Abstract
Aims
Two single-nucleotide polymorphisms (SNPs), rs1746048 and rs501120, from genome wide association studies of coronary artery disease (CAD) map to chromosome 10q11 ∼80 kb downstream of chemokine CXCL12. Therefore, we examined the relationship between these two SNPs and plasma CXCL12 levels.
Methods and Results
We tested the association of two SNPs with plasma CXCL12 levels in a two-stage study (n= 2939): first in PennCath (n= 1182), a Caucasian, angiographic CAD case–control study, and second in PennCAC (n= 1757), a community-based study of CAD risk factors. Plasma CXCL12 levels increased with age and did not vary by gender. There was no linkage disequilibrium between these two SNPs and SNPs within CXCL12 gene. However, CAD risk alleles at rs1746048 (C allele, P= 0.034; CC 2.33 ± 0.49, CT 2.27 ± 0.46, and TT 2.21 ± 0.52 ng/mL) and rs501120 (T allele, P= 0.041; TT 2.34 ± 0.49, CT 2.28 ± 0.46, and CC 2.23 ± 0.53 ng/mL) were associated with higher plasma levels of CXCL12 in age and gender adjusted models. In Stage 2, we confirmed this association (rs501120, T allele, P= 0.007), and meta-analysis strengthened this finding (n= 2939, P= 6.0 × 10−4). Finally, in exploratory analysis, the rs1746048 risk allele tended to have higher transcript levels of CXCL12 in human natural killer cells and the liver.
Conclusion
Coronary artery disease risk alleles downstream of CXCL12 are associated with plasma protein levels of CXCL12 and appear to be related to CXCL12 transcript levels in two human cell lines. This implicates CXCL12 as potentially causal and supports CXCL12 as a potential therapeutic target for CAD.
Keywords: Myocardial infarction, Cardiovascular genomics, Chemokines, CXCL12, Inflammation
Introduction
Genome-wide association studies (GWAS) have revealed many novel loci associated with coronary artery disease (CAD) and myocardial infarction (MI).1–3 Two highly replicated single-nucleotide polymorphisms (SNPs) [rs1746048, odds ratio (OR) for MI 1.17, P-value3 8.1 × 10−9 and rs501120, OR 1.33, P-value1 7.4 × 10−8] map to chromosome (chr) 10q11 ∼80 kb downstream of chemokine (C-X-C motif) ligand 12 (CXCL12), an inflammatory chemokine expressed in cells of relevance to CAD and MI.4–6 Several replicated GWAS discoveries, like the locus in the CXCL12 region, are in genomic regions distant from the likely causal gene. Although such loci may in fact lie in novel genes (e.g. non-coding RNAs)7 or be in genetic correlation, i.e. linkage disequilibrium (LD), with distant causal genes,8 there is growing evidence that some may contain regulatory sites for disease-related genes up to 1 Mb away either 3′ or 5′.9–11 The chr10q11 CAD/MI GWAS locus lies ∼80 kb away from the CXCL12 gene; therefore, it is not clear whether the GWAS finding specifically relates to CXCL12.
CXCL12 is a biologically plausible gene as it plays a role in recruiting leucocytes in response to vascular injuries12,13 and has been implicated in atherosclerosis in rodent models. There are six known CXCL12 coding splice variants, of which only two isoforms, α (also called isoform-1) and β (also called isoform-2), predominate in the adult human, with the majority in most adult tissues being isoform α.14 CXCL12 is highly expressed in human atherosclerotic plaque4 within endothelial cells and smooth muscle cells.5,15 The receptor for CXCL12, chemokine (C-X-C motif) receptor 4 (CXCR4), is found on macrophages and vascular smooth muscle cells,5 and the α isoform is secreted by platelets and is a potent platelet agonist.4 CXCL12 was recently shown to have enhanced expression on platelets, and platelet-bound CXCL12 correlated with the degree of systemic platelet activation.16 Furthermore, a small human study found plasma CXCL12 levels to be modulated in human CAD.17 A recent report showed that there was an association of rs1746048 with increased carotid intimal–medial thickness.18 These data suggest a relationship between CXCL12 and human atherosclerotic cardiovascular disease; however, whether CXCL12 actions are atheroprotective or atherogenic in humans remain uncertain.
Thus, this study examined the hypothesis that common variation at both of these SNPs at the chr10q11 locus (rs1746048 and rs501120) is associated with plasma CXCL12 levels. Further, we tested the association of one of these SNPs (rs1746048) with CXCL12 mRNA levels and isoform expression in human tissue. Finally, we posited that the direction of association between CAD/MI risk alleles and plasma CXCL12 and tissue transcript levels may provide insight into the biological relationship of CXCL12 with atherosclerosis.
Methods
Clinical study design
PennCath study design, recruitment process, and selection criteria have been reported previously.19,20 Briefly, PennCath is a cross-sectional study of biochemical and genetic CAD risk factors in a consecutive cohort of patients undergoing cardiac catheterization at the University of Pennsylvania in an Institutional Review Board-approved protocol.19,20 All subjects gave written informed consent and the study complies with the Declaration of Helsinki. Enrolment criteria included any clinical indication for cardiac catheterization. Coronary angiograms were scored immediately by the interventional cardiologist performing the procedure.
