time points after bufalin treatment (Supplementary Fig. 10a). This could potentially explain reduced NFκB transactivation activity in bufalin treated cells, but does not explain the strong cytoplasmic retention of p65 after 15–20 min TNF stimulation. Other factors involved in p65 nuclear translocation must be affected by bufalin treatment. We note that another cardiac glycoside, digitoxin, has been shown to inhibit TNF signaling by blocking the recruitment of TRADD to the TNF receptor, and directly inhibiting IKBα degradation45. By contrast, as mentioned above, we show that IKBα degradation is not inhibited by bufalin.
The inhibition of IFNβ gene induction and TNF signalling by bufalin may have practical implications. For example, high levels of interferon production play a major role in the autoimmune disease SLE7. In SLE patients, the tolerance of autoantigen breaks down and high levels of IFN are detected in serum. This leads to aberrant activation of immature myeloid dendritic cells and downstream effector cells involved in autoimmune reactions7. To determine whether the reduction of IFN levels have clinical benefits in SLE patients, anti-interferon antibodies have been tested, and preliminary results are promising46. It is therefore possible that bufalin could be used to treat SLE patients based on its ability to inhibit IFN expression. However, a major concern is whether patients can tolerate chronic treatment at doses of bufalin necessary to inhibit IFN expression. Although bufalin has not been directly tested in humans, it is a primary component of the Chinese medicine Huachansu recently tested in cancer patients. Plasma levels of 9 nM of bufalin were detected in this study, and were well tolerated47. The IC50 for bufalin necessary to inhibit IFNβ expression is 4.3 nM (Fig. 1b), thus it is possible that bufalin could be used to treat the clinical manifestations of SLE flares. Similarly, bufalin may be used to treat inflammatory diseases caused by aberrant TNF signaling. We conclude that alterations of the intracellular ion concentrations caused by cardiac glycosides can have profound effects on intracellular signaling pathways, and these effects may provide an exciting new approach to the treatment of inflammatory and autoimmune diseases.
Materials and methods
Cells, reagents and plasmids
293T, Hela, MG63, and Namalwa cells are from ATCC, wild type MEFs are from Wen-Chen Yeh (University of Toronto, Toronto). Bufalin was purchased from Calbiochem, Digoxin, ouabain, diazoxide, nimodipine, phenamil, poly dA:dT and poly I:C are from Sigma. The ion channel ligand library, Biomol Green reagents are from Biomol. The high content small molecule library is described before48. Recombinant TNFα is from Roche. pEF-BOS Flag-RIG-I is a gift from Dr. T. Fujita (Kyoto University, Japan), and mouse ATP1a1, ATP1a3, human ATP1a1 expression constructs are purchased from OpenBiosystems. Constructs for mutant RIG-I (K270A) and mouse ATP1a1 (D376E) were generated by standard site-directed mutagenesis. TBK1 expression construct is described before32. The MAVS expression construct was generated by cloning mouse MAVS cDNA into pKH3 vector.
Antibodies and western blots
Antibodies against human IRF3, ATP1a1, STAT1, Traf6, HSP70 and p65 are from Santa Cruz, RIG-I, MDA5, PARP1, cleaved PARP1 (human specific), cleaved Caspase3, phospho-IKBα Ser 32/36, phospho-S276, S468 and S536 p65 antibodies are from Cell Signaling, β-actin antibody is from Abcam. The IKBα antibody is from IMGenex. Trex1 antibody is from BD Biosciences. Western blots were carried out according to standard protocols. About 50µg of total protein lysate (lysed in a buffer of 20 mM Tris-HCl, pH7.5, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 30 mM NaF, 1 mM glycerolphosphate, 1X proteinase inhibitor (Roche) and 1 mM Na3VO4) was denatured in sampling buffer (50 mM Tris-HCl, pH 6.8, 10% glycerol, 2% SDS, 0.02% bromophenal blue and 2.5% β-mecaptoethanol) and subjected to SDS-PAGE. Proteins were transferred to a PVDF membrane, blocked with 5% milk in Tris-buffered saline Tween 20 (TBST), and incubated with various primary antibodies. Washed membranes were incubated with HRP conjugated secondary antibody and protein bands visualized with ECL reagents (Millipore or Pierce).
