Abstract
Echoviruses have been implicated in multiple human disease syndromes, including aseptic meningitis, paralysis, and heart disease, but no animal model is available for studying the pathogenesis of infection. Production of human integrin very late antigen 2, a receptor for echovirus type 1, in transgenic mice conferred susceptibility to viral infection. Intracerebral inoculation of newborn transgenic mice with echovirus leads to paralysis and wasting. No disease was observed in infected nontransgenic mice. In paralyzed mice significant damage was observed in the outer layers of the cerebrum, and numerous condensed neuronal nuclei were present. In contrast, intracerebral inoculation of adolescent (3- to 4-week-old) transgenic mice with echovirus type 1 did not lead to paralysis but an acute wasting phenotype and myocarditis. These findings establish human very late antigen 2 transgenic mice as a model for echovirus pathogenesis.
Echoviruses, like polioviruses and coxsackieviruses, are nonenveloped RNA viruses that are members of the Enterovirus group of the family Picornaviridae. It is estimated that 10-15 million illnesses are caused each year by nonpolio enteroviruses in the United States (1). Although the vast majority of these illnesses are not serious, a wide range of potentially fatal human diseases, including aseptic meningitis, encephalitis, paralysis, type I diabetes mellitus, and myocarditis, have been associated with echovirus infections (2).
Members of the enteric cytopathogenic human orphan group of viruses (echoviruses) were initially distinguished from coxsackieviruses by their inability to replicate and cause disease in newborn mice (3). Because a small animal model is not available, much of what is known about echovirus pathogenesis is inferred from epidemiological surveillance studies and mouse models for coxsackievirus and poliovirus pathogenesis. As is the case for poliovirus, the cellular receptor is most likely a major determinant of the restricted host range of echoviruses (4-7). A receptor molecule that is both sufficient and necessary to mediate virus binding and entry has been identified for only 1 of the 31 echovirus serotypes, echovirus type 1 (EV1) (8). A cellular receptor for EV1 is human very late antigen 2 (VLA-2), a heterodimer comprising one α2 subunit and one β1 subunit (integrin α2/β1). EV1 interacts with the collagen binding I domain of human α2 integrin subunits (9). Although EV1 binding and entry are mediated solely by the α2 subunit, association of an α2 subunit with a β1 subunit is required for proper transport of α2/β1 integrin to the cell surface (10). Production of the α2 polypeptide in rodent cells is sufficient to render these cells susceptible to infection by EV1, indicating that human-α2/mouse-β1 heterodimers can serve as functional EV1 receptors (8, 11). Widespread cell and tissue synthesis of integrin α2/β1 is consistent with the hypothesis that many target organs are involved in EV1 pathogenesis (12).
Transgenic mice that produce human cell receptors for infectious agents have advanced our understanding of the pathogenesis of poliovirus, measles virus, and Listeria monocytogenes (5, 6, 13, 14). The identification of VLA-2 as an echovirus receptor makes possible the establishment of a mouse model to study echovirus pathogenesis. Transgenic mice were produced by using cloned DNAs encoding VLA-2 subunits under the control of a broadly acting human β-actin promoter. VLA-2 transgenic mice express α2 and β1 transcripts in a wide range of tissues. Primary cell lines derived from heart, lung, and kidneys of VLA-2 transgenic mice are susceptible to EV1 infection. EV1 replication was detected in the central nervous system (CNS) of neonates and heart tissue of adolescent mice after intracerebral (i.c.) or i.p. inoculation. EV1 infection of newborn mice led to hindlimb paralysis, and histopathology was observed in the cerebral cortex of paralyzed mice. In contrast, adolescent mice infected with EV1 do not display neuropathology or paralytic disease but develop disease of the heart that is characterized by cellular infiltrates and destruction of myocytes. VLA-2 transgenic mice provide the first convenient animal model for studying the pathogenesis of a prototype echovirus strain.
Methods
Mice. DNAs encoding human α2 and β1 subunits of VLA-2 were produced by PCR from cellular mRNA as described (11). β1 and α2 DNAs were digested with BamHI and XbaI, respectively, and were cloned separately into a mammalian expression vector designated pBAP (kindly provided by F. Constantini, Columbia University College of Physicians and Surgeons, New York) containing a 3-kb human β-actin promoter (15). After digestion with ClaI, DNAs containing a human β-actin promoter, simian virus 40 poly(A) signal, and either the α2 or β1 cDNAs were purified and microinjected into fertilized (C57BL6/J × CBA/J) F2 eggs. Southern blot analysis was used to identify founder mice and subsequent progeny. Transgenic α2/β1 mice were produced by crossing the offspring of α2 founder #20 and β1 founder #23.
DNA and RNA Analysis. To identify the transgene status of founder mice and their progeny, genomic DNA was isolated from tail samples, fractionated by electrophoresis in a 1% agarose gel, and transferred to a nylon-based membrane; the human α2 and β1 transgenes were detected by using randomly primed32P-labeled α2 and β1 DNA.