Stage 1 of this study focused on a subset of European Ancestry PennCath participants who had genotyping and measurement of CXCL12 levels (n= 1182). Participants were categorized for CAD and MI as follows: controls (n = 390) >50 in men and >55 in women, with no angiographic CAD; and CAD cases (n= 792) with angiographic CAD (≥1 vessel, ≥50% stenosis) divided into ‘CAD-MI’ cases with history or presentation with MI (chest pain and EKG changes with elevated cardiac enzymes; n= 406) <65 at the time of first event and ‘CAD-non-MI’ cases with no MI (n= 386).19 Replication in Stage 2 was carried out in Caucasian participants in PennCAC, a community-based study of risk factors for subclinical atherosclerosis, in whom plasma CXCL12 levels were measured and rs501120 genotyping performed (n= 1757). Details of the PennCAC study population appear in Supplementary material online and as published previously.21
Genotyping and single-nucleotide polymorphism selection
Genotyping was performed on Affymetrix 6.0 chip and Illumina ITMAT-Broad-CARE (IBC) candidate gene array22 at the Center for Applied Genomics (CAG) at Children's Hospital of Philadelphia. The Affymetrix 6.0 genotyping, quality control and analysis have been described.3 The IBC array assays ∼49 200 SNPs in ∼2150 candidate genes22 as well as SNPs from contemporary GWAS results and included 38 CXCL12 SNPs designed for cosmopolitan tag-SNP coverage at minor allele frequencies >0.05 with an r2 of at least 0.5 in HapMap populations.22
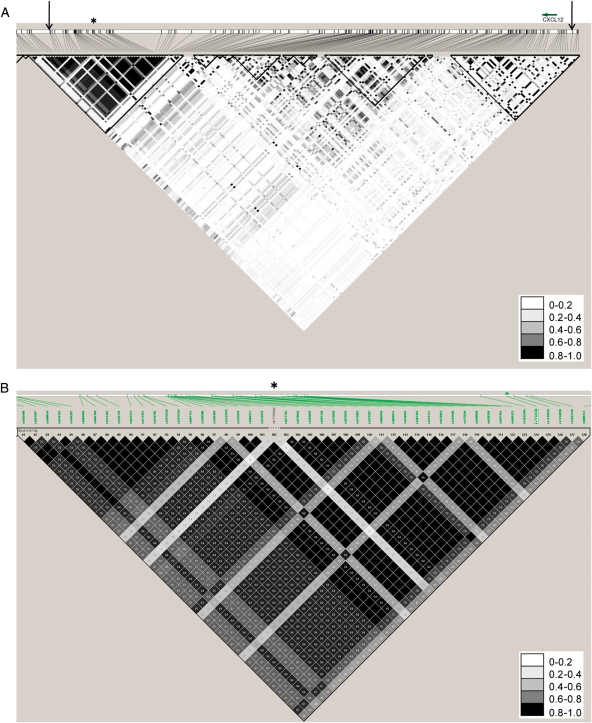
Our primary analysis focused on the association between plasma CXCL12 levels and two SNPs (rs1746048 and rs501120) ∼80 kb downstream of CXCL12 (Figure 1A). There are no other genes for 800 kb downstream (nearest gene ZNF-1) and 1 Mb upstream. These two SNPs were reported previously in the WTCCC and MI-GEN GWAS of CAD/MI, respectively,1,3 and are in almost perfect LD (Figure 1B). Linkage disequilibrium is a measure of correlation between two SNPs which may be transmitted between generations in a large block of DNA called a haplotype. Therefore, to understand whether the SNPs in the GWAS region were related to the CXCL12 gene, we first examined their LD with all the SNPs from 2 kb 3′–5′ of the CXCL12 gene (38 SNPs; see Supplementary material online, Table S1) from the IBC array and Affymetrix 6.0 array genotyped and imputed SNPs. As a descriptive analysis, we examined the correlation of plasma CXCL12 levels with established risk factors for CAD. Replication genotyping of rs501120 was performed on the IBC Version 2 candidate gene array.22 This array contained rs501120 and similar to PennCath, PennCAC had a genotype call rate exceeding 99%. Details of genotyping, quality control, and genotyped and imputed analytic methods appear in Supplementary material online.
Figure 1.
The genomic region analysed surrounding the CXCL12 region. (A) Linkage disequilibrium in PennCath in the CXCL12 region, with arrows demarcating the region from around CXCL12 to the genome-wide association studies region. The asterisk denotes the haplotype block which contains rs1746048 and rs501120. (B) A magnified view of the haplotype block containing rs1746048 and rs501120 from the HapMap CEU population. The asterisk corresponds to the same position as the PennCath linkage disequilibrium plot, and the green arrowhead denotes rs1746048.
Measurement of plasma CXCL12 levels and other biochemical parameters
CXCL12 levels were measured in previously unthawed plasma samples, stored at −80°C, using a commercial, indirect sandwich ELISA (Quantikine Immunoassay; R + D Systems, Minneapolis, MN, USA).17 Blood samples were spun within an hour at 10 000 g for 15 min at 4°C upon collection and then were frozen at −80°C until the CXCL12 ELISA was performed. Immediately after thaw prior to running the assays, all samples had a second spin for 2 min at 4°C at 10 000 g to remove debris before assay. Pooled plasma samples were included on all assay plates. The intra- and inter-assay coefficients of variation were 3.1 and 9.1%, respectively. The specificity of this assay using Western blotting and recombinant proteins confirms that the predominant isoform, isoform-α, but not isoform-β, is detected by this ELISA (personal communication with R&D, Christopher Larson). In both studies, total and high-density lipoprotein (HDL) cholesterol, triglycerides (TG), glucose, and creatinine levels were measured enzymatically on a Cobas Fara II (Roche Diagnostic Systems Inc., NJ, USA). Low-density lipoprotein (LDL) cholesterol was calculated by the Friedewald formula except when TG levels were >400 mg/dL.
Whole-transcript expression analysis of CXCL12 in human tissues
We also explored the association between rs1746048 and expression levels of CXCL12. First, we reviewed the literature for patterns of CXCL12 tissue and cell-specific expression of known isoforms.12 We examined the most appropriate expression data (exon arrays for coverage of specific CXCL12 isoforms) in liver RNA samples (n= 35) (source of secreted protein systemically) and in natural killer (NK) cells (n= 7) (found in atherosclerotic lesions)23 from an ongoing study using Affymetrix Human Exon 1.0 ST Array.24 These tissues express only the CXCL12 α and β isoforms.14 These samples also were genotyped on the Illumina HumanHap550v3 Array which contains rs1746048. The Affymetrix Human Exon Array 1.0 chips were normalized using the methods RMA-sketch, quantile normalization, and median polish as implemented by the apt-probeset-summarize command line program from Affymetrix. Expression values expressed as log-transformed were then analysed for association between rs1746048 genotype and expression of the probesets in CXCL12.
Statistical analysis
The primary goal of the study was to examine the relationship between the two published GWAS CAD/MI SNPs at 10q11 and plasma CXCL12 levels, which were approximately normal in distribution in both PennCath and PennCAC (see Supplementary material online, Figure S1). The directionality of association with CXCL12 levels was unknown a priori so a two-sided P-value of ≤0.05 was considered statistically significant. Because these SNPs were in almost perfect LD (r2 of 1.0 in HapMap and 0.98 in PennCath), we did not correct for multiple testing.25 Given a fixed sample size of 1182, we had ≥80% power (calculated using http://pngu.mgh.harvard.edu/~purcell/gpc/),26 at a significance level of 0.05, to detect variation of 2.0% or greater in plasma CXCL12 levels in our Stage 1 discovery sample.
Linear regression models of SNP associations with CXCL12 levels were built sequentially by first including just SNPs, then adding gender, age at first CAD, and CAD status, and finally also including multiple CAD risk factors including tobacco, hypertension, hyperlipidaemia, and diabetes. Secondary analyses, performed to complement the main analysis, examined the association of plasma CXCL12 with established CAD risk factors using linear regression. Continuous parameters were compared between groups by t-test for normally distributed values. Genotypes were coded as 0, 1, and 2, counting the number of risk alleles, and linear regression was performed in PLINK software27 applying an additive genetic model (trend test), adjusting for covariates as outlined in results tables. For linear regression, the β-coefficient is reported. There was no evidence for population stratification28 using a genomic control measure (λ = 1.009 in PennCath and 1.0008 in PennCAC), and details of this calculation appear in Supplementary material online. Haplotype structure and LD blocks were examined in PennCath using Haploview.29 The degree of LD between the two GWAS SNPs and other SNPs in the CXCL12 locus (±∼2 kb of the gene to the margins of the recombinant hotspots) was estimated in SNAP30 and Haploview.29 Statistical analyses of genotyped SNPs and plasma CXCL12 levels were performed using STATA 10 (College Station, TX, USA) and PLINK v1.06.27
Meta-analysis was conducted using a weighted Z-score method using software package METAL (http://www.sph.umich.edu/csg/abecasis/Metal), which accounts for the direction of association relative to a consistent reference allele. In this method, P-values for each study are first converted to a Z-score. Then, a weighted sum of Z-scores is calculated where each statistic is weighted by the square root of the effective sample size for each study. The resulting sum is divided by the square root of the total effective sample size to obtain an overall Z-statistic, which evaluates the association across different studies. A heterogeneity test applied a one-degree of freedom χ2 test to determine whether observed effect sizes were homogeneous across meta-analysed samples.
