Abstract
It has been debated for decades how cancer cells acquire metastatic capability. It is unclear whether metastases are derived from distinct subpopulations of tumor cells within the primary site with higher metastatic potential, or whether they originate from a random fraction of tumor cells. Here we show, by gene expression profiling, that human primary breast tumors are strikingly similar to the distant metastases of the same patient. Unsupervised hierarchical clustering, multidimensional scaling, and permutation testing, as well as the comparison of significantly expressed genes within a pair, reveal their genetic similarity. Our findings suggest that metastatic capability in breast cancer is an inherent feature and is not based on clonal selection.
Metastases are the main cause of death in breast cancer. They arise, after the spread of cells from a primary tumor via the blood circulation, as solid tumors in distant organs (1, 2). The prevailing model of metastasis suggests that metastatic capacity is acquired late in tumorigenesis and is a nonrandom and highly selective process (3, 4). This genetic selection model, based on in vitro culturing of tumor cell lines subsequently transplanted into mice, encompasses the escape, survival, and proliferation of a cryptic minority of tumor cells from subpopulations with increased metastatic capacity in the primary site (3-6). Such a model implies that a metastasis arising from a selected subclone would be molecularly distinct from its primary tumor. The subpopulation concept by Fidler is widely accepted, although the metastatic process has also been described as a stochastic event, giving primary tumor cells an equal metastatic potential (7-9).
It has been shown that activation of a single gene, which in turn affects a process essential for metastasis, can be sufficient for inducing metastasis in vitro (10, 11). This would imply that one gene, when activated early in its development, can empower the metastatic process once a primary tumor with additional genetic changes has been established. In human breast cancer, it has recently been shown that expression profiles can predict the risk of development of distant metastases even for small primary tumors (12, 13). These findings suggest that the capacity to metastasize might be acquired relatively early in multistep tumorigenesis (14), thereby challenging the subpopulation concept. If this inherent model is correct, a metastasis might then be genetically similar, if not identical, to that of the primary tumor from which it originated. To test this hypothesis, we compared pairs of human primary breast carcinomas and their metastases, developed years later at distant sites, by gene expression profiling.
Materials and Methods
Breast Tumors and Metastases. Tumor samples from breast cancer patients with a surgically removed distant metastasis were selected from the fresh-frozen tissue bank of the Netherlands Cancer Institute. The tumor and metastatic material was snap-frozen in liquid nitrogen within 1 h after surgery. Before and after cutting sections for RNA isolation, one slide was prepared for a hematoxylin and eosin staining to select only samples with 50% or more tumor cells. Patient histories and tissue sections were studied carefully to assure that only patients with distant metastases, and not second primary tumors, were included. Patients had no previous malignancies. Estrogen-receptor α (ER-α) expression was determined by immunohistochemistry; a tumor was deemed to be ER-α negative when <10% of the tumor cells showed staining.
RNA Isolation and Amplification. From the mammary and metastatic tumors, 30 sections of 30-μm thickness were used for total RNA isolation with RNAzol Bee (Campro Scientific, Amersfoort, The Netherlands). Isolated total RNA was subsequently DNase-treated by using the Qiagen RNase-free DNase kit and RNeasy spin columns (Qiagen, West Sussex, U.K.) and dissolved in RNase-free H2O. Four micrograms of total RNA was used to generate cDNA by using superscript II and an oligo(dT) primer containing a T7 polymerase recognition site. cRNA was generated by in vitro transcription using T7 RNA polymerase (Megascript T7 kit, Ambion, Huntingdon, U.K.). Amplification yields were 1,000- to 2,000-fold.