Detection of transgene mRNAs was performed by using Northern blot hybridization. Total RNA was isolated with TRIzol Reagent (Invitrogen) from homogenized tissues immediately after dissection. About 15 μg of total RNA was fractionated by electrophoresis in a 1% formamide agarose gel and the RNAs were transferred to a solid-support nylon-based membrane. Randomly primed32P-labeled α2 and β1 cDNA fragments were used to detect transgene mRNAs.
Virus. Echovirus type 1 Farouk strain (EV1) was obtained from the American Type Culture Collection and propagated in monolayers of HeLa S3 cells. Virus was concentrated by centrifugation for 3.5 h at 27,000 rpm in a SW28 rotor at 8°C. Pelleted virus was resuspended overnight at 25°C in 100 mM NaCl/10 mm Tris (pH 8)/1 mM EDTA/0.5% SDS. Resuspended virus was dialyzed against PBS (pH 7.4) overnight at 4°C. Newborn (1- to 2-day-old) or adolescent (3- to 4-week-old) mice were inoculated i.c. or i.p. with 109 plaque-forming units (PFU) of EV1.
To determine virus titers in mouse tissues at various times after infection, mice were killed every 24 h and dissected organs were washed in PBS (pH 7.4), weighed, snap-frozen in liquid N2, and stored at -80°C. Tissues were homogenized in 700 μl of PBS, subjected to three freeze-thaw cycles, and centrifuged to remove cellular debris. Titers of EV1 in tissue supernatants were determined by plaque assay on HeLa cell monolayers. At least three mice were infected for each time point.
Histology and in Situ Analysis. Killed mice were first perfused with PBS and then perfused with freshly prepared 4% paraformaldehyde. Excised organs were stored in 4% paraformaldehyde for 1-7 days at 4°C and either processed for paraffin sectioning or frozen in O.C.T. compound (Tissue-Tek, Torrance, CA) on dry ice. Paraffin sections of 8 μm in thickness were stained by using hematoxylin/eosin. Digoxigenin-labeled RNA probes were used to detect EV1 (+) and (-) strand RNA in tissue sections. EV1-specific hybridization probes were produced by RT-PCR amplification of a 320-bp DNA fragment corresponding to position 3303-3622 of the EV1 genome. The blunt-ended DNA product was cloned in both sense and antisense orientations in a pBluescript II SK phagemid linearized with SmaI. Sense or antisense RNA was synthesized in vitro from a BamHI-linearized fragment by using bacteriophage T7 RNA polymerase. Digoxigenin-UTP was randomly incorporated into each hybridization probe during RNA synthesis. Digoxigenin labeled antisense RNA synthesized from pSPT18-Neo was used as a negative control hybridization probe. In situ hybridization analysis was carried out on frozen sections of heart tissue and paraffin embedded brain tissue. Digoxigenin-labeled hybridization probes were diluted to a concentration of 100 ng/ml in hybridization solution containing 50% formamide. Tissue sections were hybridized with digoxigenin-labeled RNA probes for 10 min at 65°C and then overnight at 42°C. After washing tissue sections, hybridized probes were detected by using a DIG Nucleic Acid Detection Kit (Roche Molecular Biochemicals, Mannheim, Germany) according to the manufacturer's instructions.
Results
Production and Characterization of VLA-2 Transgenic Mice. To approximate the widespread expression of VLA-2 observed in human tissues, we placed human α2 and β1 transgenes under the control of a broadly acting human β-actin promoter sequence (Fig. 1A). Introduction of pBAP-α2 DNA alone or with pBAP-β1 DNAs into mouse L cells rendered them susceptible to infection with EV1 (not shown), indicating that these plasmids encode functional EV1 receptor molecules.
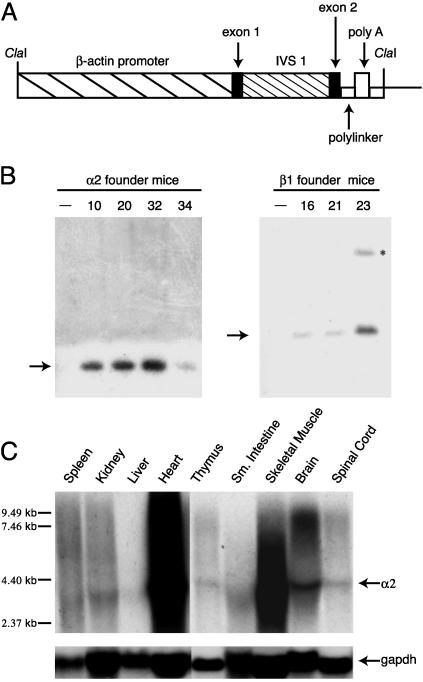
Fig. 1.