For expression data, we explored the association of rs1746048 with groups' mean normalized expression levels (after log transformation) across all probesets specific for either CXCL12-α or CXCL12-β (see Supplementary material online, Figure S2) using ANOVA. In addition, the association of rs1746048 alleles with individual probesets was assessed using Kruskal–Wallis tests, with correction for multiple testing for all probesets (n= 18).
Results
Sample characteristics in PennCath and in PennCAC
The characteristics of Stage 1 PennCath CAD cases (n= 792) as well as controls (n= 390) are shown in Table 1. By design, controls were older than cases and as expected had lower levels of CAD risk factors. The CAD cases were younger (P<0.001) than non-MI cases and differed in their risk factor profile and degrees of angiographic coronary stenosis as expected (Table 1). Table 2 shows demographic characteristics of the Stage 2 replication sample, PennCAC. The sample was a similar age and distribution of CAD risk factors; however, there were more diabetics in the community sample as expected based on the design.
Table 1.
Characteristics of PennCath study sample
| Coronary artery disease (n= 792) | Controls (n= 390) | P-valuea | |
|---|---|---|---|
| Age (years)b | 54 (46–59) | 61 (55–70) | <0.001 |
| Gender (%male) | 68.3% | 46.8% | <0.001 |
| Myocardial infarction (%) | 51.3% | 0 | <0.001 |
| Type 2 diabetes | 23.7% | 11.7% | <0.001 |
| Hypertension | 61.2% | 49.7% | 0.02 |
| Tobacco use | 46.2% | 35.3% | <0.001 |
| Family history | 50.6% | 27.8% | <0.001 |
| BMI | 29 ± 6 | 30 ± 6 | 0.08 |
| Total cholesterol (mg/dL) | 176 ± 39 | 178 ± 44 | 0.06 |
| LDL cholesterol (mg/dL) | 105 ± 33 | 107 ± 37 | 0.04 |
| HDL cholesterol (mg/dL) | 44 ± 12 | 45 ± 14 | <0.001 |
| Triglycerides (mg/dL)b | 133 (88–191) | 96 (68–139) | <0.001 |
| Creatinine (mg/dL) | 1.03 ± 0.4 | 1.01 ± 0.3 | 0.44 |
| Angiographic % stenosis | |||
| LM | 9 (0–11) | 0 (0–0) | 0.8 |
| LAD | 80 (38–100) | 0 (0–10) | 0.4 |
| LCX | 60 (9–97) | 0 (0–11) | 0.8 |
| RCA | 80 (11–100) | 0 (0–10) | 0.7 |
| CXCL12 levels (ng/mL) | 2.33 ± 0.5 | 2.34 ± 0.5 | 0.36 |
aData are displayed as mean ± SD or frequency (%) unless otherwise noted.
bMedian (IQR).
Table 2.
Characteristics of PennCAC study sample
| Caucasian samplea (n= 1757) | |
|---|---|
| Age (years)b | 54 (47–61) |
| Gender (%male) | 65.8% |
| Type 2 diabetes | 44.7% |
| Hypertension | 45.6% |
| Tobacco use | 10.9% |
| BMI | 30.5 ± 5.8 |
| Total cholesterol (mg/dL) | 192 ± 41 |
| LDL cholesterol (mg/dL) | 113 ± 34.5 |
| HDL cholesterol (mg/dL) | 48.7 ± 14.5 |
| Triglycerides (mg/dL)b | 124 (88–178) |
| Creatinine (mg/dL) | 0.99 ± 1.0 |
aData are displayed as mean ± SD or frequency (%) unless otherwise noted.
bMedian (IQR).
Association of CXCL12 levels with demographic and cardiovascular risk factors in PennCath
In PennCath, CXCL12 levels increased with age after adjusting for gender, CAD case–control status, and even other factors, but they did not vary by gender. Otherwise, there were no notable associations with any other demographic or CAD risk factors (see Supplementary material online, Table S2). We did not find association between plasma CXCL12 levels and the presence of CAD in PennCath, but we note that this sample was underpowered to detect modest associations. CXCL12 plasma level association with CAD risk factors was broadly similar in PennCAC (data not shown).
The coronary artery disease/myocardial infarction genome-wide association studies single-nucleotide polymorphisms are not in linkage disequilibrium with single-nucleotide polymorphisms in the CXCL12 gene
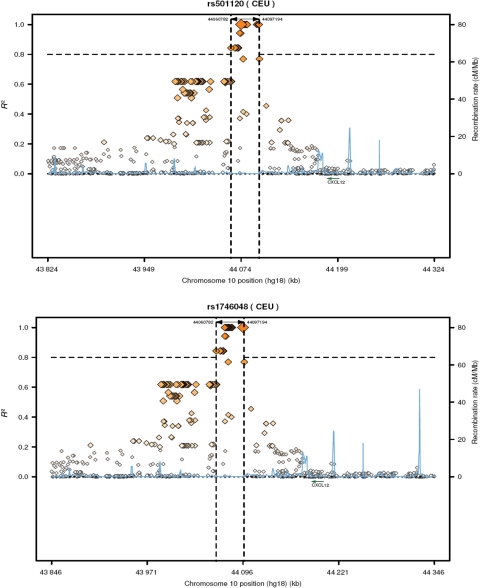
Data from HapMap suggest limited LD between published SNPs in the CAD GWAS region and SNPs in the CXCL12 gene. We examined the LD structure in PennCath (Figure 1A and B) between SNPs in the haplotype block of the GWAS CAD region (positions 44 070 368–44 106 370) including rs1746048 and rs501120, and SNPs within and ± 2 kb 3′–5′ of the CXCL12 gene (38 SNPs; see Supplementary material online, Table S1). The CXCL12 region spanned from positions 44 183 440 to 44 202 567 between the two recombinant hotspots that flank the gene (Figure 2). There was no LD (r2 < 0.1) between SNPs in the CXCL12 region and either rs1746048 or rs501120 in the GWAS region.
Figure 2.
Two published coronary artery disease single-nucleotide polymorphisms and surrounding regions, including two recombinant hotspots around CXCL12. Each single-nucleotide polymorphism is shown with linkage disequilibrium (r2) and recombination rates at these loci using HapMap CEU data; vertical dashed lines demarcate a linkage disequilibrium block in which r2 > 0.8.
Published genome-wide association studies coronary artery disease risk alleles are associated with higher plasma CXCL12 levels
Table 3 shows the crude relationship between the number of alleles at rs1746048 and rs501120 and plasma CXCL12 levels in PennCath and for rs501120 in PennCAC. The second column shows the published risk allele, OR, and P-value for CAD/MI in WTCCC and MI-GEN.1,3 Notably, CAD risk alleles at both SNPs (C allele at rs1746048 and T allele at rs501120) in PennCath and rs501120T in PennCAC were related to higher plasma levels of CXCL12. At both SNPs, there appeared to be a genotype dose–response (Table 3).