cRNA Labeling and Hybridization. Two micrograms of cRNA from one breast cancer primary tumor or breast cancer metastasis was labeled in a reverse transcriptase reaction with Cy3 or Cy5 (CyDye, Amersham Biosciences) and mixed with the same amount of reverse color Cy-labeled cRNA from a reference pool that consisted of pooled cRNA of equal amounts from 60 primary breast tumors. The breast tissue reference was chosen to be closely related to the tumors and metastases, so that we would be able to identify small expression level changes between the primary and metastatic breast tumor group. For each tumor and metastasis, two hybridizations were performed by using a reversal fluorescent dye. To monitor the consistency of the array experiments, “self-self” experiments were performed by using the hybridized tumor or metastasis tissue as reference sample. Labeled cDNAs were heated to 100°C for 5 min and added to preheated hybridization buffer (Slide hyb buffer 1, Ambion) and hybridized at 42°C to 18,336 human cDNA microarrays (Central Microarray Facility, Netherlands Cancer Institute). Fluorescent images of the microarrays were obtained by using the Scanarray 4000 microarray scanner (Perkin-Elmer). Fluorescent intensities of the images were quantified by using IMAGENE 4.2 (Biodiscovery, Marina Del Rey, CA) and corrected for background noise.
Microarray Slides. cDNA microarray slides were manufactured at the Central Microarray Facility (Netherlands Cancer Institute). Sequence-verified clones were obtained from Research Genetics (Huntsville, AL) and were spotted by using the Microgrid II arrayer (Biorobotic, Cambridge, U.K.) with a complexity of 19,200 spots per glass slide (GeneID list and information http://microarrays.nki.nl).
Analysis and Statistics. Fluorescence intensities of scanned images were quantified and normalized, and ratios were calculated and compared to the intensities of the reference pool (15). Confidence levels were assigned to measurements by using the Rosetta error model (16). To determine genes that discriminate between primary tumors and metastases, we used a supervised classification method with a nearest prototype classifier and a leave-one-out cross-validation method (12).
Gene clustering and tumor clustering were performed by using an unsupervised hierarchical clustering algorithm (Pearson correlation coefficient) using the GENESIS program (17). Pairwise similarity among tumors and metastases was calculated based on the Xdef values across all 18,336 genes, a value to decrease the uncertainty of array measurements such as low spot intensities by significance corrected expression data.
Mapping by multidimensional scaling was performed in such way that the intertumor distances in the lower-dimensional space correspond as well as possible to the intertumor distances in the original (i.e., 18,336) space. We used the Pearson correlation (1 - r2) between two tumor profiles as measure of distance between tumors.
The within-pair-between-pair scatter ratio (WPBPSR) measures the ratio of the dissimilarities between matched pairs with respect to the dissimilarities between randomly matched tumors. To determine the statistical significance of the WPBPSR, a permutation test was performed. During each iteration of this test, the given tumors and metastases were randomly paired and the WPBPSR was computed for this random pairing; this procedure was repeated 10,000 times.
Additional Microarray Information. The description of this microarray study followed the Minimum Information About a Microarray Experiment (MIAME) guidelines (18). The original data and detailed protocols for RNA isolation, amplification, labeling and hybridization are available at www.nki.nl/nkidep/pa/microarray.
Results
Despite the fact that only a few cancer patients have distant metastases surgically removed, we were able to select eight pairs of primary breast carcinomas and their matching distant metastases. The interval between the surgical removal of the primary tumors and metastases varied from 1.6 to 15 years (median 3.6 years). Two of the patients developed a metastasis to the lung, one patient showed metastatic spread to the skin of the arm, one patient showed metastatic spread to a distant (supraclaviculair) lymph node, and four patients showed metastatic spread to the ovary (Table 1).
Table 1. Patient characteristics.
| Therapy for primary tumor
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Patient | Localization metastasis* | Interval to metastasis, y | Tumor diameter, mm | LN status/diagnosis | ER-α status | Chemo | Horm | Mastectomy |
| 1 | Ovary | 3.9 | 15 | - | + | + | ||
| 2 | Ovary | 2.9 | 15 | - | + | + | + | |
| 3 | Lymph node† | 3.3 | 30 | + | - | + | - | |
| 4 | Ovary | 6.3 | 10 | + | + | + | + | |
| 5 | Lung | 3.0 | 24 | + | - | + | + | - |
| 6 | Arm | 4.4 | 23 | - | - | + | + | - |
| 7 | Lung | 1.6 | 70 | - | - | + | + | |
| 8 | Ovary | 15.0 | 13 | - | + | + | + | + |
LN, lymph node; Chemo, chemotherapy; Horm, hormonal treatment.