Generation and characterization of human α2 and β1 integrin transgenic mice. (A) Schematic diagram of plasmid pBAP consisting of 3 kb of human β-actin regulatory sequence, noncoding exon 1, intron 1, exon 2, and a polylinker into which α2 and β1 cDNAs were inserted. The polyadenylation signal is from simian virus 40. IVS, intervening sequence. (B) Southern blot hybridization analysis of genomic DNA from α2 (Left) and β1 (Right) transgenic founder mice. The hybridization probe was a HindIII fragment of α2 DNA (Left)ora BamHI fragment of β1 DNA (Right). Transgenic founder number is at top of each lane. Nontransgenic litter mates designated by -. Arrows indicate DNA fragments detected by α2 or β1 probes. Asterisk indicates incomplete digestion product. (C) Northern blot hybridization analysis of human α2 mRNA from mouse tissues of founder #20 offspring. The hybridization probe was a32P-labeled α2 (Top) or gapdh (Bottom) cDNA fragment. Arrows to the right of blots indicate α2 and gapdh mRNA. Molecular weight markers are to the left of the blot.
We identified four α2 and three β1 founder mice from independent microinjections of pBAP-α2 or pBAP-β1 DNAs into fertilized F2 eggs. Transgenic founders identified by Southern blot analysis of genomic tail DNA were mated with inbred CBA/J mice (Fig. 1B).
Using Northern blot analysis, we observed widespread expression of a 3.5-kb α2 transcript (Fig. 1C) and a 2.4-kb β1 transcript (not shown) in organs of all transgenic founder progeny, with the highest levels in heart, skeletal muscle, and brain (Fig. 1C). Transgene mRNAs were not detected in nontransgenic tissues. Levels of human α2 and β1 proteins in tissue homogenates and in cell lines established from transgenic organs correlated with mRNA levels. Cell lines established from α2 and α2β1 transgenic organs were susceptible to EV1 infection. Progeny of founder lines that expressed the highest levels of α2 and/or β1 transgene mRNA and protein were used in subsequent infection studies.
Age-Dependent Susceptibility of Transgenic Mice to EV1-Induced Disease. Newborn α2/β1 and α2 transgenic mice were inoculated i.c. with 109 PFU of EV1 to determine their susceptibility to infection (Table 1). Whereas no clinical symptoms were observed in nontransgenic mice when inoculated i.c. with 109 PFU of EV1, infected α2/β1 or α2 transgenic mice developed spastic hindlimb paralysis and failed to thrive (Table 1). We also observed wasting in some transgenic mice that were not paralyzed after infection. Paralysis was initially observed 3-4 days after infection and was usually fatal. No clinical symptoms were observed in either transgenic or nontransgenic mice when inoculated with 107 PFU of EV1 (not shown). Intraperitoneal inoculation of newborn transgenic mice did not lead to paralysis but wasting. Virus was detected by in situ hybridization in the CNS of mice inoculated i.p., indicating that EV1 is neurotropic in this transgenic mouse line (not shown).
Table 1. Age-dependent susceptibility to EV1-induced disease.
| Genotype | Age | n | Paralyzed | Wasting |
|---|---|---|---|---|
| α2/β1 | 24–48 h | 72 | 46/72 | 12/72 |
| α2 | 24–48 h | 52 | 12/52 | 14/52 |
| –/– | 24–48 h | 28 | 0/28 | 0/28 |
| α2/β1 | 3–4 wk | 81 | 0/81 | 26/81 |
| α2 | 3–4 wk | 107 | 0/107 | 41/107 |
| –/– | 3–4 wk | 42 | 0/42 | 0/42 |
Newborn and adolescent mice were inoculated i.c. with 109 PFU of EV1. n, number of mice infected.
Previous studies have shown that immunocompetent adult mice are not susceptible to CNS disease caused by measles virus or reovirus (14, 16). To determine whether susceptibility to EV1-induced paralysis was age-dependent, adolescent (3- to 4-week-old) and adult (6- to 8-week-old) mice were inoculated i.c. with 109 PFU of EV1. Paralysis was not detected in any of the adolescent or adult mice infected with EV1, but some adolescent mice at 2-6 days after infection did develop a fatal wasting disease characterized by ruffled fur, hunched back, and weight loss (Table 1). None of the nontransgenic mice infected with EV1 showed any signs of disease (Table 1). Adolescent transgenic mice were also susceptible to EV1-induced disease after i.p. inoculation (not shown).