Table 3.
Plasma levels of CXCL12 vary by single-nucleotide polymorphisms from genome-wide association studies of coronary artery disease/myocardial infarction in PennCath and PennCAC
| SNP (study, RAF) | Published risk allele, OR for CAD/MI (P-value) | Plasma CXCL12 levels by number of risk alleles in PennCath | Plasma CXCL12 levels by number of risk alleles in PennCAC |
|---|---|---|---|
| rs501120 (WTCCC, 0.87; PennCath, 0.87; PennCAC, 0.86) | T, 1.33 (10−8)a | TTï (n = 874) 2.34 ± 0.49 | TT‖ (n = 993) 2.52 ± 0.57 |
| TCï (n = 271) 2.28 ± 0.46 | TC‖ (n = 345) 2.49 ± 0.54 | ||
| CCï (n = 28) 2.23 ± 0.53 | CC‖ (n = 35) 2.41 ± 0.46 | ||
| rs1746048b (MIGEN, 0.86; PennCath, 0.87; PennCAC, 0.86) | C, 1.17 (10−9)c | CC¶ (n = 874) 2.33 ± 0.49 | N/A |
| CT¶ (n = 276) 2.27 ± 0.46 | N/A | ||
| TT¶ (n = 27) 2.21 ± 0.52 | N/A | ||
RAF, risk allele frequency.
bBased on quantitative trait means for rs1632484, which has r2 = 0.98 in PennCath and r2 = 1.0 in HapMap with rs1746048. The allelic dosage effects are based upon rs1632484 because it was directly genotyped in PennCath, whereas rs501120 was imputed.
cData derived from Myocardial Infarction Genetics Consortium Study.3
ïP for trend = 0.038.
‖P for trend = 0.005.
¶P for trend = 0.036.
Table 4 shows the association between rs1746048 and rs501120 and plasma CXCL12 levels in adjusted models. In PennCath, both published GWAS CAD SNPs (rs1746048, β-coefficient 0.060, P= 0.034; rs501120, β-coefficient 0.055, P= 0.041) were associated with plasma levels of CXCL12 even after cardiovascular risk factor adjustment. In PennCAC, we replicated the association of rs501120 with plasma CXCL12 levels (rs501120, β-coefficient 0.065, P= 0.007) (Table 4). There was no difference with gender (interaction P= 0.68 and 0.66 for rs1746048 and rs501120, respectively). Finally, we performed a weighted Z-score statistic meta-analysis across PennCath and PennCAC with an effective sample size of n= 2939 and demonstrated a stronger combined association (Z = 3.43, P= 6.0 × 10−4; Table 4). Across studies, Cochran's Q-statistic was 0.005 (P= 0.94), indicating a lack of heterogeneity across the two studies. Overall, these data suggest that CAD/MI risk alleles at the chr10q11 (∼80 kb downstream of the gene) are associated with higher plasma CXCL12 levels.
Table 4.
Associations of single-nucleotide polymorphisms from published genome-wide association studies of coronary artery disease/myocardial infarction with plasma CXCL12 levels in PennCath and PennCAC
| SNP | Published risk allele, OR for CAD/MI (P-value) | Linear regression models | PennCath covariate effecta for plasma CXCL12 (P-value) | PennCAC covariate effecta for plasma CXCL12 (P-value) | Meta-analysis Z-score (P-value)b |
|---|---|---|---|---|---|
| rs501120 | T, 1.33 (10−8)c | Model 1d | 0.059 (0.038) | 0.067 (0.007) | — |
| Model 2e | 0.055 (0.041) | 0.065 (0.007) | 3.43 (6.0 × 10−4) | ||
| Model 3f | 0.057 (0.048) | 0.063 (0.008) | — | ||
| rs1746048 | C, 1.17 (10−9)g | Model 1d | 0.061 (0.036) | N/A | N/A |
| Model 2e | 0.060 (0.034) | N/A | N/A | ||
| Model 3f | 0.059 (0.044) | N/A | N/A | ||
aβ and P-value are for additive test based on the risk allele, C for rs1746048 and T for rs501120.
bOn the basis of the sample size of n = 2939, summary statistics derived from model adjusted for age and gender in both PennCath and PennCAC.
dModel 1: unadjusted association.
eModel 2: adjusted for age, gender, and CAD status in PennCath and adjusted for age and gender in PennCAC.
fModel 3: further adjusted for type 2 diabetes, hypertension, and tobacco use.
gData derived from Myocardial Infarction Genetics Consortium Study.3
The genome-wide association studies coronary artery disease risk allele at rs1746048 is associated with higher CXCL12 transcript levels
Tables 5 and 6 and Supplementary material online, Figure S2, suggest that in NK cells and the liver, the rs1746048C risk allele tends to have higher CXCL12 transcript levels. Notably, these tissues express CXCL12 isoforms α and β, but not other splice variants.14 The rs1746048C allele was associated with higher mean expression levels across probesets specific for CXCL12-α, but not β, marginally in NK cells (P= 0.09) and in the liver (P= 0.003) (Table 5; see Supplementary material online, Figure S3A). When we examined individual probes within CXCL12, after correction for number of probes tested (P< 0.05 after Bonferroni correction for 18 probesets), rs1746048C was associated with higher levels of CXCL12-α probeset 3286617 in NK cells [CC normalized expression 3.7 (SD 0.41), CT 1.9 (0.44), TT 1.5 (0); P= 0.002] and CXCL12-α probeset 3286619 in the liver [CC 5.36 (0.77), CT 3.71 (0.63), TT 3.11 (0); P= 0.001] (Table 6; see Supplementary material online, Figure S2). Overall, these two human expression experiments provide preliminary evidence that the rs1746048C CAD/MI risk allele may be associated with higher CXCL12 transcript levels.
Table 5.
The genome-wide association studies coronary artery disease risk allele at rs1746048 is associated with higher CXCL12 transcript levels in human tissue: group means for probes marking isoforms α and β
| SNP | Published risk allele, OR for CAD/MI (P-value) | Expression data seta | Genotype (n) | Normalizedb expression value (SD), α-isoform probes* | P-value, α-isoform* | Normalizedc expression value (SD), β-isoform probes* | P-value, β-isoform* |
|---|---|---|---|---|---|---|---|
| rs1746048 | C, 1.17 (10−9)d | NK | CC (3) | 3.44 (0.59) | 0.09 | 3.12 (0.53) | 0.76 |
| CT (3) | 2.83 (0.56) | 3.32 (0.45) | |||||
| TT (1) | 2.76 (0) | 2.83 (0) | |||||
| Liver | CC (25) | 3.82 (0.27) | 0.003 | 4.82 (0) | 0.17 | ||
| CT (11) | 3.69 (0.24) | 5.0 (0.29) | |||||
| TT (1) | 3.41 (0) | 4.89 (0.31) |
aThese data are derived from seven individuals with NK cell expression on Affymetrix Human ST Exon 1.0 Array and genotyped on the Illumina 550kV3 chip. The liver data are derived from 35 individuals with liver tissue expression on Affymetrix Human ST Exon 1.0 Array and genotyped on the Illumina 550kV3 chip. These tissues express CXCL12 isoforms α and β, but not other splice variants.14
bFour Affymetrix probe IDs are specific to CXCL12-α isoform: 3286616, 3286617, 3286618, and 3286619.
cFive Affymetrix probe IDs are specific to CXCL12-β isoform: 3286605, 3286606, 3286607, 3286608, and 3286609.
dData derived from Myocardial Infarction Genetics Consortium Study.3
*P-value derived from ANOVA testing of normalized transcript expression of isoform group probe means using genotypes 0, 1, and 2 for number of copies of risk alleles.