Surgically removed
Supraclavicular
To study the gene expression profiles of matching primary breast and metastatic tumors and gain insight into specific changes associated with breast cancer progression, we used human 18,336 cDNA microarrays. To identify genes that could discriminate primary tumors from metastases, we used a supervised classification method. The top ranked genes that separate the two classes best in a nearest prototype classifier (12) were determined and used in a cross-validation procedure. At each validation iteration in this procedure, a matched pair was left out and subsequently classified. No classifier, employing an incremental number of genes, could be determined because of low performance; in fact, the performance resembled random classification (data not shown). Moreover, because a low number of samples with a high number of genes would more easily give a classifier by chance provides an additional argument that the primary and metastatic breast tumors tested here do not differ by a general subset of genes.
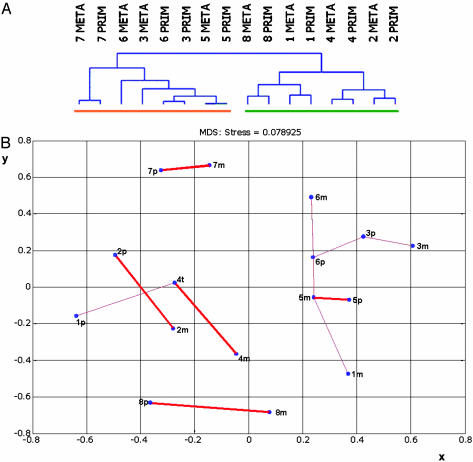
To further scrutinize our hypothesis, we looked for similarity between the primary tumors and matching metastases. Unsupervised hierarchical clustering grouped the tumors on the basis of their similarity measured over all 18,336 cDNAs on the array. Six of the eight pairs clustered next to each other (Fig. 1A and Fig. 3, which is published as supporting information on the PNAS web site). In these pairs, the primary tumors had a higher similarity to its affiliated metastasis than to other tumors, indicating that the gene expression profiles of the primary and matching distant metastatic breast tumors are highly similar. Two of the primary tumors did not cluster with its distant metastasis, but had a higher similarity to each other (Prim3 and Prim6, Fig. 1 A). The division of the dendrogram into the two main branches is explained by the notion that four tumors and matching metastases display the highly dominant ER-α expression profile and four pairs do not (12, 19, 20).
Fig. 1.
(A) Unsupervised hierarchical clustering of 16 matching primary breast and metastatic tumors from eight patients, measured over 18,336 genes. The dendrogram has two large branches; the orange bar represents ER-α-negative tumors, the green bar represents ER-α-positive tumors. Alignment of six matching pairs was established, grouping of two pairs in one subbranch. META, metastasis; PRIM, primary tumor. (B) Two-dimensional representation of a multidimensional scaling analysis of eight matching primary and metastatic tumors using 18,336 genes. x and y axes; distance in arbitrary units. A thick red line indicates two-way-pairing, and a thin red line indicates one-way pairing. Np(n = 1-8), patient number primary tumor; nm(n = 1-8), patient number distant metastasis.
Next, a multidimensional scaling analysis was applied. By using this tool, the relations measured over all genes on the array between all tumors and metastases can be visualized in two dimensions. By doing so, the genetic similarities or dissimilarities between tumors and metastases are depicted as distance. The gene expression profiles of tumor and metastasis of patient 5 are the most similar, as shown by the shortest distance (Fig. 1B). A two-way pairing of the primary and metastatic tumor from the same patient was established in five cases (Fig. 1B, thick red line). This means that the gene expression profiles of these five tumor and metastasis pairs are so similar, measured over all genes, that they only match each other in the matrix. As observed in the hierarchical clustering analysis, the tumor samples of patients 3 and 6 form a subgroup in the multidimensional scaling and have a lower similarity to their matching metastases than to each other (Fig. 1B, 3p-3m, thin red line). However, in this subgroup, metastasis 3m does form a one-way pairing with primary tumor 3p, and metastasis 6m forms a one-way pairing with tumor 6p. The primary and metastatic tumor of patient 1 did not establish a pair, and three additional one-way pairings were formed.
