Abstract
Tandem mass spectrometry (MS/MS) of intact, noncovalently-bound protein-ligand complexes can yield structural information on the site of ligand binding. Fourier transform ion cyclotron resonance (FT-ICR) top-down MS of the 29 kDa carbonic anhydrase-zinc complex and adenylate kinase bound to adenosine triphosphate (ATP) with collisionally activated dissociation (CAD) and/or electron capture dissociation (ECD) generates product ions that retain the ligand and their identities are consistent with the solution phase structure. Increasing gas phase protein charging from electrospray ionization (ESI) by the addition of supercharging reagents, such as m-nitrobenzyl alcohol and sulfolane, to the protein analyte solution improves the capability of MS/MS to generate holo-product ions. Top-down proteomics for protein sequencing can be enhanced by increasing analyte charging.
Keywords: electrospray ionization, noncovalent complexes, supercharging, protein-ligand binding, adenylate kinase, carbonic anhydrase
1. Introduction
Although most proteomic approaches currently incorporate a “bottom-up” strategy in which proteins are digested into smaller-sized peptides, and protein identifications are derived from the mass spectrometry (MS)-based analysis of the enzymatically cleaved peptides, there is value in the direct measurement and fragmentation of intact proteins, or “top-down” sequencing [1]. The molecular mass of an intact protein defines the native covalent state of a gene's product including the effects of post-transcriptional/translational modifications, and associated heterogeneity that are modulated by the actions of other gene products. Moreover, the fragmentation pattern of large gas phase proteins can generate sufficient information for identification from sequence databases, particularly when combined with accurate mass measurements of both the intact molecule and its product ions [2].
The development of electrospray ionization (ESI) and the ready generation of multiply charged molecules opened up the prospects for tandem mass spectrometry and top-down MS [3] (but this was predicted by Fenn, as he speculated that the ability of ESI to produce multiply charged ions “could make a most important contribution to the practice of tandem mass spectrometry of MS-MS in sequencing biopolymers [4]”). Soon after Fenn's development of ESI, it was shown that the enhanced efficiency for collisionally activated dissociation (CAD) of multiply charged molecules could generate sequence-informative product ions for biomolecules as large as 66 kDa serum albumin proteins [5], 80 kDa transferrin [6] and to beyond 200 kDa [7]. CAD-MS/MS of multiply charged proteins yield multiply charged products [8]. Although the multiply charged products could be interpreted with data from low resolving power analyzers, the determination of precursor and product charge state is much more amenable with higher resolving power instruments (e.g., Fourier transform ion cyclotron resonance (FT-ICR) and orbitraps). Further, data from instruments armed with electron capture dissociation (ECD) [9-12] or electron transfer dissociation (ETD) [13] can yield much more substantial sequence coverage for larger proteins. Nature Methods recently listed top-down proteomics among “methods to watch” and an emerging method essential for characterizing various protein variants and posttranslational modifications, with potentially high impact in biomedical research [14].
An additional potential application of top-down MS is the elucidation of noncovalent ligand binding sites to targeted proteins. Ligands are represented by a variety of molecules including metal ions, small molecules, DNA/RNA, or other proteins that interact specifically with a host protein (or other type of biomolecule) to form functional complexes. These complexes are the fundamental machines for almost all cellular activities and processes. Understanding such complexes on a molecular level is important for knowing how they are function on a biological level. It also aids efforts in drug design and drug discovery for the development of more effective therapeutics.
ESI-MS has been used to measure complex stoichiometry for a variety of protein-ligand complexes and to determine the relative or absolute solution binding affinity of an association [15-17], even for weakly bound complexes with solution dissociation constants (kd) in the millimolar range [18, 19].