Luciferase reporter assay and chemical treatment
Approximately 40,000 293T cells were seeded in each well of a 96 well plate, and co-transfected with IFNβ-110 firefly luciferase reporter and renilla luciferase plasmids32. After 24 hrs, cells were treated with various chemicals at the indicated concentrations, and Sendai virus infection was initiated one hour later. After another 24hrs, cells were lysed and subjected to Dual-Glo luciferase assay analysis (Promega) with an Analyst AD plate reader. IC50 for bufalin, ouabain and digoxin to inhibit IFNβ expression was calculated using Masterplex software by the four-parameter logistic fit method.
In vitro RNA binding and ATPase assay
Double strand RNA corresponding to GFP sequences (67bp of the 3’ end) was generated by in vitro transcription with T7 RNA polymerase. About 200ng of dsRNA was incubated with 0.5 µg of recombinant Flag-tagged RIG-I protein in a 20µl buffer of 20 mM Tris-HCl, pH8.0, 1.5 mM MgCl2, 1.5mM DTT and 5% glycerol for 15min at room temperature. RNA:protein complexes were separated in a 0.8% agarose gel with 0.5X TBE running buffer, and run for 1.5 hrs at 100 volts. For the ATPase assay, the same RNA:protein complexes were formed by incubation at 37°C for 15 min, then ATP added to a final concentration of 1mM and further incubated for 15 min. The released free phosphates were measured with the BIOMOL GREEN kit (Biomol) according to the manufacturer instructions.
Biotin-labeled dsRNA pull down assay
For the biotin-labeled dsRNA pull down assay, we generated dsRNA (GFP sequences) by in vitro T7 RNA polymerase transcription in the presence of biotin-11-UTP (Ambion). About 8 µg of this dsRNA was transfected into 2 million 293T/RIG-I stable cells treated with and without 1µM bufalin. 6 hrs later, cell lysates were prepared and subjected to NeutrAvidin beads (Pierce) pull down for 1 hr at 4 °C. Bound protein was separated by SDS-PAGE and transferred to PVDF membrane. Binding of these RNAs by RIG-I was monitored by probing the membrane with an anti-RIG-I antibody.
Virus infection, RNA preparation, microarray analysis and immunofluorescent staining
Concentrated Sendai virus stock (Cantell strain, Charles River Lab) was added to cultured cells at a concentration of 200–300 HAU/ml and incubated for the indicated time before harvesting the cells for protein or RNA analysis. Total RNA was extracted with Trizol reagent (invitrogen). RT-PCR and real time quantitative PCR were conducted according to standard protocols. For microarray experiments, RNA was biotin-labeled with the Illumina TotalPrep RNA Amplification Kit (Ambion) and subjected to the Illumina HumanRef-8 v3.0 BeadChip microarray analysis. Immunofluorescent staining was conducted according to standard protocols: cells were fixed with 4% formaldehyde for 10 min, washed with PBS and permeabilized with 0.1% Triton X-100 in PBS, and incubated with primary antibodies over night. Cells were extensively washed before incubating with FITC-conjugated secondary antibody, mounted with DAPI containing media, and subjected to epifluorescent or confocal microscopy.
Lentivirus mediated shRNA knockdown
shRNA constructs were generated by cloning sequences 5’-ccggaaagactgaaagaatac-3’ targeting human ATP1a1 mRNA, or 5’-gtgattcgaaatggagagaaa-3’ targeting mouse ATP1a1 mRNA into the pLKO.1 TRC cloning vector, a construct with scramble sequences was used as control. Packaging of lentivirus was achieved by co-transfecting 293T cells with targeting plasmid together with pLP1, pLP2 and pLP-VSVG plasmids according to the Viralpower Lentivirus expression system from Invitrogen. The supernatants from transfected cells was harvested and used to infect new cells. Knockdown cells were pooled by puromycin selection and the efficiency of knocking-down was verified by western blot.
Supplementary Material
Fig.7.
Fig.8.
Fig.9.
Acknowledgments
This work is supported by NIH grant 5R01AI020642-26 (to T.M.). S.C. is supported by the postdoctoral fellowship from Juvenile Diabetes Research Foundation. We thank Xiaoyun Sun (Brandeis University) for help with the microarray analysis. We also thank Sze-Ling Ng for critical reading of this manuscript and Stefanie Schalm for helpful discussions.
Footnotes
Author Contributions
J.Y., S.C. and T.M. conceived the research, J.Y. and S.C. conducted experiments, J.Y. and T.M. wrote the paper.