Table 6.
The genome-wide association studies coronary artery disease risk allele at rs1746048 is associated with higher CXCL12 transcript levels in human tissue: strongest individual probe associations
| SNP | Published risk allele, OR for CAD/MI (P-value) | Affymetrix Human Exon 1.0 ST array probeset ID (type of probe) | Affymetrix Human Exon 1.0 ST array exon cluster IDa | Expression data setb | Genotype (n) | Normalized expression value (SD) | P-value* | P-value** |
|---|---|---|---|---|---|---|---|---|
| rs1746048 | C, 1.17 (10−9)c | 3286617 (core, α) | 605625 | NK | CC (3) | 3.72 (0.41) | 9.5 × 10−5 | 0.002 |
| CT (3) | 1.9 (0.44) | |||||||
| TT (1) | 1.5 (0) | |||||||
| 3296619 (core, α) | 605625 | Liver | CC (25) | 5.36 (0.77) | 4.8 × 10−5 | 0.001 | ||
| CT (11) | 3.71 (0.63) | |||||||
| TT (1) | 3.11 (0) |
aThe exon cluster ID 605625 marks an exon in CXCL12 isoform-α (splice variant 1), but not isoform β.
bThese data are derived from seven individuals with NK cell expression on Affymetrix Human ST Exon 1.0 Array and genotyped on the Illumina 550kV3 chip. The liver data are derived from 35 individuals with liver tissue expression on Affymetrix Human ST Exon 1.0 Array and genotyped on the Illumina 550kV3 chip. These tissues express CXCL12 isoforms α and β, but not other splice variants.14
cData derived from Myocardial Infarction Genetics Consortium Study.3
*P-value derived from non-parametric testing of normalized transcript expression using genotypes 0, 1, and 2 for number of copies of risk alleles.
**P-value after Bonferroni correction (18 probesets tested).
Discussion
Recent GWAS and replication studies1–3 have implicated a locus on chr10q11, marked by rs1746048 and rs501120, in CAD and MI. This region on chr10q11 at 44.2 Mb is ∼80 kb downstream of the closest gene, CXCL12, an inflammatory chemokine which is a plausible biological candidate for the GWAS signal. We demonstrate that the CAD/MI risk alleles at this GWAS locus are associated with higher plasma CXCL12 levels in two independent samples and also appear to relate to higher CXCL12 mRNA in human NK cells and the liver. This implicates CXCL12 as being the causal gene for the replicated GWAS signal at 10q11 and also suggests that CXCL12 may be pro-atherogenic because the CAD/MI risk alleles appear to be related to higher CXCL12 transcript and plasma levels. As SNPs at the GWAS locus are not in LD with SNPs within or around the CXCL12 gene itself, this downstream region may contain distant regulatory elements that affect plasma protein levels likely through transcriptional regulation.
The chr10q11 locus where the CAD/MI signal resides downstream of CXCL12 was discovered and replicated in case–control studies of ∼25 000 participants.2,3 PennCath GWAS CAD data were used for replication in one of these studies.3 In PennCath, the direction and strength of rs1746048 and rs501120 association with CAD/MI was consistent with other studies, but did not reach significance in PennCath alone consistent with lack of power in this small GWAS sample. We emphasize that analysis of chr10q11 SNPs with CAD/MI in PennCath was not a focus of the current paper, given that this relationship has already been shown in three large-scale studies1–3, including one that contains the PennCath CAD data.3
Many novel GWAS findings that are replicated lack functional or mechanistic data linking a gene in the region to disease phenotype.31 In fact, of the novel loci for CAD and MI, about half do not reside within a gene, and a fifth of them occur in gene deserts. Therefore, it is of utmost importance to follow up newly discovered loci to confirm the suspected gene and understand the biological consequences of common variation at these loci.32 Our approach uses one strategy in applying a measureable phenotype in blood, the protein product of the suspected gene, as well as tissue transcript levels, to test whether GWAS variation modulates this intermediate phenotype.33
Other than CXCL12 there are no known genes 3′ downstream for almost 800 kb and 5′ upstream for 1 Mb of the chr10q11 CAD GWAS region. Several genetic and molecular explanations are possible including a GWAS signal that identifies a novel proximate gene or long distance LD with remote causal genes.8 There is also growing evidence that some of these GWAS loci may contain regulatory sites for genes up to 1 Mb away.9–11,34–36 Our analysis of LD structure in this region using PennCath Affymetrix 6.0 genotyped and imputed data and IBC SNP data shows that there is no meaningful LD between SNPs in the GWAS locus and SNPs in or around CXCL12 itself. This argues against LD between the GWAS locus and CXCL12 as the explanation for the CAD/MI signal.
In the absence of apparent LD, our finding that the GWAS SNPs rs1746048 and rs501120 are associated with plasma CXCL12 levels suggests that regulatory sites in the GWAS region may modulate CXCL12 expression. In fact, the CAD risk allele at rs1746048 may be associated with higher CXCL12 mRNA levels in human tissues. The rs1746048 and rs501120 SNPs are within 22 kb of each other, are in strong LD (r2 = 0.98 in PennCath and 1.0 in HapMap) within the same haplotype block. Thus, there may be functional variation with biological effect which remains to be discovered through sequencing within this haplotype block. Indeed, this GWAS block on chr10q11 has several regions which are highly conserved across species. Notably, a search of publicly available databases (TRANSFAC 7.0, www.generegulation.org, http://genome.ucsc.edu/, and PROMO, http://alggen.lsi.upc.es/) for putative transcription factor (TF)-binding sites within the GWAS haplotype block identified several predicted TF-binding sites including HEY1 (chr10:44074342–44075197), NFKB (chr10:44075584–4407632), and FOSL2 (chr10:44094447–44094712). Although, the GWAS SNPs do not lie directly in these predicted TF-binding sites, a number of SNPs in LD with these CAD-associated SNPs fall within potential binding sites. It is possible that haplotypic variation marked by common variation at rs1746048 and rs501120 may affect regional affinity for binding of various TF. Furthermore, rs501120 is within a predicted histone methylation region which might alter transcriptional activity in the region. Direct evidence, however, that variation in the CAD/MI GWAS region regulates TF binding or CXCL12 expression remains to be empirically tested.