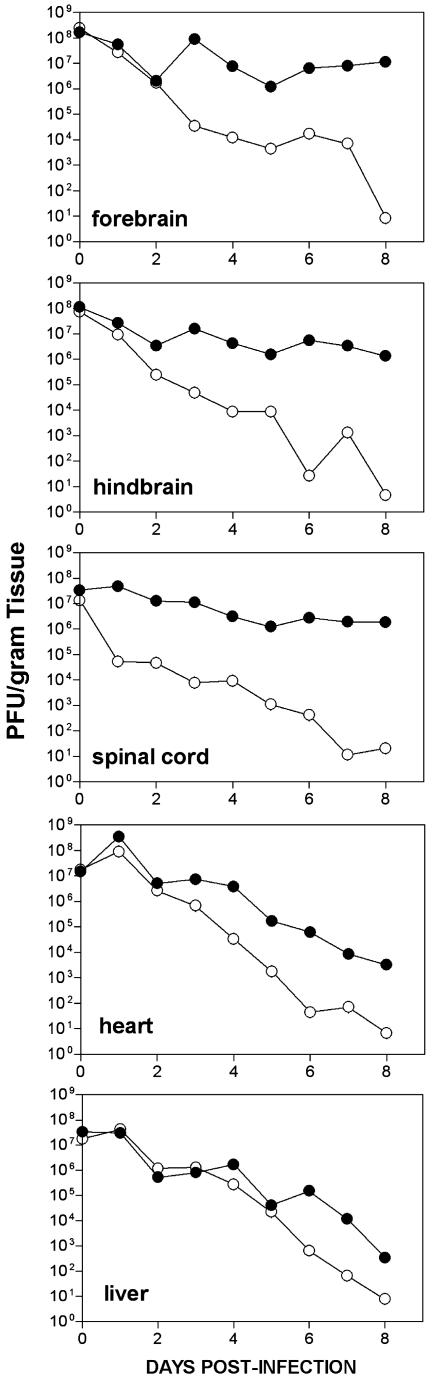
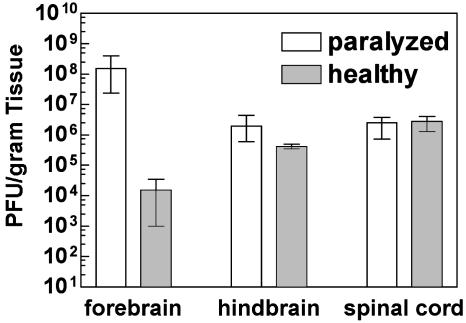
EV1 Replication in Transgenic Mice. To determine whether EV1 replication correlated with disease symptoms, virus titers in individual organs were measured at different times after infection. After i.c. inoculation of 1- to 2-day-old mice, virus titers were >105 PFU/g of tissue in the CNS of α2β1 transgenic mice (Fig. 2). At late times after infection, viral titers in transgenic CNS tissues were 103- to 106-fold higher than in nontransgenic tissues. Although little difference occurred in viral titers in hindbrains and spinal cords of healthy and paralyzed transgenic mice, viral titers in the forebrains of paralyzed mice were at least 103-fold higher than in healthy mice at the same time after infection (Fig. 3). Viral titers in heart, liver (Fig. 2), kidney, lung, spleen, and small intestine (not shown) of transgenic mice decreased over time, indicating that little or no viral replication occurred in these tissues. Virus was nearly cleared by 8 days after infection in all nontransgenic mouse tissues we examined (Fig. 2).
Fig. 2.
Time course of virus replication in organs of newborn mice infected intracerebrally with EV1. At the times indicated, mice were killed and viral titers in organs of α2/β1 (•) or nontransgenic (○) mice were determined by plaque assay of tissue homogenates. Each time point is a geometric mean of three or more samples.
Fig. 3.
Virus titers in CNS tissues of healthy and paralyzed α2/β1 transgenic mice 8 days after infection. Viral titers were determined by plaque assay. Geometric mean for each data set was calculated by using three or more samples. Maximum and minimum error bars are shown.
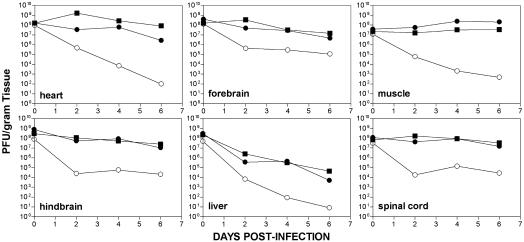
After i.c. inoculation of adolescent mice, EV1 titers rose slightly over time in heart and skeletal muscle, suggesting that viral replication occurs in these tissues (Fig. 4). Virus in the brain and spinal cord was maintained at or near input levels, whereas titers in spleen, lung, kidney, and small intestine decreased significantly by day 6 after infection (Fig. 4 and not shown). Titers decreased substantially over time in all tissues that we examined from adolescent nontransgenic mice (Fig. 4).
Fig. 4.
Time course of virus replication in organs of adolescent mice infected intracerebrally with EV1. Viral titers in organ homogenates of α2/β1 transgenic (•), α2 transgenic (▪), and nontransgenic (○) mice were determined by plaque assay. Each time point is a geometric mean of three or more samples.
Virus recovered from the brain or heart of sick mice did not differ in virulence or tropism (not shown). These findings indicate that mouse adapted variants of EV1 were not selected in transgenic mice.