References
- 1.Sen GC. Viruses and interferons. Annu Rev Microbiol. 2001;55:255–281. doi: 10.1146/annurev.micro.55.1.255. [DOI] [PubMed] [Google Scholar]
- 2.Honda K, Takaoka A, Taniguchi T. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity. 2006;25:349–360. doi: 10.1016/j.immuni.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 3.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ablasser A, et al. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia-Sastre A, Biron CA. Type 1 interferons and the virus-host relationship: a lesson in detente. Science. 2006;312:879–882. doi: 10.1126/science.1125676. [DOI] [PubMed] [Google Scholar]
- 6.Le Bon A, Tough DF. Links between innate and adaptive immunity via type I interferon. Curr Opin Immunol. 2002;14:432–436. doi: 10.1016/s0952-7915(02)00354-0. [DOI] [PubMed] [Google Scholar]
- 7.Banchereau J, Pascual V. Type I interferon in systemic lupus erythematosus and other autoimmune diseases. Immunity. 2006;25:383–392. doi: 10.1016/j.immuni.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 8.Hall JC, Rosen A. Type I interferons: crucial participants in disease amplification in autoimmunity. Nat Rev Rheumatol. 6:40–49. doi: 10.1038/nrrheum.2009.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mandl JN, et al. Divergent TLR7 and TLR9 signaling and type I interferon production distinguish pathogenic and nonpathogenic AIDS virus infections. Nat Med. 2008;14:1077–1087. doi: 10.1038/nm.1871. [DOI] [PubMed] [Google Scholar]
- 10.Whittemore LA, Maniatis T. Postinduction turnoff of beta-interferon gene expression. Mol Cell Biol. 1990;10:1329–1337. doi: 10.1128/mcb.10.4.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacquelin B, et al. Nonpathogenic SIV infection of African green monkeys induces a strong but rapidly controlled type I IFN response. J Clin Invest. 2009;119:3544–3555. doi: 10.1172/JCI40093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maniatis T, et al. Structure and function of the interferon-beta enhanceosome. Cold Spring Harb Symp Quant Biol. 1998;63:609–620. doi: 10.1101/sqb.1998.63.609. [DOI] [PubMed] [Google Scholar]
- 13.Sun L, Liu S, Chen ZJ. SnapShot: pathways of antiviral innate immunity. Cell. 140:436–436. e2. doi: 10.1016/j.cell.2010.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ford E, Thanos D. The transcriptional code of human IFN-beta gene expression. Biochim Biophys Acta. 1799:328–336. doi: 10.1016/j.bbagrm.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 15.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 16.Cui S, et al. The C-terminal regulatory domain is the RNA 5'-triphosphate sensor of RIG-I. Mol Cell. 2008;29:169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 17.Takahasi K, et al. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol Cell. 2008;29:428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 18.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 19.Tang ED, Wang CY. MAVS self-association mediates antiviral innate immune signaling. J Virol. 2009;83:3420–3428. doi: 10.1128/JVI.02623-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujita T. A nonself RNA pattern: tri-p to panhandle. Immunity. 2009;31:4–5. doi: 10.1016/j.immuni.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 21.Myong S, et al. Cytosolic viral sensor RIG-I is a 5'-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323:1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gack MU, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 23.Gack MU, Nistal-Villan E, Inn KS, Garcia-Sastre A, Jung JU. Phosphorylation-mediated negative regulation of RIG-I antiviral activity. J Virol. 84:3220–3229. doi: 10.1128/JVI.02241-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saha SK, et al. Regulation of antiviral responses by a direct and specific interaction between TRAF3 and Cardif. EMBO J. 2006;25:3257–3263. doi: 10.1038/sj.emboj.7601220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo B, Cheng G. Modulation of the interferon antiviral response by the TBK1/IKKi adaptor protein TANK. J Biol Chem. 2007;282:11817–11826. doi: 10.1074/jbc.M700017200. [DOI] [PubMed] [Google Scholar]