For noncovalently-bound protein-ligand complexes, it was previously thought that noncovalent ligand binding would not survive the MS/MS process. For example, because of the weak interactions between a protein and its ligand, CAD or even infrared multiphoton dissociation (IRMPD) of the complex usually results in simply the separation of the ligand from the protein, revealing little new structural information. However, we have demonstrated previously that top-down MS with ECD and/or CAD can be used to determine the ligand binding sites for specific protein-ligand complexes [19, 20]. Earlier work had suggested that weak, noncovalent intermolecular bonds could be preserved upon ECD [21]. We used ECD to localize the binding site of a polyamine compound, spermine, to the 13 kDa α-synuclein protein that has been implicated in Parkinson's disease. Spermine was retained by the c-/z•-products to localize spermine binding to the C-terminal region of the protein. Thus, although the solution binding association for the α-synuclein/spermine complex is relatively weak (kd ~ 10-3 M), ligand binding is retained in the gas phase and even upon ECD [19].
A factor that governs whether tandem MS of noncovalent protein-ligand complexes can yield any structural information is the relative stability of the gas phase complex. For example, the strength of electrostatic interactions is significantly enhanced in the absence of solvent. Recently, we reported the unusually stable gas phase complex formed between proteins and di- and triphosphate nucleotides, such as the ribonuclease A-cytidine triphosphate (RNase A-CTP) complex [22]. With covalent-like strength, enhanced gas phase electrostatic interactions can be sustained in CAD-MS/MS experiments.
This report demonstrates the enhanced efficiency of top-down MS of noncovalent protein-ligand complexes upon increasing the multiple charging of the gas phase complex. John Fenn's pioneering work paved the path to exploit the benefits of multiple charging by mass spectrometry. Enhanced multiple charging of native proteins and protein complexes can be induced by the addition of “supercharging” reagents, such as m-nitrobenzyl alcohol (m-NBA) [23, 24] and sulfolane [25]. Because electron capture cross sections increase quadratically with charge [26], addition of one more charge can dramatically enhance the efficiency of ECD/ETD. We show that increasing the multiple charging of protein-ligand complexes by the addition of supercharging reagents improves the prospects for gaining ligand binding site information.
2. Experimental
2.1. Materials
ATP, adenylate kinase (AK; myokinase, from chicken muscle, product number M5520), carbonic anhydrase II (bovine), m-NBA, and sulfolane were purchased from Sigma-Aldrich (St. Louis, MO). All protein samples were desalted and concentrated with 20 mM ammonium acetate buffer (pH 6.8) using centrifugal filter devices (10 kDa MWCO, Amicon Ultra; Millipore Corporation, Billerica, MA). After desalting, ATP was added to a 5 μM AK solution in 20 mM ammonium acetate. The solution protein and ligand ratio was kept at 1:1.
2.2. Top-down MS experiments
A nanoESI source and Au/Pd coated borosilicate glass capillaries (Proxeon Biosystems, Odense, Denmark), with flow rate around 50 nL/min, were coupled to an 7-Tesla LTQ-FT Ultra mass spectrometry (Thermo Fisher Scientific, San Jose, CA) to acquire positive ionization mode ESI-MS spectra. Instrument parameters for data acquisition have been described previously [20]. In the source region of the LTQ-FT Ultra, the capillary temperature was set to 210°C, the capillary voltage was +45V, and the tube lens was set to +225V. The resolution of the FT-ICR measurements was established to be 200,000 at 400 mass-to-charge ratio (m/z). Top-down MS/MS were accomplished with CAD or activated ECD (aiECD) in which ECD is coupled with infrared laser-heating of the product ions to dissociate hydrogen bonds retained upon ECD and thereby enhance product ion yield. Product ion mass measurement accuracy was generally better than 7 ppm.
3. Results and discussion
The multiple charging properties of ESI were exploited for MS/MS of intact proteins relatively early in the recent history of electrospray ionization, as protein ions can be dissociated effectively to generate sequence-informative products. This “top-down” strategy works well for proteins in denaturing, acidic solutions that promote high analyte charge. However, ESI charging for proteins and noncovalently-bound protein complexes in physiological pH solutions is generally lower, and this characteristic conspires to lower the efficiency for generating sequence-bearing product ions in tandem MS experiments.