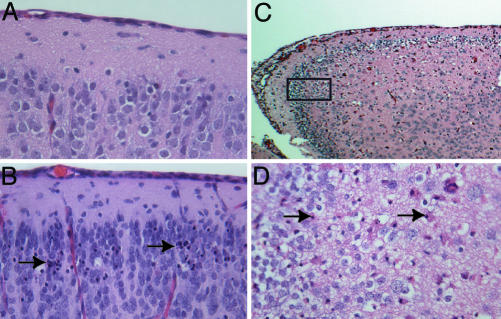
Neuropathology Associated with EV1 Infection of Newborn Transgenic Mice. The results described above suggest that paralysis of newborn transgenic mice may be a consequence of virus replication in the CNS. Histopathological examination of newborn transgenic mice that developed paralysis after i.c. inoculation of EV1 revealed abnormalities in the cerebral cortex (Fig. 5 B-D). Vacuolization and necrotic cells of neuronal morphology were observed in a region of the cortex near the hippocampus. The basophilic cytoplasm and nuclei of neuronal cells are normally stained blue with hematoxylin/eosin, but many neurons with condensed nuclear material and eosinophilic staining cytoplasm were observed in brains of paralyzed mice. Serial sectioning of embedded brain tissue was used to localize the most prominent areas of necrosis and vacuolization to the outer granular and pyramidal cell layers of the retrosplenial and motor cortices. Isolated examples of necrotic cells in other areas of the cerebral cortex and hindbrain of both healthy and sick animals were occasionally observed. Sick animals also had mild inflammation of the pia matter, but this finding was not as consistent or impressive as the necrotic cellular lesions in the cerebral cortex. We detected no abnormal lesions in the brains of infected newborn nontransgenic mice (Fig. 5A).
Fig. 5.
Neuropathology associated with EV1 infection in transgenic mice. (A-D) Hematoxylin/eosin-stained tissue sections. (A) Brain of nontransgenic mouse 4 days after infection (×40). (B) Brain of α2/β1 transgenic mouse 4 days after infection (×40). (C and D) Brain of α2 transgenic mouse 7 days after infection ×10 (C) and ×40 (D). Boxed area in C designates the area shown at higher magnification in D. Arrows in B and D indicate necrotic neurons.
Adolescent transgenic mice did not develop paralytic disease, despite the presence of high viral titers in the CNS. No histological abnormalities were observed in brain and spinal cord of such animals (not shown).
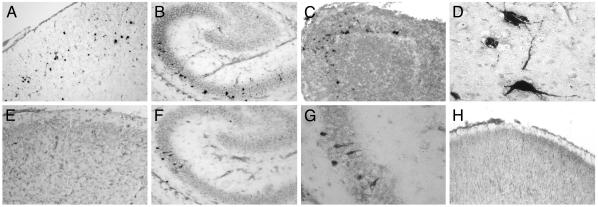
In situ hybridization was performed to determine whether sites of histopathology and virus replication coincided. Viral RNA was consistently detected in the same general area of the cerebral cortex as where histopathology was observed (Fig. 6 A and E). Viral RNA was also detected in regions of the brain where no pathology was observed, including the hippocampus (Fig. 6 B, F, and G), olfactory bulb (Fig. 6C), brainstem (Fig. 6D), and spinal cord (not shown). Viral RNA was not detected when tissues from infected nontransgenic mice were examined by using probes specific to viral RNA, or when a control hybridization probe was used (Fig. 6H and not shown).
Fig. 6.
Detection of EV1 RNA in brain-tissue sections of infected newborn transgenic mice by in situ hybridization analysis. (A and E) Detection of (+) strand (A) or (-) strand (E) viral RNA in the cerebral cortex of an α2/β1 transgenic mouse day 3 after infection (×10). (B, F, and G) Detection of (+) strand (B) and (-) strand viral RNA (F and G) in the hippocampus of an α2/β1 transgenic mouse day 4 after infection. [Original magnification, ×10 (B and F) and ×40 (G)]. (C) Detection of (+) strand viral RNA in the olfactory bulb of an α2/β1 transgenic mouse on day 5 after infection (×10). (D) Detection of (+) strand viral RNA in brainstem neurons of an α2/β1 transgenic mouse day 3 after infection (×40). (H) Cerebral cortex of a nontransgenic mouse day 3 after infection hybridized with digoxigenin-labeled (-) strand viral RNA (×10).