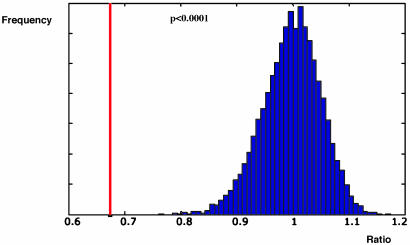
To ascertain whether the similarity we observed between primary and metastatic tumors was not a result of chance, a computational analysis was performed to establish a WPBPSR (see Materials and Methods). Subsequently, we determined the statistical significance of this WPBPSR for the eight given pairs by a permutation test. During each iteration of this test, repeated 10,000 times, we randomized the labels of the 16 primary tumors and metastases, and the WPBPSR was computed for each random pairing. The similarity between matching primary and metastatic tumor pairs was shown to be significantly higher than the similarity between random pairs (WPBPSR of 0.67 versus 1.0 ± 0.05; P < 0.0001) (Fig. 2). This finding demonstrates that the similarity within the pairs of primary and metastatic tumors was not due to chance, but rather that the expression profiles of primary breast carcinomas are similar to their corresponding metastatic lesions.
Fig. 2.
Permutation test of the within-pair-between-pair-scatter ratio (WPBPSR). Blue, null hypothesis distribution. Distribution after randomization of the labels of the primary and metastatic tumors, repeated 10,000 times (WPBPSR = 1 ± 0.05). The red line represents the WPBPSR of the eight matching primary and metastatic tumor pairs (WPBPSR = 0.67; P < 0.001).
To further confirm the genetic similarity within a pair, we selected genes that were significantly expressed in primary and metastatic tumor pairs as computed by the Rosetta error model (P < 0.01 in at least two experiments) (12, 16). Of the 18,336 genes on the array, 17,748-18,271 genes did not show a difference in expression level between the matching pairs. On average, >92% of these genes were coregulated between primary and matching metastatic tumors as compared to the reference (data not shown). Only 2-44 significantly expressed genes within each of the matching primary breast and metastatic tumor pairs were antiregulated. None of these genes were antiregulated in all eight tested primary and metastatic tumor pairs (data not shown), indicating that these genes are not involved in a common pathway for metastasis in these tumors.
Discussion
The data presented here show that gene expression profiles of primary breast tumors are maintained in their distant metastases, even if metastases develop after a long interval. For example, the profile of the metastasis in the ovary of patient 8 is virtually indistinguishable from that of the primary breast tumor, which was surgically treated 15 years before. Furthermore, the microenvironment of a distant metastasis, embedded in a different organ, apparently does not influence the overall gene expression profile to such an extent that we can distinguish a distant metastasis from its matching primary tumor. Only primary tumor 3, of the eight pairs tested, showed a higher similarity to primary tumor 6 than to its own metastasis. However, this could be caused by a relatively low percentage of tumor cells in the snap-frozen tissues of these primary tumors (both 50%), compared with their metastases (80% and 90%, respectively). Genes expressed in the 50% nontumor cells may influence the gene expression profiles of these two primary tumors (21), resulting in similarity.
The maintenance of the overall gene expression is exemplified by our inability to establish a pattern in genetic expression changes between the primary tumors and metastases. This stands in contrast to the prevalent model, which predicts that the acquisition of metastatic potential is determined in subpopulations of the primary tumor and is a rare event in cancer progression (3-6). Our findings support the notion that the genetic changes in primary tumors that favor metastasis occur early in tumor progression (13, 14), and are consistent with our recent report that disease outcome of breast cancer patients can be predicted by a “good” or “poor” prognosis signature of the primary tumor (12, 13).