We have demonstrated that the addition of charge-promoting agents (i.e., supercharging) can increase the ESI multiple charging of native proteins and protein complexes, and the supercharged protein complexes retain noncovalent binding with its ligand partners. Although the mechanism of supercharging of native proteins is not well understood [23-25], it is clear that the protein complexes are not sufficiently denatured upon supercharging that noncovalent ligand binding is disrupted. Also, this increased charging results in more effective MS/MS for determining ligand binding sites.
3.1 CAD of Zn-bound carbonic anhydrase
Carbonic anhydrase II (CA-II) is a 29 kDa zinc metalloenzyme that catalyzes the hydration of carbon dioxide to carbonic acid. A divalent zinc ion is an essential cofactor for CA-II [27]. The high resolution structure from x-ray crystallography shows the zinc coordination to be almost tetrahedral, with the active site Zn2+ ion coordinated to three histidyl residues (His-94, His-96 and His-119) and a water molecule [28]. Zinc binding is pH dependant, and its affinity is on the picomolar scale at pH 7 [29].
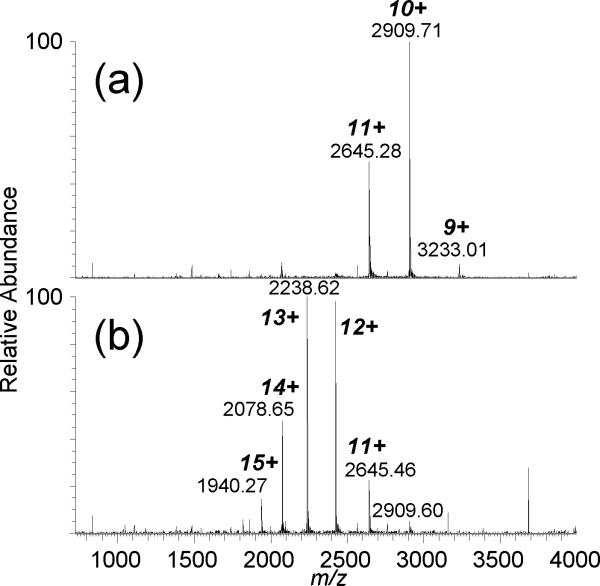
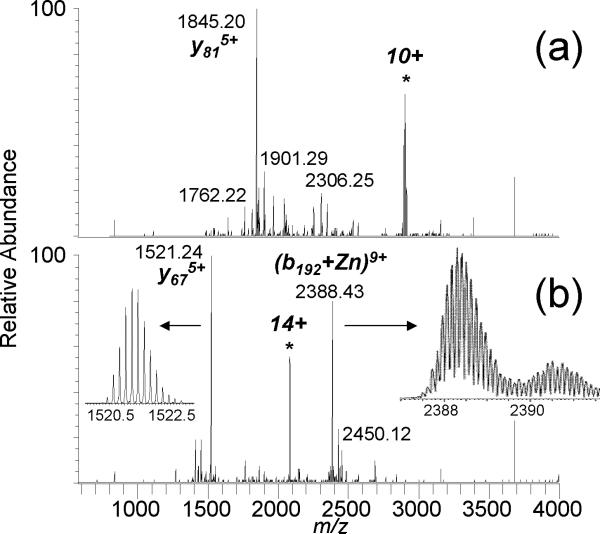
Top-down mass spectrometry has been demonstrated for characterizing copper binding [30] and the binding of a platinum anticancer drug [31] to small proteins. We attempt to use top-down MS to localize metal binding to a larger protein, CA-II. For zinc-bound CA-II, the addition of m-NBA increases charging by nearly 50% at pH 6.8, with maximum charging increasing from 11+ to 16+ with 0.5% v/v m-NBA (Figure 1) and to 19+ with 1% m-NBA [23]. There is sufficient zinc present in the commercial sample and/or in the buffers and sample vessels that the holo-form ((M+Zn+nH)n+2) is readily measured without the additional incorporation of zinc. CAD of the 10+ Zn-protein complex yields product ions that do not cover the zinc-binding domain, with the y818+ product ion from the cleavage on the N-terminal side of Pro-179 as the most abundant; dissociation near proline residues (or the “proline effect”) has been noted for protein top-down MS experiments [32, 33], including for CA-II [1]. No products retaining zinc were measured. However, with 0.5% m-NBA, CAD of the supercharged 14+ CA-II/Zn complex generates the y675+/(b192 + Zn)9+ complementary product ion pair, which in sum (mass and charge) account for the entire protein molecule (Figure 2). Again, the proline effect is observed from cleavage of the Tyr-192/Pro-193 amide bond as the primary product, but the b192 fragment retains zinc and is consistent with the expected zinc binding sites. Other product ions retaining zinc include b191+Zn and b135+Zn. CAD of the supercharged 13+ CA-II/Zn complex generates the y675+/(b192 + Zn)8+ product ion pair (data not shown).