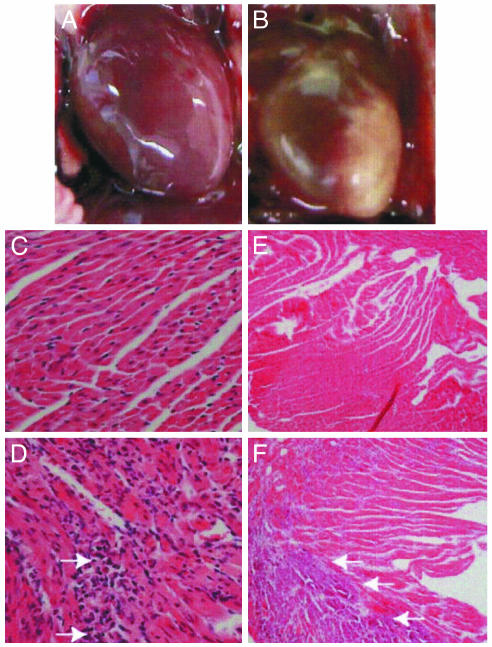
Cardiopathology Associated with EV1 Infection of Adolescent Transgenic Mice. High viral titers were detected in heart tissue of sick transgenic mice, and, on examination of these mice, significant necrosis of heart tissue of both α2 and α2/β1 mice was found (Fig. 7B and not shown). Lesions appeared to be limited to the left ventricular wall. Heart tissue of infected nontransgenic mice appeared normal (Fig. 7A).
Fig. 7.
Cardiopathology associated with EV1 infection of adolescent transgenic mice. (A) Nontransgenic heart 6 days after infection. (B) Transgenic heart 6 days after infection. (C-F) Hematoxylin/eosin-stained heart-tissue sections. (C and D) Nontransgenic (C) and transgenic (D) heart tissue sections at day 2 after infection (×40). Foci of inflammation are noted with arrows. (E and F) Nontransgenic (E) and transgenic (F) heart tissue sections at day 6 after infection (×10). Arrows indicate areas of significant necrosis.
When heart tissue was examined for histopathology, damage to the myocardium progressed with time after infection. Foci of cellular infiltrate in the myocardium of sick animals 2 days after infection were detected (Fig. 7D), and by day 6 widespread necrosis involving primarily the left ventricular wall and intraventricular septum had occurred (Fig. 7F). The morphology of necrotic areas was characteristic of calcification. No inflammation of either the pericardium or endocardium was noted in animals with significant myocarditis. Abnormal pathology was not observed in pancreas, liver, lung, or skeletal muscle of mice with myocarditis. None of the nontransgenic heart tissues examined showed any signs of myocarditis (Fig. 7 C and E). No signs of myocarditis were observed in EV1-infected transgenic neonates (not shown).
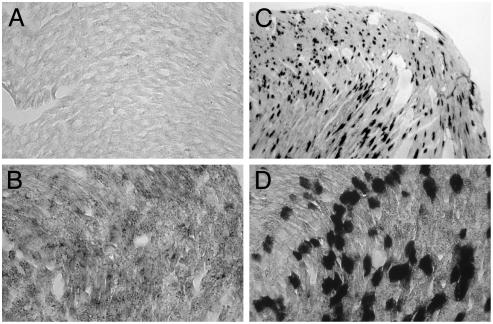
Sites of viral replication were detected by in situ hybridization in the hearts of animals 2 days after infection. Both (-) and (+) strand viral RNAs were detected in the myocardium of α2β1 mice (Fig. 8 B-D). Although the areas of greatest tissue damage were limited to the left ventricular wall and the intraventricular septum, foci of viral RNA were detected in both ventricular walls and the intraventricular septum. Signals for detection of (-) strand RNA were less intense, presumably because the ratio of (-)to(+) strands is as low as 1:70 during picornavirus infections (17). We detected no hybridization by using control RNA probes. Heart tissue of nontransgenic mice was negative for viral RNA at 2 days after infection (Fig. 8A). Viral RNA from the inoculum was not detected in heart tissue of transgenic mice killed 1 h after infection (not shown). These observations indicate that EV1 infects and replicates in susceptible cells before viral RNA and tissue damage can be detected.
Fig. 8.
Detection of viral RNA in infected transgenic heart tissue by In situ hybridization analysis. (A) Nontransgenic heart tissue section 2 days after infection hybridized with digoxigenin-labeled (+) strand viral RNA. (B-D) Detection of (-) strand (B) and (+) strand (C and D) viral RNA in transgenic heart tissue sections at day 2 after infection. [Original magnifications, ×4(C) and ×40 (A, B, and D).]
Discussion
Although echoviruses were so named because the viruses were not associated with disease, they are now known to cause significant worldwide morbidity and mortality (3). Little is known about how echoviruses cause disease. To address this shortcoming, we have developed a transgenic mouse model to study the pathogenesis of a prototype echovirus strain, EV1. Transgenic mice that produce human α2/β1 integrin or the α2 subunit alone are susceptible to EV1 infection and display significant age-dependent pathology.
Production of α2 and β1 alone or together in transgenic mice had no adverse or noticeable effects. As expected, transcriptional regulation of α2 and β1 transgenes by a human β-actin promoter lead to expression in many tissues, mimicking the widespread distribution of VLA-2 in humans. The level of stable transgene mRNA varied among different tissues. Highest levels of transgene mRNA were detected in heart, skeletal muscle, and brain. As observed in transgenic mouse models of measles and poliomyelitis, transgene expression levels and sites of expression are major factors in determining disease progression and outcome (4-6, 14, 18). Identification of the cell types that produce human α2 and β1 integrins should provide a better understanding of EV1 pathogenesis in this mouse model.