So far, basic knowledge on the acquired metastatic phenotype is largely based on “single gene” overexpressing cell lines injected into mice (10, 22, 23). Recently, however, it has been shown that a human breast cancer-derived cell line with established metastatic capacity does possess our previously described “poor prognosis signature” (12), and also that subpopulations of cells display a profile predicting the site of metastasis (24). The reported bone-specific capacity is correlated with overexpression of a set of genes. Apparently, metastatic outgrowth requires additional subtle genetic alterations. The human tumors and metastases in our study, however, do not display a metastasis-site specific pattern, which may relate to the variety of metastasis locations (25).
The overall gene expression approach described here cannot exclude that there are single cells, or even subpopulations, with distinctive expression profiles in a primary tumor. These differences may confer metastatic potential, which in turn gives advantage in escaping the primary tumor and undergoing the full multistep metastatic process (3, 4, 6). However, one would then expect to be able to detect differences in the gene expression profile of a metastasis exclusively grown out of advantageous cells with metastatic capacities when compared to the overall bulk expression of its matching primary tumor. Clearly, there was no such distinction in our matching pairs, showing that the metastatic outgrowth at distant sites did not result in major changes in the gene expression of the tumor.
The small number of differentially expressed genes within the matching primary tumor and metastasis pairs we observed did not reveal a metastasis-specific gene set. It cannot be ruled out that this is due to the small number of samples, although close analysis of these antiregulated genes revealed mostly tissue-specific genes from the site of metastasis (data not shown). If the differentially expressed genes would be responsible for metastasis development, then a different set of genes is involved in the metastatic spread in all eight cases.
Our results should be distinguished from the recent report that primary and metastatic adenocarcinomas can be discriminated by a gene expression signature associated with metastasis (26). This signature was established by comparing different tumor types and unmatched primary and metastatic tumors, which might be the reason for the identification of a tissue-independent classifier. Furthermore, the reported similarity by hierarchical clustering of two primary breast tumors and their paired local lymph node metastases by Perou et al. (20) is in line with our results, but distinctive, because distant metastases were not studied.
Recently, predictive expression profiles of primary breast tumors for (neo-) adjuvant systemic treatment have been established, identifying cancer patients whose tumors are sensitive to a specific drug (20, 27). Our finding of a genetic similarity between a primary breast tumor and its distant metastasis suggests that therapy recommendations based on the expression profile of the primary tumor are a rational approach toward preventing the outgrowth of micrometastases.
Supplementary Material
Acknowledgments
We thank A. Velds for array bioinformatics support, A. Floore for advice on array experiments, and R. Kortlever, G. Hart, and R. Bernards for critically reading the manuscript. This work was supported by the Dutch Cancer Society.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ER-α, estrogen-receptor α; WPBPSR, within-pair-between-pair scatter ratio.
References
- 1.Chambers, A. F., Groom, A. C. & MacDonald, I. C. (2002) Nat. Rev. Cancer 2, 563-572. [DOI] [PubMed] [Google Scholar]
- 2.Harris, J. R., Lippman, M. E., Morrow, M. & Osborne, C. K. (2000) in Diseases of the Breast, ed. Freeman, S. (Lippincott Williams & Wilkins, Philadelphia), pp. 749-751.