Figure 1.
ESI-MS of bovine carbonic anhydrase II (bound to zinc) in (a) 20 mM ammonium acetate (pH 6.8) and (b) 20 mM ammonium acetate with 0.5% v/v m-NBA.
Figure 2.
ESI CAD-MS/MS mass spectra of the (a) 10+ zinc-bound CA-II and the (b) 14+ zinc-bound CA-II (with m-NBA).
Our preliminary CAD-MS/MS analysis of the native CA-II/Zn complex did not yield high sequence coverage overall compared to top-down MS of the higher charged denatured protein. Increasing charge of the CA-II/Zn complex did not significantly improve the efficiency for product ion generation; CAD of the 10+ complex yielded 28 products with charge states 1+-7+ that could be assigned, whereas the 14+ yielded 29 products ranging in charge from 2+ to 10+. However, the higher charging allowed for the formation of product ions that retain the zinc ligand (e.g., b192+Zn). Additional MS3 and perhaps ECD experiments could provide means to better pinpoint the zinc ligand binding site for CA-II.
3.2 CAD and ECD of adenylate kinase-ATP complex
Kinases are phosphotransferase enzymes that transfer phosphate groups from adenosine-5’-triphosphate (ATP) to proteins or single nucleotides and are key players in cell signaling. Adenylate kinase (AK; or myokinase) belongs to a family of nucleoside monophosphate kinases and maintains the cellular equilibrium concentration of adenylate nucleotides by catalyzing the reversible transfer of a phosphate group from ATP to AMP to generate two ADPs.
We had previously reported that CAD-MS/MS of the gas phase RNase A-CTP complex results in fragmentation of the CTP ligand to form free CMP and RNase A retaining a diphosphate group (pp) as the primary products [22]. Although the RNase A-CTP complex has a solution kd of ca. 10-6 M, the near covalent-like strength of gas phase electrostatic interactions provided the opportunity to identify and characterize binding sites of protein-nucleic acid ligand complexes, such as protein kinases binding to ATP. For the 22 kDa adenylate kinase-ATP complex, the CAD data suggests the ATP-binding region to be residues 121-140, consistent with crystal structures and photoaffinity labeling experiments [20].
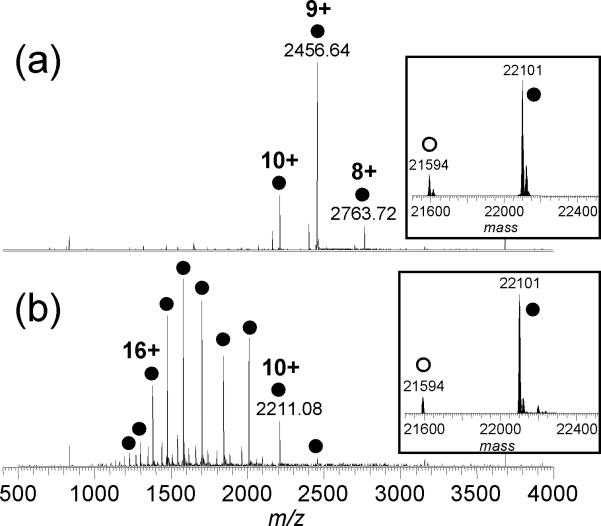
Increasing the charge states of the precursors improved the dissociation efficiency for both CAD and ECD of the AK-ATP complex. The maximum charge state observed for AK-ATP increased from 10+ to 18+ upon addition of 200 mM sulfolane (Figure 3). A small amount of the apo-form was observed using the sample preparation conditions described in the Experimental section. The holo/apo ratio did not change significantly with sulfolane-induced supercharging (Figure 3, inset), suggesting that minimal solution denaturation of the complex (to release the ATP ligand) occurs upon addition of sulfolane.