The virus strain, mouse strain, immune status, and age at time of infection are critical determinants of coxsackie B virus disease in mice (19-21). The prototype Farouk strain of EV1 used in this report is the best-studied EV1 strain. EV1-Farouk was isolated from an asymptomatic patient, and its virulence may be attenuated. This property may explain why high levels of virus are needed to cause disease in α2 or α2/β1 transgenic mice. We are currently testing the virulence of other clinical isolates of EV1 in VLA-2 mice to address this possibility. The genetic background of the C57Bl6/J × CBA/J transgenic mice used in this study may also play a role in determining susceptibility to infection by EV1.
Although EV1 is not often associated with neurovirulence in humans, it does harbor a 5′-oligopyrimidine tract sequence present in serotypes that frequently cause CNS infections (22). Intracerebral infection of neonatal mice with 109 PFU of EV1 led to spastic hindlimb paralysis similar to that reported in mice infected with coxsackie B virus (23). Consistent with this clinical picture, neuronal damage in the motor cortices was observed in paralyzed transgenic mice. The cause of neuronal death is not known. Because we detected EV1 RNA in neurons, the cytopathic effect could be a consequence of lytic EV1 infection. EV1 infection may induce the synthesis of cytokines that are deleterious to immature neurons of neonates or as in poliovirus infections, EV1 may trigger an apoptotic cascade leading to cell death (24). In contrast to infection of neonates, i.c. inoculation of 3- to 4-week-old mice did not lead to neuropathology and paralysis. The results of epidemiological studies have shown that infants and agammaglobulinemic patients are more susceptible to CNS disease caused by echoviruses (25). Immune status and decreased susceptibility of neurons in older mice may determine protection from CNS disease.
After inoculation with EV1, adolescent transgenic mice do not develop CNS disease but rather fatal acute myocarditis. The cellular infiltrate we detected in the myocardium 2 days after infection may play a role in disease progression. It is not known why adolescent mice but not neonates or adults develop myocarditis. No age-dependent difference in α2 mRNA levels was detected in heart tissue. In mice infected with coxsackievirus B3 the immune system both prevents viral replication and exacerbates myocardial damage (26-28). Age-dependent immune status may be a critical determinant of myocardial disease during EV1 infection of α2 and α2/β1 transgenic mice. Development of myocarditis in these transgenic mice may also depend on host proteins. Coxsackievirus B3 interaction with p56lck and cleavage of dystrophin by the viral 2A proteinase are believed to contribute to disease progression in the heart (29-31). Coxsackie B viruses and EV1 are highly related, suggesting common mechanisms of pathogenesis (32, 33). The development of a transgenic mouse model to study EV1 infection should provide valuable insight into how echoviruses and other enteroviruses cause CNS and heart disease.
Acknowledgments
We thank the Columbia University Cancer Center histology service and transgenic facility for technical help. This work was supported by National Institutes of Health Fellowship AI10238 and National Institutes of Health Grant AI48690.
This paper was submitted directly (Track II) to the PNAS office. Abbreviations: i.c., intracerebral; EV1, echovirus type 1; VLA-2, very late antigen 2; PFU, plaque-forming units.
References
- 1.Strikas, R. A., Anderson, L. J. & Parker, R. A. (1986) J. Infect. Dis. 153, 346-351. [DOI] [PubMed] [Google Scholar]
- 2.Grist, N. R., Bell, E. J. & Assaad, F. (1978) Prog. Med. Virol. 24, 114-157. [PubMed] [Google Scholar]
- 3.Dalldorf, G., Enders, J. F., Hammon, W. M., Sabin, A. B., Syverton, J. T. & Melnick, J. L. (1955) Science 122, 1187-1188.13274079 [Google Scholar]
- 4.Ida-Hosonuma, M., Iwasaki, T., Taya, C., Sato, Y., Li, J., Nagata, N., Yonekawa, H. & Koike, S. (2002) J. Gen. Virol. 83, 1095-1105. [DOI] [PubMed] [Google Scholar]