CXCL12 is unusual among chemokines in playing a role in developmental signalling and haematopoiesis as well as stem cell mobilization and angiogenesis.15,37–39 CXCL12 is a plausible candidate for human atherosclerosis, in that its protein product is directly involved in trafficking of leucocytes that are involved in the development and complications of atherosclerosis. It is also expressed in cells directly relevant to atherogenesis,40 including endothelial cells, smooth muscle cells,5 leucocytes, and platelets.4,16 Animal and human studies provide conflicting findings, however, with both beneficial and detrimental effects described for CXCL12 depending on vascular phenotype and acuity. For example, in mice with established atherosclerosis, blockade of CCR4, the CXCL12 receptor, accelerated atherosclerosis with expansion of plaque neutrophils and macrophages, suggesting an anti-atherogenic function for CXCL12.6 In contrast, in a chronic model of atherosclerosis, apolipoprotein (apoE)−/− mice were shown to have smaller plaque areas and smooth muscle cell (SMC) content after repopulation with CXCR4−/− bone marrow or transfer of a lentivirus encoding an CXCL12-α antagonist,13 suggesting that CXCL12-α might be pro-atherogenic. Yet, CXCL12 may serve a protective function during the time of acute vascular injury.13,41 Xiao et al.42 examined the relationship of both CXCL12 genotype variation (rs2297630 in intron 3, no LD to rs501120 or rs1746048) and plasma levels of CXCL12/SDF1-α with EPC number and function. They found an association of rs2297630 with EPCs and vascular responses consistent with their hypothesis that CXCL12 regulates EPC in acute vascular injuries. CXCL12 knockout is lethal38 in mice and conditional deletion studies in atherosclerosis have not been reported, thus definitive data in rodent models of atherosclerosis and vascular injury are lacking.
In a single-centre, small human study, plasma levels of CXCL12 were lower in patients with CAD compared with healthy controls also suggesting an anti-atherogenic role.17 In contrast, a G/A variant in the 3′ UTR of CXCL12 (rs1801157, no LD to GWAS SNPs) was associated with lower plasma CXCL12 levels and lower carotid atherosclerosis in a longitudinal cohort of HIV-1-infected patients.43 A more recent study demonstrated that the GWAS rs501120T risk allele was related to higher carotid intimal–medial thickness, but lower plasma CXCL12 levels, in contrast to our findings.18 However, the sample size in this study was smaller (n= 738) than ours and the inverse association with plasma CXCL12 levels was not replicated.
Our findings provide support for CXCL12 as an atherogenic gene in humans. The published CAD/MI ORs for rs1746048 and rs501120 suggest ∼13% increased odds of CAD for the risk alleles.1–3 We found that these same alleles were associated with ∼8% increase in plasma CXCL12 levels and an apparent increase in CXCL12 transcript levels in human tissues. We cannot exclude that CXCL12 induction might be an indirect physiological response to counter atherogenic stimuli regulated by the chr10q11 CAD GWAS SNPs. Although biologically relevant trans-actions of the 10q11 locus across the genome cannot be excluded, we doubt significant cis-effects on genes other than CXCL12; CXCL12 is the closest gene to the GWAS signal and, in available exon expression data sets (www.duke.edu, SNPexpress), the strongest chr10 mRNA transcript association for the GWAS SNPs are with probesets in CXCL12. In fact, consistent with our findings, the Duke data set suggests that the rs1746048C CAD-related allele may be related to increased expression of CXCL12 mRNA in peripheral blood mononuclear cells (β = 30.1, P= 0.0016). Although additional expression data sets are available in the public domain (e.g. http://eqtl.uchicago.edu/cgi-bin/gbrowse/eqtl/), none have adequate coverage of CXCL12 exons and isoforms of relevance.
Our findings should be interpreted cautiously in terms of directionality and causality. Our primary study was cross-sectional in nature, confounded by hospital-based recruitment and relatively small (Stage 1, n= 1182; Stage 2, n= 1757); however, it is the largest effort to date examining chr10q11 variation and plasma CXCL12 levels. We focused primarily on two SNPs in high LD and acknowledge that we did not correct for multiple testing, although this is arguably not needed when r2 (0.98 for these two SNPs) is high.25 We acknowledge that our sample processing for plasma was not ideal for platelet depletion and that residual platelets in storage might have influenced findings. Platelet contamination, however, is likely to be very modest44 and should be random (non-differential), thus tending to bias our results towards the null rather than contribute to false-positive findings. Finally, although we have replicated the plasma CXCL12 association and provided preliminary evidence for association with increased CXCL12 mRNA levels, much work needs to be done to understand the specific mechanisms of actions of the GWAS locus and to define directionality of effect for gain of function and loss of function at the GWAS locus and CXCL12 in CAD and MI.
Supplementary material
Supplementary material is available at European Heart Journal online.
Funding
This work was supported by the following: (i) recruitment of the PennCath study sample was supported by the Cardiovascular Institute of the University of Pennsylvania; (ii) genotyping was performed at the Center for Applied Genomics at the Children's Hospital of Philadelphia and supported by GlaxoSmithKline through an Alternate Drug Discovery Initiative research alliance award (to M.R. and D.J.R.) with the University of Pennsylvania School of Medicine; (iii) N.N.M. is supported by K23 HL-097151-01 and is a recipient of the ACC Young Investigator Award for the Metabolic Syndrome; (iv) M.R. is also supported by RO1 HL-073278 and P50 HL-083799-SCCOR. PennCAC was supported in part by a Clinical and Translational Science Award (UL1RR024134) from the National Center for Research Resources (NCRR) of the National Institute of Health. There are no significant relationships with industry.
Conflict of interest: none declared.
Acknowledgements
We wish to thank Andrew Edmondson and Christine Hinkle for their assistance in genomic analysis and interpretation of transcript results.