Figure 3.
ESI mass spectra of AK in the presence of ATP (10 mM ammonium acetate, pH 6.6) (a) without and (b) with 200 mM sulfolane. The peaks marked with open circles represent multiply charged molecules of the apo-protein, and those marked with filled circles represent the 1:1 AK-ATP complex. The corresponding mass spectra converted to the mass domain are shown in the insets.
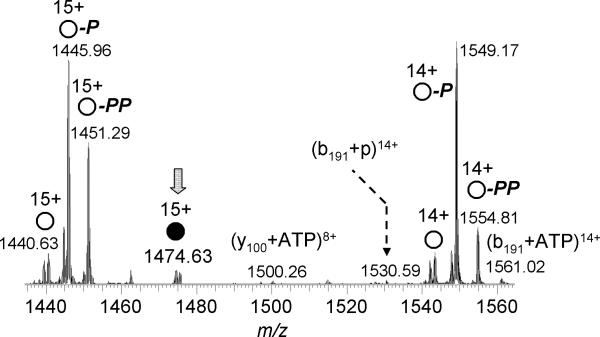
As for the RNase A-CTP complex [22], CAD of AK-ATP releases AMP to generate the AK-pp complex, i.e., AK bound to a diphosphate group, as well as b-/y-product ions from the polypeptide backbone that retain the diphosphate group (Figure 4) [20]. By mapping the CAD-generated product ions measured for the holo AK-ATP and AK-pp complexes onto the AK primary sequence, the putative ATP binding sites can be inferred.
Figure 4.
ESI CAD-tandem mass spectrum of the 14+-charged AK-ATP 1:1 complex. Product ions of the intact apo-protein are marked by open circles, and those of the holo-protein are marked with the filled circle. Retention of one phosphate group by the product ion is represented by “p”, and retention of two phosphates is labeled with “pp.” The loss or gain of a phosphate group is labeled with “- p” or “+ p”, respectively.
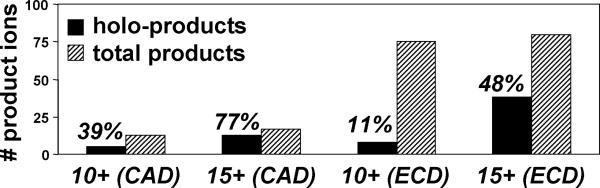
Comparison of the CAD and ECD data generated for the 10+ (without sulfolane, [20]) and the 15+ (with sulfolane) AK-ATP complex showed somewhat improved sequence coverage for the higher charged precursor, but with a much higher proportion of holo-product formation (Figure 5). The total number of product ions and sequence coverage generated by CAD and ECD for the 10+ and 15+ complex are similar. However, the relative proportion of those products that retain either the diphosphate group or even the intact ATP molecule was significantly enhanced with the higher charged 15+ complex; for example, 48% of the 80 total product ions (42% sequence coverage) generated by ECD of 15+ AK-ATP retained either pp or ATP, compared to only 11% for the 10+ complex. Moreover, most of the ECD-formed holo-products for the 15+ complex retained the intact ATP, whereas no intact ATP was observed for ECD of the 10+ complex.
Figure 5.
The number of product ions generated by CAD and ECD of the AK-ATP complex. MS/MS of the higher charged precursor yield a high proportion of holo-products (percentage listed in parentheses).
Despite the improved CAD/ECD performance for the higher charged complex, this did not improve the ability to localize the site(s) of ATP binding for adenylate kinase. The previous CAD/ECD data from the 10+ complex narrowed down ATP binding to residues 121-140 [20]. ECD products from the 15+ complex brackets ATP binding to a region between amino acids 94 and 150. Combined with the CAD data, the binding region is again suggested to be near residues 121-140. The overall improvement in ECD performance for supercharged native complexes should be more valuable for complexes that are less stable in the gas phase than the electrostatically-held protein-nucleic acid species.