- 26.Kawai T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 27.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhong B, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 29.Schroder M, Baran M, Bowie AG. Viral targeting of DEAD box protein 3 reveals its role in TBK1/IKKepsilon-mediated IRF activation. EMBO J. 2008;27:2147–2157. doi: 10.1038/emboj.2008.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prassas I, Diamandis EP. Novel therapeutic applications of cardiac glycosides. Nat Rev Drug Discov. 2008;7:926–935. doi: 10.1038/nrd2682. [DOI] [PubMed] [Google Scholar]
- 31.Hemmi H, et al. The roles of two IkappaB kinase-related kinases in lipopolysaccharide and double stranded RNA signaling and viral infection. J Exp Med. 2004;199:1641–1650. doi: 10.1084/jem.20040520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fitzgerald KA, et al. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 33.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 34.Saito T, et al. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci U S A. 2007;104:582–587. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langer GA. Ionic basis of myocardial contractility. Annu Rev Med. 1977;28:13–20. doi: 10.1146/annurev.me.28.020177.000305. [DOI] [PubMed] [Google Scholar]
- 36.Lingrel JB. The physiological significance of the cardiotonic steroid/ouabain-binding site of the Na,K-ATPase. Annu Rev Physiol. 72:395–412. doi: 10.1146/annurev-physiol-021909-135725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohtsubo M, Noguchi S, Takeda K, Morohashi M, Kawamura M. Site-directed mutagenesis of Asp-376, the catalytic phosphorylation site, and Lys-507, the putative ATP-binding site, of the alpha-subunit of Torpedo californica Na+/K(+)-ATPase. Biochim Biophys Acta. 1990;1021:157–160. doi: 10.1016/0005-2736(90)90028-m. [DOI] [PubMed] [Google Scholar]
- 38.Simpson CD, et al. Inhibition of the sodium potassium adenosine triphosphatase pump sensitizes cancer cells to anoikis and prevents distant tumor formation. Cancer Res. 2009;69:2739–2747. doi: 10.1158/0008-5472.CAN-08-2530. [DOI] [PubMed] [Google Scholar]
- 39.Izquierdo I. Nimodipine and the recovery of memory. Trends Pharmacol Sci. 1990;11:309–310. doi: 10.1016/0165-6147(90)90229-2. [DOI] [PubMed] [Google Scholar]
- 40.Trube G, Rorsman P, Ohno-Shosaku T. Opposite effects of tolbutamide and diazoxide on the ATP-dependent K+ channel in mouse pancreatic beta-cells. Pflugers Arch. 1986;407:493–499. doi: 10.1007/BF00657506. [DOI] [PubMed] [Google Scholar]
- 41.Garvin JL, Simon SA, Cragoe EJ, Jr, Mandel LJ. Phenamil: an irreversible inhibitor of sodium channels in the toad urinary bladder. J Membr Biol. 1985;87:45–54. doi: 10.1007/BF01870698. [DOI] [PubMed] [Google Scholar]
- 42.Xie Z, Cai T. Na+-K+--ATPase-mediated signal transduction: from protein interaction to cellular function. Mol Interv. 2003;3:157–168. doi: 10.1124/mi.3.3.157. [DOI] [PubMed] [Google Scholar]
- 43.Gee P, et al. Essential role of the N-terminal domain in the regulation of RIG-I ATPase activity. J Biol Chem. 2008;283:9488–9496. doi: 10.1074/jbc.M706777200. [DOI] [PubMed] [Google Scholar]
- 44.Cyert MS. Regulation of nuclear localization during signaling. J Biol Chem. 2001;276:20805–20808. doi: 10.1074/jbc.R100012200. [DOI] [PubMed] [Google Scholar]
- 45.Yang Q, et al. Cardiac glycosides inhibit TNF-alpha/NF-kappaB signaling by blocking recruitment of TNF receptor-associated death domain to the TNF receptor. Proc Natl Acad Sci U S A. 2005;102:9631–9636. doi: 10.1073/pnas.0504097102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ronnblom L, Elkon KB. Cytokines as therapeutic targets in SLE. Nat Rev Rheumatol. 6:339–347. doi: 10.1038/nrrheum.2010.64. [DOI] [PubMed] [Google Scholar]
- 47.Meng Z, et al. Pilot study of huachansu in patients with hepatocellular carcinoma, nonsmall-cell lung cancer, or pancreatic cancer. Cancer. 2009;115:5309–5318. doi: 10.1002/cncr.24602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen S, et al. A small molecule that directs differentiation of human ESCs into the pancreatic lineage. Nat Chem Biol. 2009;5:258–265. doi: 10.1038/nchembio.154. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.