References
- 1.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann HE, Barrett JH, Konig IR, Stevens SE, Szymczak S, Tregouet DA, Iles MM, Pahlke F, Pollard H, Lieb W, Cambien F, Fischer M, Ouwehand W, Blankenberg S, Balmforth AJ, Baessler A, Ball SG, Strom TM, Braenne I, Gieger C, Deloukas P, Tobin MD, Ziegler A, Thompson JR, Schunkert H. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kathiresan S, Voight BF, Purcell S, Musunuru K, Ardissino D, Mannucci PM, Anand S, Engert JC, Samani NJ, Schunkert H, Erdmann J, Reilly MP, Rader DJ, Morgan T, Spertus JA, Stoll M, Girelli D, McKeown PP, Patterson CC, Siscovick DS, O'Donnell CJ, Elosua R, Peltonen L, Salomaa V, Schwartz SM, Melander O, Altshuler D, Merlini PA, Berzuini C, Bernardinelli L, Peyvandi F, Tubaro M, Celli P, Ferrario M, Fetiveau R, Marziliano N, Casari G, Galli M, Ribichini F, Rossi M, Bernardi F, Zonzin P, Piazza A, Yee J, Friedlander Y, Marrugat J, Lucas G, Subirana I, Sala J, Ramos R, Meigs JB, Williams G, Nathan DM, MacRae CA, Havulinna AS, Berglund G, Hirschhorn JN, Asselta R, Duga S, Spreafico M, Daly MJ, Nemesh J, Korn JM, McCarroll SA, Surti A, Guiducci C, Gianniny L, Mirel D, Parkin M, Burtt N, Gabriel SB, Thompson JR, Braund PS, Wright BJ, Balmforth AJ, Ball SG, Hall AS, Linsel-Nitschke P, Lieb W, Ziegler A, Konig I, Hengstenberg C, Fischer M, Stark K, Grosshennig A, Preuss M, Wichmann HE, Schreiber S, Ouwehand W, Deloukas P, Scholz M, Cambien F, Li M, Chen Z, Wilensky R, Matthai W, Qasim A, Hakonarson HH, Devaney J, Burnett MS, Pichard AD, Kent KM, Satler L, Lindsay JM, Waksman R, Epstein SE, Scheffold T, Berger K, Huge A, Martinelli N, Olivieri O, Corrocher R, McKeown P, Erdmann E, Konig IR, Holm H, Thorleifsson G, Thorsteinsdottir U, Stefansson K, Do R, Xie C, Siscovick D. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2009;41:334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abi-Younes S, Sauty A, Mach F, Sukhova GK, Libby P, Luster AD. The stromal cell-derived factor-1 chemokine is a potent platelet agonist highly expressed in atherosclerotic plaques. Circ Res. 2000;86:131–138. doi: 10.1161/01.res.86.2.131. [DOI] [PubMed] [Google Scholar]
- 5.Zeiffer U, Schober A, Lietz M, Liehn EA, Erl W, Emans N, Yan ZQ, Weber C. Neointimal smooth muscle cells display a proinflammatory phenotype resulting in increased leukocyte recruitment mediated by P-selectin and chemokines. Circ Res. 2004;94:776–784. doi: 10.1161/01.RES.0000121105.72718.5C. [DOI] [PubMed] [Google Scholar]
- 6.Zernecke A, Bot I, Djalali-Talab Y, Shagdarsuren E, Bidzhekov K, Meiler S, Krohn R, Schober A, Sperandio M, Soehnlein O, Bornemann J, Tacke F, Biessen EA, Weber C. Protective role of CXC receptor 4/CXC ligand 12 unveils the importance of neutrophils in atherosclerosis. Circ Res. 2008;102:209–217. doi: 10.1161/CIRCRESAHA.107.160697. [DOI] [PubMed] [Google Scholar]
- 7.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, Hinds DA, Pennacchio LA, Tybjaerg-Hansen A, Folsom AR, Boerwinkle E, Hobbs HH, Cohen JC. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomlinson I, Webb E, Carvajal-Carmona L, Broderick P, Kemp Z, Spain S, Penegar S, Chandler I, Gorman M, Wood W, Barclay E, Lubbe S, Martin L, Sellick G, Jaeger E, Hubner R, Wild R, Rowan A, Fielding S, Howarth K, Silver A, Atkin W, Muir K, Logan R, Kerr D, Johnstone E, Sieber O, Gray R, Thomas H, Peto J, Cazier JB, Houlston R. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39:984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 9.Lettice LA, Heaney SJ, Purdie LA, Li L, de Beer P, Oostra BA, Goode D, Elgar G, Hill RE, de Graaff E. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 10.Nobrega MA, Ovcharenko I, Afzal V, Rubin EM. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- 11.Qin Y, Kong LK, Poirier C, Truong C, Overbeek PA, Bishop CE. Long-range activation of Sox9 in Odd Sex (Ods) mice. Hum Mol Genet. 2004;13:1213–1218. doi: 10.1093/hmg/ddh141. [DOI] [PubMed] [Google Scholar]
- 12.Shirozu M, Nakano T, Inazawa J, Tashiro K, Tada H, Shinohara T, Honjo T. Structure and chromosomal localization of the human stromal cell-derived factor 1 (SDF1) gene. Genomics. 1995;28:495–500. doi: 10.1006/geno.1995.1180. [DOI] [PubMed] [Google Scholar]
- 13.Zernecke A, Schober A, Bot I, von Hundelshausen P, Liehn EA, Mopps B, Mericskay M, Gierschik P, Biessen EA, Weber C. SDF-1alpha/CXCR4 axis is instrumental in neointimal hyperplasia and recruitment of smooth muscle progenitor cells. Circ Res. 2005;96:784–791. doi: 10.1161/01.RES.0000162100.52009.38. [DOI] [PubMed] [Google Scholar]
- 14.Yu L, Cecil J, Peng SB, Schrementi J, Kovacevic S, Paul D, Su EW, Wang J. Identification and expression of novel isoforms of human stromal cell-derived factor 1. Gene. 2006;374:174–179. doi: 10.1016/j.gene.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Walter DH, Haendeler J, Reinhold J, Rochwalsky U, Seeger F, Honold J, Hoffmann J, Urbich C, Lehmann R, Arenzana-Seisdesdos F, Aicher A, Heeschen C, Fichtlscherer S, Zeiher AM, Dimmeler S. Impaired CXCR4 signaling contributes to the reduced neovascularization capacity of endothelial progenitor cells from patients with coronary artery disease. Circ Res. 2005;97:1142–1151. doi: 10.1161/01.RES.0000193596.94936.2c. [DOI] [PubMed] [Google Scholar]
- 16.Stellos K, Bigalke B, Langer H, Geisler T, Schad A, Kogel A, Pfaff F, Stakos D, Seizer P, Muller I, Htun P, Lindemann S, Gawaz M. Expression of stromal-cell-derived factor-1 on circulating platelets is increased in patients with acute coronary syndrome and correlates with the number of CD34+ progenitor cells. Eur Heart J. 2009;30:584–593. doi: 10.1093/eurheartj/ehn566. [DOI] [PubMed] [Google Scholar]
- 17.Damas JK, Waehre T, Yndestad A, Ueland T, Muller F, Eiken HG, Holm AM, Halvorsen B, Froland SS, Gullestad L, Aukrust P. Stromal cell-derived factor-1alpha in unstable angina: potential antiinflammatory and matrix-stabilizing effects. Circulation. 2002;106:36–42. doi: 10.1161/01.cir.0000020001.09990.90. [DOI] [PubMed] [Google Scholar]
- 18.Kiechl S, Laxton RC, Xiao Q, Hernesniemi JA, Raitakari OT, Kahonen M, Mayosi BM, Jula A, Moilanen L, Willeit J, Watkins H, Samani NJ, Lehtimaki TJ, Keavney B, Xu Q, Ye S. Coronary artery disease-related genetic variant on chromosome 10q11 is associated with carotid intima-media thickness and atherosclerosis. Arterioscler Thromb Vasc Biol. 2010;30:2678–2683. doi: 10.1161/ATVBAHA.110.213785. [DOI] [PubMed] [Google Scholar]