- 3.Fidler, I. J. & Kripke, M. L. (1977) Science 197, 893-895. [DOI] [PubMed] [Google Scholar]
- 4.Poste, G. & Fidler, I. J. (1980) Nature 283, 139-146. [DOI] [PubMed] [Google Scholar]
- 5.Price, J. E., Carr, D. & Tarin, D. (1984) J. Natl. Cancer Inst. 73, 1319-1326. [PubMed] [Google Scholar]
- 6.Fidler, I. J. & Hart, I. R. (1982) Science 217, 998-1003. [DOI] [PubMed] [Google Scholar]
- 7.Eccles, S. A., Heckford, S. E. & Alexander, P. (1980) Br. J. Cancer 42, 252-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giavazzi, R., Alessandri, G., Spreafico, F., Garattini, S. & Mantovani, A. (1980) Br. J. Cancer 42, 462-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Milas, L., Peters, L. J. & Ito, H. (1983) Clin. Exp. Metastasis 1, 309-315. [DOI] [PubMed] [Google Scholar]
- 10.Clark, E. A., Golub, T. R., Lander, E. S. & Hynes, R. O. (2000) Nature 406, 532-535. [DOI] [PubMed] [Google Scholar]
- 11.Pozzatti, R., Muschel, R., Williams, J., Padmanabhan, R., Howard, B., Liotta, L. & Khoury, G. (1986) Science 232, 223-227. [DOI] [PubMed] [Google Scholar]
- 12.van `t Veer, L. J., Dai, H., van de Vijver, M. J., He, Y. D., Hart, A. A., Mao, M., Peterse, H. L., van der Kooy, K., Marton, M. J., Witteveen, A. T., et al. (2002) Nature 415, 530-536. [DOI] [PubMed] [Google Scholar]
- 13.van de Vijver, M. J., He, Y. D., van't Veer, L. J., Dai, H., Hart, A. A., Voskuil, D. W., Schreiber, G. J., Peterse, J. L., Roberts, C., Marton, M. J., et al. (2002) N. Engl. J. Med. 347, 1999-2009. [DOI] [PubMed] [Google Scholar]
- 14.Bernards, R. & Weinberg, R. A. (2002) Nature 418, 823. [DOI] [PubMed] [Google Scholar]
- 15.Yang, Y. H., Dudoit, S., Luu, P., Lin, D. M., Peng, V., Ngai, J. & Speed, T. P. (2002) Nucleic Acids Res. 30, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hughes, T. R., Marton, M. J., Jones, A. R., Roberts, C. J., Stoughton, R., Armour, C. D., Bennett, H. A., Coffey, E., Dai, H., He, Y. D., et al. (2000) Cell 102, 109-126. [DOI] [PubMed] [Google Scholar]
- 17.Sturn, A., Quackenbush, J. & Trajanoski, Z. (2002) Bioinformatics 18, 207-208. [DOI] [PubMed] [Google Scholar]
- 18.Brazma, A., Hingamp, P., Quackenbush, J., Sherlock, G., Spellman, P., Stoeckert, C., Aach, J., Ansorge, W., Ball, C. A., Causton, H. C., et al. (2001) Nat. Genet. 29, 365-371. [DOI] [PubMed] [Google Scholar]
- 19.Gruvberger, S., Ringner, M., Chen, Y., Panavally, S., Saal, L. H., Borg, A., Ferno, M., Peterson, C. & Meltzer, P. S. (2001) Cancer Res. 61, 5979-5984. [PubMed] [Google Scholar]
- 20.Perou, C. M., Sørlie, T., Eisen, M. B., van de Rijn, M., Jeffrey, S. S., Rees, C. A., Pollack, J. R., Ross, D. T., Johnsen, H., Akslen, L. A., et al. (2000) Nature 406, 747-752. [DOI] [PubMed] [Google Scholar]
- 21.Radinsky, R. (1995) Cancer Metastasis Rev. 14, 323-338. [DOI] [PubMed] [Google Scholar]
- 22.Roberts, D. D. (1996) FASEB J. 10, 1183-1191. [PubMed] [Google Scholar]
- 23.del Peso, L., Hernandez-Alcoceba, R., Embade, N., Carnero, A., Esteve, P., Paje, C. & Lacal, J. C. (1997) Oncogene 15, 3047-3057. [DOI] [PubMed] [Google Scholar]
- 24.Kang, Y., Siegel, P. M., Shu, W., Drobnjak, M., Kakonen, S. M., Cordon-Cardo, C., Guise, T. A. & Massague, J. (2003) Cancer Cell 3, 537-549. [DOI] [PubMed] [Google Scholar]
- 25.van't Veer, L. J. & Weigelt, B. (2003) Nat. Med. 9, 999-1000. [DOI] [PubMed] [Google Scholar]
- 26.Ramaswamy, S., Ross, K. N., Lander, E. S. & Golub, T. R. (2003) Nat. Genet. 33, 49-54. [DOI] [PubMed] [Google Scholar]
- 27.Lønning, P. E., Sorlie, T., Perou, C. M., Brown, P. O., Botstein, D. & Borresen-Dale, A. L. (2001) Endocr. Relat. Cancer 8, 259-263. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.