4. Conclusion
The ability to increase analyte charge beyond that realized under “normal” electrospray conditions is advantageous for the analysis of noncovalently-bound protein-ligand complexes. ESI-produced ions for native protein complexes are usually found in a m/z region higher than that found for denatured proteins. For the 2 MDa 70S ribosome [34] and the 4 MDa hepatitis B virus capsid [35], multiply charged ions are found beyond m/z 27,000. Moving the charge distribution to lower m/z by increasing charge improves detection efficiency for measuring larger protein complexes, analogous to Fenn's original observation of ESI-MS of large biomolecules [4].
Moreover, increasing analyte charge allows for more effective tandem MS of protein complexes. Our preliminary analysis of CA-II/Zn and AK-ATP suggests that CAD and ECD probing of the higher charged complexes generated by supercharging agents yields more products that retain the ligand (i.e., holo-product ions). This is a useful feature for developing experimental strategies to define the ligand binding domains of proteins. Also, the data suggests that many structural aspects of the intermolecular associations formed initially in solution are preserved upon transition to the gas phase and are stable even upon activation and dissociation events in the MS/MS experiment.
Acknowledgments
Support from the National Institutes of Health (RR 20004) and the NIH/NCRR High-End Instrumentation Program (S10 RR023045) are acknowledged. This article is dedicated to John Fenn, who pioneered the virtues of multiple charging for protein MS analysis. He inspired us “to continue stumbling for a while along the road ahead, kicking over stones here and there, driven by curiosity to find out what may be hidden under the next one [36].” We should all strive to be as curious as John Fenn, as one can't always predict what hidden jewels can be discovered by turning over stones on the road of scientific exploration.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kelleher NL, Lin HY, Valaskovic GA, Aaserud DJ, Fridriksson EK, McLafferty FW. Top down versus bottom up protein characterization by tandem high-resolution mass spectrometry. J. Am. Chem. Soc. 1999;121:806–812. [Google Scholar]
- 2.Mortz E, O'Connor PB, Roepstorff P, Kelleher NL, Wood TD, McLafferty FW, Mann M. Sequence tag identification of intact proteins by matching tanden mass spectral data against sequence data bases. Proc. Natl. Acad. Sci. USA. 1996;93:8264–8267. doi: 10.1073/pnas.93.16.8264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 4.Meng CK, Mann M, Fenn JB. Of protons or proteins, Z. Phys. D - Atoms Molec. Clusters. 1988;10:361–368. [Google Scholar]
- 5.Loo JA, Edmonds CG, Smith RD. Tandem mass spectrometry of very large molecules: Serum albumin sequence information from multiply charged ions formed by electrospray ionization. Anal. Chem. 1991;63:2488–2499. doi: 10.1021/ac00021a018. [DOI] [PubMed] [Google Scholar]
- 6.Thevis M, Ogorzalek Loo RR, Loo JA. Mass spectrometric characterization of transferrins and their fragments derived by reduction of disulfide bonds. J. Am. Soc. Mass Spectrom. 2003;14:635–647. doi: 10.1016/S1044-0305(03)00199-5. [DOI] [PubMed] [Google Scholar]
- 7.Han X, Jin M, Breuker K, McLafferty FW. Extending top-down mass spectrometry to proteins with masses greater than 200 kilodaltons. Science. 2006;314:109–112. doi: 10.1126/science.1128868. [DOI] [PubMed] [Google Scholar]