- 19.Reilly MP, Li M, He J, Ferguson JF, Stylianou IM, Mehta NN, Burnett MS, Devaney JM, Knouff CW, Thompson JR, Horne BD, Stewart AF, Assimes TL, Wild PS, Allayee H, Nitschke PL, Patel RS, Martinelli N, Girelli D, Quyyumi AA, Anderson JL, Erdmann J, Hall AS, Schunkert H, Quertermous T, Blankenberg S, Hazen SL, Roberts R, Kathiresan S, Samani NJ, Epstein SE, Rader DJ. Identification of ADAMTS7 as a novel locus for coronary atherosclerosis and association of ABO with myocardial infarction in the presence of coronary atherosclerosis: two genome-wide association studies. Lancet. 2011;377:383–392. doi: 10.1016/S0140-6736(10)61996-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lehrke M, Millington SC, Lefterova M, Cumaranatunge RG, Szapary P, Wilensky R, Rader DJ, Lazar MA, Reilly MP. CXCL16 is a marker of inflammation, atherosclerosis, and acute coronary syndromes in humans. J Am Coll Cardiol. 2007;49:442–449. doi: 10.1016/j.jacc.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 21.Shen H, Bielak LF, Ferguson JF, Streeten EA, Yerges-Armstrong LM, Liu J, Post W, O'Connell JR, Hixson JE, Kardia SL, Sun YV, Jhun MA, Wang X, Mehta NN, Li M, Koller DL, Hakonarson H, Keating BJ, Rader DJ, Shuldiner AR, Peyser PA, Reilly MP, Mitchell BD. Association of the vitamin D metabolism gene CYP24A1 with coronary artery calcification. Arterioscler Thromb Vasc Biol. 2010;30:2648–2654. doi: 10.1161/ATVBAHA.110.211805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keating BJ, Tischfield S, Murray SS, Bhangale T, Price TS, Glessner JT, Galver L, Barrett JC, Grant SF, Farlow DN, Chandrupatla HR, Hansen M, Ajmal S, Papanicolaou GJ, Guo Y, Li M, Derohannessian S, de Bakker PI, Bailey SD, Montpetit A, Edmondson AC, Taylor K, Gai X, Wang SS, Fornage M, Shaikh T, Groop L, Boehnke M, Hall AS, Hattersley AT, Frackelton E, Patterson N, Chiang CW, Kim CE, Fabsitz RR, Ouwehand W, Price AL, Munroe P, Caulfield M, Drake T, Boerwinkle E, Reich D, Whitehead AS, Cappola TP, Samani NJ, Lusis AJ, Schadt E, Wilson JG, Koenig W, McCarthy MI, Kathiresan S, Gabriel SB, Hakonarson H, Anand SS, Reilly M, Engert JC, Nickerson DA, Rader DJ, Hirschhorn JN, Fitzgerald GA. Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PLoS One. 2008;3:e3583. doi: 10.1371/journal.pone.0003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whitman SC, Rateri DL, Szilvassy SJ, Yokoyama W, Daugherty A. Depletion of natural killer cell function decreases atherosclerosis in low-density lipoprotein receptor null mice. Arterioscler Thromb Vasc Biol. 2004;24:1049–1054. doi: 10.1161/01.ATV.0000124923.95545.2c. [DOI] [PubMed] [Google Scholar]
- 24.Hakonarson H, Grant SF, Bradfield JP, Marchand L, Kim CE, Glessner JT, Grabs R, Casalunovo T, Taback SP, Frackelton EC, Lawson ML, Robinson LJ, Skraban R, Lu Y, Chiavacci RM, Stanley CA, Kirsch SE, Rappaport EF, Orange JS, Monos DS, Devoto M, Qu HQ, Polychronakos C. A genome-wide association study identifies KIAA0350 as a type 1 diabetes gene. Nature. 2007;448:591–594. doi: 10.1038/nature06010. [DOI] [PubMed] [Google Scholar]
- 25.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 27.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 29.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 30.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Donnelly P. Progress and challenges in genome-wide association studies in humans. Nature. 2008;456:728–731. doi: 10.1038/nature07631. [DOI] [PubMed] [Google Scholar]
- 32.Pearson TA, Manolio TA. How to interpret a genome-wide association study. JAMA. 2008;299:1335–1344. doi: 10.1001/jama.299.11.1335. [DOI] [PubMed] [Google Scholar]
- 33.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 34.Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, Wang Q, Bekiranov S, Sementchenko V, Fox EA, Silver PA, Gingeras TR, Liu XS, Brown M. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 35.Helledie T, Grontved L, Jensen SS, Kiilerich P, Rietveld L, Albrektsen T, Boysen MS, Nohr J, Larsen LK, Fleckner J, Stunnenberg HG, Kristiansen K, Mandrup S. The gene encoding the Acyl-CoA-binding protein is activated by peroxisome proliferator-activated receptor gamma through an intronic response element functionally conserved between humans and rodents. J Biol Chem. 2002;277:26821–26830. doi: 10.1074/jbc.M111295200. [DOI] [PubMed] [Google Scholar]
- 36.Lefterova MI, Zhang Y, Steger DJ, Schupp M, Schug J, Cristancho A, Feng D, Zhuo D, Stoeckert CJ, Jr, Liu XS, Lazar MA. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008;22:2941–2952. doi: 10.1101/gad.1709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma Q, Jones D, Borghesani PR, Segal RA, Nagasawa T, Kishimoto T, Bronson RT, Springer TA. Impaired B-lymphopoiesis, myelopoiesis, and derailed cerebellar neuron migration in CXCR4- and SDF-1-deficient mice. Proc Natl Acad Sci USA. 1998;95:9448–9453. doi: 10.1073/pnas.95.16.9448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagasawa T, Hirota S, Tachibana K, Takakura N, Nishikawa S, Kitamura Y, Yoshida N, Kikutani H, Kishimoto T. Defects of B-cell lymphopoiesis and bone-marrow myelopoiesis in mice lacking the CXC chemokine PBSF/SDF-1. Nature. 1996;382:635–638. doi: 10.1038/382635a0. [DOI] [PubMed] [Google Scholar]
- 39.Zou YR, Kottmann AH, Kuroda M, Taniuchi I, Littman DR. Function of the chemokine receptor CXCR4 in haematopoiesis and in cerebellar development. Nature. 1998;393:595–599. doi: 10.1038/31269. [DOI] [PubMed] [Google Scholar]
- 40.Zernecke A, Shagdarsuren E, Weber C. Chemokines in atherosclerosis. An update. Arterioscler Thromb Vasc Biol. 2008;28:1897–1908. doi: 10.1161/ATVBAHA.107.161174. [DOI] [PubMed] [Google Scholar]
- 41.Schober A, Knarren S, Lietz M, Lin EA, Weber C. Crucial role of stromal cell-derived factor-1alpha in neointima formation after vascular injury in apolipoprotein E-deficient mice. Circulation. 2003;108:2491–2497. doi: 10.1161/01.CIR.0000099508.76665.9A. [DOI] [PubMed] [Google Scholar]
- 42.Xiao Q, Ye S, Oberhollenzer F, Mayr A, Jahangiri M, Willeit J, Kiechl S, Xu Q. SDF1 gene variation is associated with circulating SDF1alpha level and endothelial progenitor cell number: the Bruneck Study. PLoS One. 2008;3:e4061. doi: 10.1371/journal.pone.0004061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coll B, Alonso-Villaverde C, Parra S, Montero M, Tous M, Joven J, Masana L. The stromal derived factor-1 mutated allele (SDF1–3′A) is associated with a lower incidence of atherosclerosis in HIV-infected patients. AIDS. 2005;19:1877–1883. doi: 10.1097/01.aids.0000183516.22266.dd. [DOI] [PubMed] [Google Scholar]
- 44.Wiesner T, Bugl S, Mayer F, Hartmann J, Kopp H-G. Differential changes in platelet VEGF, Tsp, CXCL12, and CXCL4 in patients with metastatic cancer. Clin Exp Metastas. 2010;27:141–149. doi: 10.1007/s10585-010-9311-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.