- 5.Ren, R. B., Costantini, F., Gorgacz, E. J., Lee, J. J. & Racaniello, V. R. (1990) Cell 63, 353-362. [DOI] [PubMed] [Google Scholar]
- 6.Ren, R. & Racaniello, V. R. (1992) J. Virol. 66, 296-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zimmermann, H., Eggers, H. J. & Nelsen-Salz, B. (1997) Virology 233, 149-156. [DOI] [PubMed] [Google Scholar]
- 8.Bergelson, J. M., St. John, N., Kawaguchi, S., Chan, M., Stubdal, H., Modlin, J. & Finberg, R. W. (1993) J. Virol. 67, 6847-6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.King, S. L., Kamata, T., Cunningham, J. A., Emsley, J., Liddington, R. C., Takada, Y. & Bergelson, J. M. (1997) J. Biol. Chem. 272, 28518-28522. [DOI] [PubMed] [Google Scholar]
- 10.Chan, B. M., Matsuura, N., Takada, Y., Zetter, B. R. & Hemler, M. E. (1991) Science 251, 1600-1602. [DOI] [PubMed] [Google Scholar]
- 11.Zhang, S. & Racaniello, V. R. (1997) Virology 235, 293-301. [DOI] [PubMed] [Google Scholar]
- 12.Zutter, M. M. & Santoro, S. A. (1990) Am. J. Pathol. 137, 113-120. [PMC free article] [PubMed] [Google Scholar]
- 13.Lecuit, M., Vandormael-Pournin, S., Lefort, J., Huerre, M., Gounon, P., Dupuy, C., Babinet, C. & Cossart, P. (2001) Science 292, 1722-1725. [DOI] [PubMed] [Google Scholar]
- 14.Oldstone, M. B., Lewicki, H., Thomas, D., Tishon, A., Dales, S., Patterson, J., Manchester, M., Homann, D., Naniche, D. & Holz, A. (1999) Cell 98, 629-640. [DOI] [PubMed] [Google Scholar]
- 15.Gunning, P., Leavitt, J., Muscat, G., Ng, S. Y. & Kedes, L. (1987) Proc. Natl. Acad. Sci. USA 84, 4831-4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mann, M. A., Knipe, D. M., Fischbach, G. D. & Fields, B. N. (2002) Virology 303, 222-231. [DOI] [PubMed] [Google Scholar]
- 17.Novak, J. E. & Kirkegaard, K. (1991) J. Virol. 65, 3384-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rall, G. F., Manchester, M., Daniels, L. R., Callahan, E. M., Belman, A. R. & Oldstone, M. B. (1997) Proc. Natl. Acad. Sci. USA 94, 4659-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khatib, R., Chason, J. L., Silberberg, B. K. & Lerner, A. M. (1980) J. Infect. Dis. 141, 394-403. [DOI] [PubMed] [Google Scholar]
- 20.Tracy, S., Hofling, K., Pirruccello, S., Lane, P. H., Reyna, S. M. & Gauntt, C. J. (2000) J. Med. Virol. 62, 70-81. [DOI] [PubMed] [Google Scholar]
- 21.Woodruff, J. F. (1970) J. Infect. Dis. 121, 164-181. [DOI] [PubMed] [Google Scholar]
- 22.Romero, J. R. & Rotbart, H. A. (1995) J. Virol. 69, 1370-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roberts, G. B. & Boyd, J. F. (1987) J. Infect. 15, 45-56. [DOI] [PubMed] [Google Scholar]
- 24.Girard, S., Couderc, T., Destombes, J., Thiesson, D., Delpeyroux, F. & Blondel, B. (1999) J. Virol. 73, 6066-6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKinney, R. E., Jr., Katz, S. L. & Wilfert, C. M. (1987) Rev. Infect. Dis. 9, 334-356. [DOI] [PubMed] [Google Scholar]
- 26.Gebhard, J. R., Perry, C. M., Harkins, S., Lane, T., Mena, I., Asensio, V. C., Campbell, I. L. & Whitton, J. L. (1998) Am. J. Pathol. 153, 417-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henke, A., Huber, S., Stelzner, A. & Whitton, J. L. (1995) J. Virol. 69, 6720-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horwitz, M. S., La Cava, A., Fine, C., Rodriguez, E., Ilic, A. & Sarvetnick, N. (2000) Nat. Med. 6, 693-697. [DOI] [PubMed] [Google Scholar]
- 29.Badorff, C., Lee, G. H., Lamphear, B. J., Martone, M. E., Campbell, K. P., Rhoads, R. E. & Knowlton, K. U. (1999) Nat. Med. 5, 320-326. [DOI] [PubMed] [Google Scholar]
- 30.Liu, P., Aitken, K., Kong, Y. Y., Opavsky, M. A., Martino, T., Dawood, F., Wen, W. H., Kozieradzki, I., Bachmaier, K., Straus, D., et al. (2000) Nat. Med. 6, 429-434. [DOI] [PubMed] [Google Scholar]
- 31.Xiong, D., Lee, G. H., Badorff, C., Dorner, A., Lee, S., Wolf, P. & Knowlton, K. U. (2002) Nat. Med. 8, 872-877. [DOI] [PubMed] [Google Scholar]
- 32.Huttunen, P., Santti, J., Pulli, T. & Hyypiä, T. (1996) J. Gen. Virol. 77, 715-725. [DOI] [PubMed] [Google Scholar]
- 33.Hyypiä, T., Hovi, T., Knowles, N. J. & Stanway, G. (1997) J. Gen. Virol. 78, 1-11. [DOI] [PubMed] [Google Scholar]