- 8.Loo JA, Edmonds CG, Smith RD. Primary sequence information from intact proteins by electrospray ionization tandem mass spectrometry. Science. 1990;248:201–204. doi: 10.1126/science.2326633. [DOI] [PubMed] [Google Scholar]
- 9.Ge Y, Lawhorn BG, ElNaggar M, Strauss E, Park JH, Begley TP, McLafferty FW. Top down characterization of larger proteins (45 kDa) by electron capture dissociation mass spectrometry. J. Am. Chem. Soc. 2002;124:672–678. doi: 10.1021/ja011335z. [DOI] [PubMed] [Google Scholar]
- 10.Horn DM, Zubarev RA, McLafferty FW. Automated de novo sequencing of proteins by tandem high-resolution mass spectrometry. Proc. Natl. Acad. Sci. USA. 2000;97:10313–10317. doi: 10.1073/pnas.97.19.10313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McLafferty FW, Horn DM, Breuker K, Ge Y, Lewis MA, Cerda B, Zubarev RA, Carpenter BK. Electron capture dissociation of gaseous multiply charged ions by Fourier-transform ion cyclotron resonance. J. Am. Soc. Mass Spectrom. 2001;12:245–249. doi: 10.1016/S1044-0305(00)00223-3. [DOI] [PubMed] [Google Scholar]
- 12.Ouvry-Patat SA, Torres MP, Gelfand CA, Quek HH, Easterling M, Speir JP, Borchers CH. Top-down proteomics on a high-field Fourier transform ion cyclotron resonance mass spectrometer. Methods Mol. Biol. 2009;492:215–231. doi: 10.1007/978-1-59745-493-3_12. [DOI] [PubMed] [Google Scholar]
- 13.Bunger MK, Cargile BJ, Ngunjiri A, Bundy JL, Stephenson JL., Jr. Automated proteomics of E. coli via top-down electron-transfer dissociation mass spectrometry. Anal. Chem. 2008;80:1459–1467. doi: 10.1021/ac7018409. [DOI] [PubMed] [Google Scholar]
- 14.Doerr A. Special feature - methods to watch: Top-down mass spectrometry. Nature Methods. 2008;5:24. [Google Scholar]
- 15.Heck AJR, van den Heuvel RHH. Investigation of intact protein complexes by mass spectrometry. Mass Spectrom. Rev. 2004;23:368–389. doi: 10.1002/mas.10081. [DOI] [PubMed] [Google Scholar]
- 16.Loo JA. Studying noncovalent protein complexes by electrospray ionization mass spectrometry. Mass Spectrom. Rev. 1997;16:1–23. doi: 10.1002/(SICI)1098-2787(1997)16:1<1::AID-MAS1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 17.Loo JA. Electrospray ionization mass spectrometry: A technology for studying noncovalent macromolecular complexes. Int. J. Mass Spectrom. 2000;200:175–186. [Google Scholar]
- 18.Loo JA, Holsworth DD, Root-Bernstein RS. Use of electrospray ionization mass spectrometry to probe antisense peptide interactions. Biol. Mass Spectrom. 1994;23:6–12. doi: 10.1002/bms.1200230103. [DOI] [PubMed] [Google Scholar]
- 19.Xie Y, Zhang J, Yin S, Loo JA. Top-down ESI-ECD-FT-ICR mass spectrometry localizes noncovalent protein-ligand binding sites. J. Am. Chem. Soc. 2006;128:14432–14433. doi: 10.1021/ja063197p. [DOI] [PubMed] [Google Scholar]
- 20.Yin S, Loo JA. Elucidating the site of protein-ATP binding by top-down mass spectrometry. J. Am Soc. Mass Spectrom. 2010;21 doi: 10.1016/j.jasms.2010.01.002. in press. [DOI] [PubMed] [Google Scholar]
- 21.Zubarev RA. Electron-capture dissociation tandem mass spectrometry. Curr. Opin. Biotechnol. 2004;15:12–16. doi: 10.1016/j.copbio.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 22.Yin S, Xie Y, Loo JA. Mass spectrometry of protein-ligand complexes: Enhanced gas-phase stability of ribonuclease-nucleotide complexes. J. Am. Soc. Mass Spectrom. 2008;19:1199–1208. doi: 10.1016/j.jasms.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lomeli SH, Yin S, Ogorzalek Loo RR, Loo JA. Increasing charge while preserving noncovalent protein complexes for ESI-MS. J. Am. Soc. Mass Spectrom. 2009;20:593–596. doi: 10.1016/j.jasms.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sterling HJ, Williams ER. Origin of supercharging in electrospray ionization of noncovalent complexes from aqueous solution. J. Am. Soc. Mass Spectrom. 2009;20:1933–1943. doi: 10.1016/j.jasms.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lomeli SH, Peng IX, Yin S, Ogorzalek-Loo RR, Loo JA. New reagents for increasing ESI multiple charging of proteins and protein complexes. J. Am Soc. Mass Spectrom. 2010;21:127–131. doi: 10.1016/j.jasms.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zubarev RA, Horn DM, Fridriksson EK, Kelleher NL, Kruger NA, Lewis MA, Carpenter BK, McLafferty FW. Electron capture dissociation for structural characterization of multiply charged protein cations. Anal. Chem. 2000;72:563–573. doi: 10.1021/ac990811p. [DOI] [PubMed] [Google Scholar]
- 27.DiTusa CA, Christensen T, McCall KA, Fierke CA, Toone EJ. Thermodynamics of metal ion binding. 1. Metal ion binding by wild-type carbonic anhydrase. Biochemistry. 2001;40:5338–5344. doi: 10.1021/bi001731e. [DOI] [PubMed] [Google Scholar]
- 28.Saito R, Sato T, Ikai A, Tanaka N. Structure of bovine carbonic anhydrase II at 1.95Å resolution. Acta Cryst. 2004;D60:792–795. doi: 10.1107/S0907444904003166. [DOI] [PubMed] [Google Scholar]
- 29.Caprita R, Caprita A, Ilia G, Ciucanu I. The influence of the pH on the apparent dissociation constant for Zn2+-carbonic anhydrase isoenzyme II. Rev. Chim. 2006;57:1112–1114. [Google Scholar]
- 30.Erales J, Gontero B, Whitelegge J, Halgand F. Mapping of a copper-binding site on the small cp12 chloroplastic protein of Chlamydomonas reinhardtii using top-down mass spectrometry and site-directed mutagenesis. Biochem. J. 2009;419:75–82. doi: 10.1042/BJ20082004. [DOI] [PubMed] [Google Scholar]
- 31.Hartinger CG, Tsybin YO, Fuchser J, Dyson PJ. Characterization of platinum anticancer drug protein-binding sites using a top-down mass spectrometric approach. Inorg. Chem. 2008;47:17–19. doi: 10.1021/ic702236m. [DOI] [PubMed] [Google Scholar]
- 32.Loo JA, Edmonds CG, Smith RD. Primary sequence information from intact proteins by electrospray ionization tandem mass-spectrometry. Science. 1990;248:201–204. doi: 10.1126/science.2326633. [DOI] [PubMed] [Google Scholar]
- 33.Loo JA, Edmonds CG, Smith RD. Tandem mass-spectrometry of very large molecules .2. Dissociation of multiply charged proline-containing proteins from electrospray ionization. Anal. Chem. 1993;65:425–438. doi: 10.1021/ac00052a020. [DOI] [PubMed] [Google Scholar]
- 34.McKay AR, Ruotolo BT, Ilag LL, Robinson CV. Mass measurements of increased accuracy resolve heterogeneous populations of intact ribosomes. J. Am. Chem. Soc. 2006;128:11433–11442. doi: 10.1021/ja061468q. [DOI] [PubMed] [Google Scholar]
- 35.Uetrecht C, Versluis C, Watts NR, Roos WH, Wuite GJL, Wingfield PT, Steven AC, Heck AJR. High-resolution mass spectrometry of viral assemblies: Molecular composition and stability of dimorphic hepatitis B virus capsids. Proc. Natl. Acad. Sci. USA. 2008;105:9216–9220. doi: 10.1073/pnas.0800406105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fenn JB. Research in retrospect: Some biograffiti of a journeyman chemist. Ann. Rev. Phys. Chem. 1996;47:1–41. doi: 10.1146/annurev.physchem.47.1.1. [DOI] [PubMed] [Google Scholar]